Abstract
This work reports an amperometric biosensor for the determination of miRNA-21, a relevant oncogene. The methodology involves a competitive DNA-target miRNA hybridization assay performed on the surface of magnetic microbeads (MBs) and amperometric transduction at screen-printed carbon electrodes (SPCEs). The target miRNA competes with a synthetic fluorescein isothiocyanate (FITC)-modified miRNA with an identical sequence for hybridization with a biotinylated and complementary DNA probe (b-Cp) immobilized on the surface of streptavidin-modified MBs (b-Cp-MBs). Upon labeling, the FITC-modified miRNA attached to the MBs with horseradish peroxidase (HRP)-conjugated anti-FITC Fab fragments and magnetic capturing of the MBs onto the working electrode surface of SPCEs. The cathodic current measured at −0.20 V (versus the Ag pseudo-reference electrode) was demonstrated to be inversely proportional to the concentration of the target miRNA. This convenient biosensing method provided a linear range between 0.7 and 10.0 nM and a limit of detection (LOD) of 0.2 nM (5 fmol in 25 μL of sample) for the synthetic target miRNA without any amplification step. An acceptable selectivity towards single-base mismatched oligonucleotides, a high storage stability of the b-Cp-MBs, and usefulness for the accurate determination of miRNA-21 in raw total RNA (RNAt) extracted from breast cancer cells (MCF-7) were demonstrated.
1. Introduction
Breast cancer (BC) is one of the three most common invasive cancers in females, with an estimated 1.5 million new cases per year [1], and one of the leading causes of cancer mortality among women worldwide [2]. Effective management of BC depends on early diagnosis and proper monitoring of the response of patients to therapy, which implies the need for identification of sensitive and specific biomarkers useful for early detection and disease monitoring [1].
Accumulated evidence in the past several years has highlighted the great promise of miRNAs as diagnostic, prognostic, or predictive biomarkers in the clinical management of patients with BC [3]. These miRNAs are short, non-coding single-stranded RNA (ssRNA) molecules, 19–23 nucleotides in length [4], acting at the post-transcriptional level by partial binding to the mRNA of genes [1,5,6]. However, the determination of miRNAs demands highly sensitive and selective methodologies due to their low abundance and high sequence similarity in family members. Moreover, their short length makes it difficult to use traditional DNA methods (primers and probe selection for amplification and sandwich assays) for their determination [7,8,9,10,11,12].
Current strategies for miRNA determination include miRNA microarrays [13,14,15,16], Northern blotting [17,18], and in situ hybridization and quantitative reverse-transcription polymerase chain reaction (qRT-PCR) [19,20,21]. However, the wide applicability of these strategies is hindered by their high cost, poor sensitivity, and the need for tedious analysis protocols. Furthermore, the requirement for skilled technicians and sophisticated and non-portable instruments restricts their use to central laboratories. Therefore, there is a pressing need to develop simple, sensitive, rapid, portable, and low-cost methodologies for the determination of miRNAs. Within this context, electrochemical biosensors have demonstrated very interesting features compatible with current clinic demands in terms of portability and simplicity for the determination of miRNAs.
On the other hand, it has been widely demonstrated that magnetic microbeads (MBs) offer very attractive characteristics in the development of electrochemical biosensors, such as lower limits of detection, faster assay kinetics, and smaller matrix effects [22,23,24,25]. These advantages have been coupled in many cases with those provided by planar disposable electrodes (versatility of fabrication and mass production at low cost) to develop different competitive electrochemical biosensing platforms for the quantification of miRNAs. Nevertheless, most of these platforms involve different preparation steps and require the use of expensive affinity reagents [26,27,28,29,30,31] and multiple probes in connection with amplification strategies [28,32].
With the aim of simplifying these strategies, we describe in this work the first competitive direct hybridization assay implemented onto the surface of MBs for the amperometric determination of miRNAs. Additional advantages of this methodology compared to our previous developments include simplicity, one-step incubation, lower cost, and not requiring the use of expensive affinity/amplification reagents and/or multiple probes. The method implies the competition of the target miRNA with a synthetic fluorescein isothiocyanate (FITC)-miRNA for the hybridization with a biotinylated DNA capture probe (b-Cp) immobilized onto Strep-MBs. As a model miRNA target, miRNA-21, a biomarker for many types of cancer and also cardiovascular disease [1,33], was chosen to exemplify the technique. Amperometric transduction was performed at screen-printed carbon electrodes (SPCEs) using the H2O2/Hydroquinone (HQ) system after labeling the FITC-miRNA immobilized onto the b-Cp-MBs with an anti-FITC-HRP. In this study, we show that this method allows the accurate determination of miRNA-21 in breast cancer cells without reverse transcription to cDNA, amplification, preconcentration, or purification steps.
2. Experimental
2.1. Apparatus and Electrodes
Amperometric measurements were made with a CH Instruments (Austin, TX, USA) 812B potentiostat controlled by CH Instruments software. Screen-printed carbon electrodes (SPCEs) (DRP-110, DropSens, Spain), consisting of a 4-mm diameter carbon working electrode, a carbon counter electrode, and an Ag pseudo-reference electrode, were used as electrochemical transducers in conjunction with a specific cable connector (DRP-CAC, DropSens, Llanera, Spain). All measurements were performed at room temperature. A neodymium magnet (AIMAN GZ, Madrid, Spain) embedded in a homemade Teflon casing was used to magnetically capture the modified-MBs on the surface of SPCEs.
A Raypa steam sterilizer, a biological safety cabinet Telstar Biostar, an incubator shaker Thermo-shaker MT100 (Universal Labortechnik, Leipzig, Germany), a Bunsen AGT-9 vortex for homogenization of the solutions, a magnetic particle concentrator DynaMag™-2 (123.21D, Invitrogen Dynal AS, Oslo, Norway), and a Sigma 1–15 K refrigerated microcentrifuge were also employed. The quality and quantity of the extracted cellular RNA were evaluated by using a NanoDrop® ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
2.2. Reagents and Solutions
All reagents were of the highest analytical grade. Streptavidin-modified magnetic beads (Strep-MBs, 2.8 µm Ø, 10 mg·mL−1, Dynabeads M-280 Streptavidin, 11206D) were purchased from Dynal Biotech ASA.
NaCl, KCl, NaH2PO4, Na2HPO4, and Tris–HCl were purchased from Scharlab. Hydroquinone (HQ) and H2O2 (30%, w/v) were purchased from Sigma-Aldrich and ethylenediaminetetraacetic acid (EDTA) from Merck (Darmstadt, Germany). Anti-fluorescein (FITC) Fab Fragments conjugated with HRP (anti-FITC-HRP) were purchased from Roche. A blocker casein solution (BB solution, a ready-to-use, PBS solution of 1% w/v purified casein) was purchased from Thermo Scientific. All the DNA and RNA synthetic oligonucleotides (sequence described in Table 1, purchased from Sigma-Aldrich, St. Louis, MO, USA) were reconstituted upon reception in nuclease-free water to a final concentration of 100 μM, divided into small aliquots, and stored at −80 °C.

Table 1.
Oligonucleotides used in this work.
All the required buffer solutions were prepared in deionized water from a Millipore Milli-Q purification system (18.2 MΩ cm): phosphate-buffered saline (PBS) consisting of 0.01 M phosphate buffer solution containing 0.137 M NaCl and 0.0027 M KCl, pH 7.5; Binding and Washing buffer (B&W) consisting of 10 mM Tris–HCl solution containing 1 mM EDTA and 2 M NaCl, pH 7.5 (sterilized after their preparation by autoclaving at 120 °C for 15 min); PBS:BB (1:1) solution prepared by mixing equal volumes of PBS and BB solutions and 0.05 M phosphate buffer of pH 6.0.
2.3. MBs Modification
A 0.5-μL aliquot of the commercial Strep-MBs suspension was transferred into a microcentrifuge tube and washed twice with 50 μL of B&W buffer. Between washes, the particles were placed in the magnetic concentrator and, after 3 min, the supernatant was discarded. Washed MBs were incubated for 15 min at 30 °C under continuous stirring (950 rpm) with 25 μL of 2.5 nM of the b-Cp solution (prepared in B&W). After two washing steps with 50 μL of a PBS:BB (1:1) solution, the b-Cp-MBs can be stored (in 50 μL filtered PBS at 4 °C) or used directly to perform the determination. In this latter case, the b-Cp-MBs were incubated for 120 min (950 rpm, 30 °C) in 25 μL of a mixture solution containing the synthetic target miRNA (or 0.5–2.0 μg of the extracted RNAt), 2.5 nM FITC-miRNA, and 1/500 diluted anti-FITC-HRP (prepared also in PBS:BB (1:1)). After washing twice with 50 μL of PBS:BB (1:1) solution, the modified-MBs were re-suspended in 50 μL of 0.05 M sodium phosphate buffer solution (pH 6.0) to perform the amperometric detection.
All the MB manipulations carried out before the amperometric measurements were made in a laminar flow cabinet to avoid RNAse contamination and prevent miRNA degradation. Moreover, control experiments in the absence of target miRNA were performed daily to evaluate the blank signal.
2.4. Electrochemical Measurements
Amperometric measurements were made by pipetting the 50 μL of the modified MBs suspension onto the SPCE after placing it on a homemade casing of Teflon with an encapsulated neodymium magnet. In this way, the MBs were magnetically captured on the working carbon electrode in a reproducible and stable manner. Then, the SPCE/magnet holding block ensemble was immersed into an electrochemical cell containing 10 mL of 0.05 M phosphate buffer of pH 6.0 supplemented with 1.0 mM HQ (prepared just before performing the electrochemical measurement). The amperometric measurement was made at −0.20 V versus an Ag pseudo-reference electrode in stirred solutions. Once the background current was stabilized, 50 μL of a freshly prepared 0.1 M H2O2 solution were added and the cathodic current recorded until the steady-state current was reached (~100 s). All of the amperometric signals represent the difference between the steady-state and the background currents. Data shown correspond to the average of at least three replicates with the confidence intervals calculated for α = 0.05. The limits of detection (LOD) and quantification (LOQ) were estimated according to the 3 × sb/m and 10 × sb/m criteria, respectively, with sb being the standard deviation for 10 measurements made with no target miRNA and m the slope value of the calibration plot obtained with the synthetic target miRNA.
2.5. Cultured Cells and RNAt Extraction
Protocols used for cultured cells and RNAt extraction were the same as described previously [26].
Briefly, non-tumorigenic epithelial MCF-10A cells were grown at 37 °C in a humidified atmosphere containing 5% CO2 in high-glucose DMEM/Ham’s Nutrient Mixture F12 (1:1) with 2.5 mM l-glutamine (GIBCO-Invitrogen, Carlsbad, CA, USA), 5% horse serum (Gibco), 10 mg mL−1 human insulin (Sigma, St. Louis, MO, USA), 0.5 mg mL−1 hydrocortisone (Sigma), 10 ng mL−1 epidermal growth factor (EGF), and 100 ng mL−1 cholera toxin (QuadraTech Ltd., Epsom, UK). Breast cancer cells MCF-7 were cultured in high-glucose DMEM (Dulbecco’s modified Eagle’s medium) supplemented with 10% fetal bovine serum, 100 U mL−1 penicillin, 100 μg mL−1 streptomycin, and 2.5 mM l-glutamine.
For RNAt isolation, cells were washed with PBS, scraped off, and spun down. The pellet was homogenized in Tri Reagent (Molecular Research Center, Inc., Cincinnati, OH, USA) for 5 min at room temperature before chloroform extraction and the RNAt, in the upper aqueous phase, was precipitated with isopropylalcohol and washed twice in 70% EtOH. The pellet was dried out in a heating plate for 10 min at 80 °C and, then, dissolved in RNase-free water, and stored at −80 °C [26]. The absorbance ratio values measured at the appropriate wavelengths (260, 230, and 280 nm) with an ND-1000 spectrophotometer confirmed in all cases pure RNA.
3. Results and Discussion
The method reported here relies on a competitive DNA/RNA hybridization using a biotinylated DNA capture probe (b-Cp), complementary to the target miRNA, immobilized onto Strep-MBs and a fluorescein isothiocyanate (FITC)-modified synthetic RNA (FITC-miRNA) whose sequence is identical to that of the target miRNA. The b-Cp-modified MBs were incubated in the sample solution supplemented with fixed concentrations of the FITC-miRNA and anti-FITC-HRP so that the FITC-miRNA competed with the target miRNA for hybridization with the b-Cp immobilized onto the Strep-MBs. Therefore, the higher the concentration of the target miRNA, the lower the amperometric response measured using H2O2 as enzyme substrate and HQ as redox mediator due to the lower number of FITC-miRNA molecules and therefore of anti-FITC-HRP attached to the MBs (see Figure 1a).
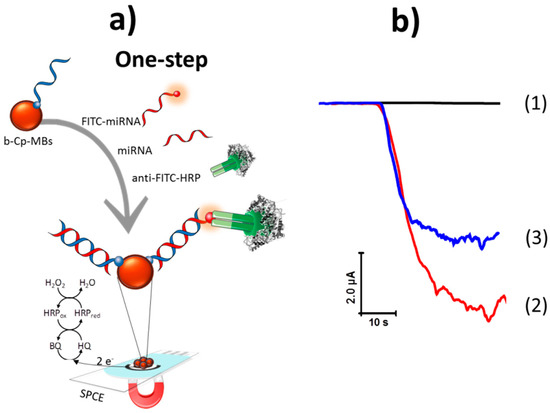
Figure 1.
(a) Schematic display of the amperometric biosensor developed for miRNA-21 determination based on a competitive DNA/RNA hybridization assay onto magnetic beads (MBs) and amperometric detection using the H2O2/ Hydroquinone (HQ) system at a screen-printed carbon electrode (SPCE). (b) Amperometric responses obtained with the developed biosensor in the absence (1) and in the presence of 2.5 nM FITC-miRNA (2) and 5.0 nM of miRNA-21 (3). HRP: horseradish peroxidase, BQ: 1,4-hydroquinone.
Figure 1b, amperogram 1, shows that no significant amperometric response was recorded in the absence of FITC-miRNA because there was no attached anti-FITC-HRP. As expected, the largest amperometric signal was obtained when the electrode was incubated in a solution containing only the synthetic FITC-miRNA (amperogram 2 in Figure 1b) in agreement with the larger number of attached anti-FITC-HRP. However, this response decreased notably in the presence of both the FITC-miRNA and the target miRNA (amperogram 3 in Figure 1b) due to their competitive hybridization with the b-Cp, leading to a lower amount of FITC-miRNA molecules and therefore of anti-FITC-HRP attached on the MBs. These results demonstrated the feasibility of the direct competitive hybridization assay onto b-Cp-MBs for the quantification of miRNAs.
All of the experimental variables involved in the preparation of the competitive hybridization-based biosensor for miRNA determination were optimized. The adopted selection criterion was the largest current ratio between the values measured at a previously optimized detection potential for the HRP/HQ/H2O2 system of −0.20 V (versus the Ag pseudo-reference electrode), [34], in the absence (S0) and in the presence of 5.0 nM (S1) synthetic miRNA-21 (S0/S1 ratio). Table 2 summarizes the tested variables, their checked ranges, and the values selected for further work.

Table 2.
Optimization of the experimental variables affecting the performance of the amperometric biosensor developed for miRNA-21 determination.
Two key variables in competitive hybridization assays are the loadings of the capture probe (b-Cp) and tracer agent (FITC-miRNA), which should be carefully optimized and kept fixed in the assays [35]. The b-Cp loading was tested between 0.5 and 50.0 nM. As it can be observed in Figure 2a, the largest S0/S1 ratio was found for 2.5 nM. For lower loadings, the responses in the absence of target miRNA were very low as a result of the small number of immobilized FITC-miRNA molecules. Conversely, larger loadings provoked a decrease in the S0/S1 ratio due to a hindered competition in the presence of large probe concentrations. This optimal low probe concentration is in agreement with the behavior expected in competitive configurations where higher sensitivity is obtained for a lower probe concentration, unlike what happens in sandwich formats [35]. The dependence of the amperometric signals with the FITC-miRNA concentration was tested over the 0.25 to 5 nM range. Data displayed in Figure 2b show that a larger S0/S1 ratio was found using a 2.5 nM concentration of FITC-miRNA, where a sufficiently large amperometric response was obtained in the presence of the target analyte.
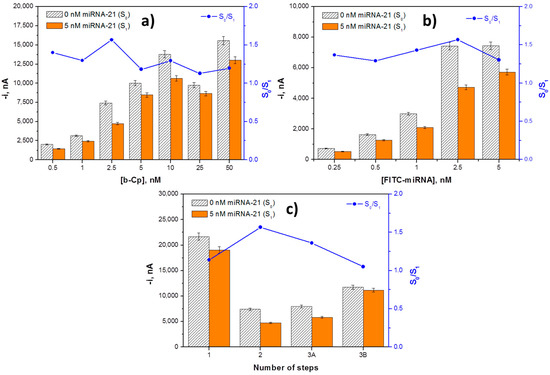
Figure 2.
Dependence of the amperometric signals measured with the competitive DNA/RNA hybridization biosensor in the absence (S0) and in the presence of 5.0 nM miRNA-21 (S1), and the corresponding S0/S1 ratio with the b-Cp (a) and FITC-miRNA (b) loadings and with the number of steps involved in the biosensor preparation (c). Error bars estimated as three times the standard deviation of three replicates.
Moreover, the sequence of the different steps involved in the biosensor preparation protocol may have an important effect on the assay performance. The different protocols evaluated, all involving 30 min incubation steps, include:
- -
- Protocol 1, single step: incubating the Strep-MBs with a mixture solution containing b-Cp, target miRNA-21, FITC-miRNA, and anti-FITC-HRP.
- -
- Protocol 2, 2 steps: (1) immobilization of the b-Cp onto the Strep-MBs and (2) incubation of the b-Cp-MBs with a mixture solution containing target miRNA-21, FITC-miRNA, and anti-FITC-HRP.
- -
- Protocol 3A, 3 steps: (1) immobilization of the b-Cp onto the Strep-MBs, (2) hybridization of the miRNA-21 onto the b-Cp-MBs, and (3) incubation of the miRNA-21/b-Cp-MBs with a mixture solution containing FITC-miRNA and anti-FITC-HRP.
- -
- Protocol 3B, 3 steps: (1) immobilization of the b-Cp onto the Strep-MBs, (2) incubation of the b-Cp-MBs with a mixture solution containing target miRNA-21 and FITC-miRNA, and (3) labeling of the FITC-miRNA attached to the b-Cp-MBs with the anti-FITC-HRP.
The amperometric responses measured for 0.0 and 5.0 nM of target miRNA-21 as well as the corresponding S0/S1 ratios using the different protocols are compared in Figure 2c. As can be observed, although protocols 1, 2, and 3A allowed for discrimination between the absence and presence of the target miRNA, the largest S0/S1 ratio was achieved following protocol 2, which was selected to implement the biosensing strategy. The poor discrimination observed using protocol 3 can be attributed to a less-favourable competition with FITC-miRNA labeled with the anti-FITC-HRP. The lack of discrimination observed using protocol 3B could be attributed to a hindered recognition of the FITC-miRNA, once immobilized on MBs, by the anti-FITC-HRP.
3.1. Analytical Characteristics
The calibration curve constructed for the amperometric determination of the synthetic target miRNA is displayed in Figure 3. As expected for a competitive hybridization assay, the larger the concentration of target miRNA the lower the amperometric response as a consequence of the lower concentration of FITC-miRNA and consequently of anti-FITC-HRP attached to the MBs. A linear dependence (r = 0.999) of the current values measured with the miRNA-21 concentration was found from 0.7 to 10.0 nM, with slope and intercept values of (−496 ± 12) nA nM−1 and (7279 ± 65) nA, respectively. The calculated LOD and LOQ values were 0.2 and 0.7 nM (5 and 17.5 fmol in 25 μL of sample), respectively.
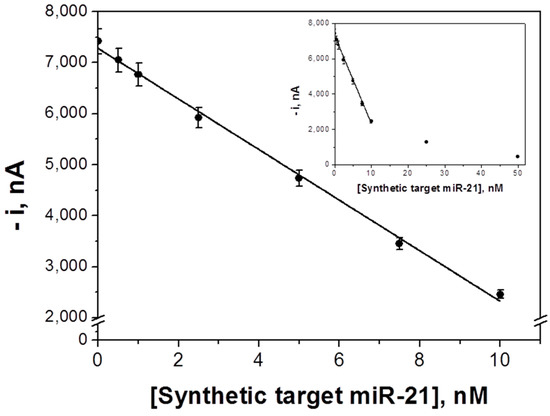
Figure 3.
Calibration plot constructed for synthetic target miRNA-21 with the competitive DNA/RNA hybridization biosensor. Inset shows the saturation effect at higher concentrations of the target miRNA. Error bars estimated as three times the standard deviation of three replicates.
The achieved LOD is similar (0.2 versus 0.4 nM) [27] or higher than those reported for other MBs-based approaches for the determination of miRNAs (6 aM [28], 0.4 pM [31], 2.4 pM [29], 40 pM [26], and 60 pM [32]). However, the methodology reported here is carried out in a single incubation step (once the MBs were modified with the b-Cp) with a very low concentration of synthetic nucleic acid probe (2.5 nM) and without any amplification strategy. In addition, no expensive affinity bioreceptors [26,27,28,29,30,31] were needed. These features greatly simplify the potential implementation of this technology in portable and automatic devices suitable to carry out the determination at affordable cost in different settings. It is also worthwhile mentioning that the sensitivity achieved is sufficient for relevant applications as are shown in Section 3.3. Moreover, it is worth mentioning that although 2 h is the optimal time, the determination could be performed in half the time (1 h) losing 66% of the sensitivity.
The amperometric responses for 5.0 nM miRNA-21 provided by 10 different biosensors prepared in the same manner exhibited a high reproducibility with a relative standard deviation (RSD) value of 3.3%, thus confirming the suitability of both the sensor fabrication and the amperometric transduction protocols used.
The storage stability of the b-Cp-MBs was evaluated by keeping them at 4 °C in microcentrifuge tubes containing 50 μL of filtered PBS. Each working day, the amperometric responses obtained with the biosensors prepared using the stored b-Cp-MBs for 0.0 and 5.0 nM synthetic miRNA-21 solutions were compared. Interestingly, no significant decrease in the resulting S0/S1 ratio (not shown) was observed during 49 days (no longer times were evaluated). This behavior creates the possibility of preparing and then storing the b-Cp-MBs for at least this amount of time until biosensor preparation is required.
3.2. Selectivity
The selectivity of the developed biosensor was checked by comparing the amperometric signals measured in the absence (S0) and in the presence of 5.0 nM miRNA-21 with those recorded in the presence of two different single-base mismatched in central (1-m(c)) or terminal (1-m(t)) position sequences, two fully non-complementary (NC) sequences (corresponding to miRNA-205 and miRNA-122), and mixtures containing the target miRNA and the two 1-m (mixture 1) or the two NC (mixture 2) sequences (each at a 5.0 nM concentration). As it is shown in Figure 4, the amperometric responses obtained with the NC sequences and the 1-m (c) were similar to those measured in the absence of target miRNA, thus proving that these sequences cannot compete with the FITC-miRNA for the b-Cp immobilized onto the MBs. Moreover, the amperometric response obtained for 1-m (t) is intermediate between the responses obtained in the absence and in the presence of target RNA, which is in agreement with a less-favorable hybridization with this sequence due to the presence of the mismatched base in the terminal position of the hybrid. The amperometric signals obtained for the mixture solutions are in agreement with the results for the individual sequences. The responses recorded for the mixtures prepared in the presence of the 1-m and the NC sequences are lower and similar to that obtained in the presence of the target miRNA alone. These results confirm the high selectivity of the developed biosensor against the NC and 1-m (c) sequences. The selectivity towards 1-m (t) can be considered acceptable. Considering the low probability that the target miRNA was in the sample with 1-m sequences at the same concentration level and with the mismatched base in terminal position [36], this fact cannot be considered as an important problem for the applicability of the biosensor. Indeed, the results achieved with the two 1-m sequences demonstrated the potential ability of the developed biosensor to detect the existence and even the position of a single mismatched base.
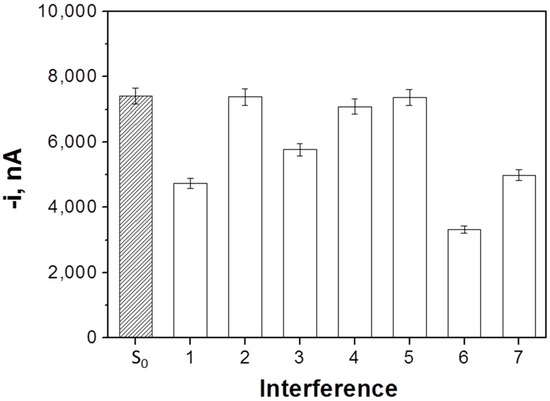
Figure 4.
Selectivity of the developed competitive DNA/RNA hybridization-based biosensor for determination of miRNA-21. Amperometric responses measured in the absence (S0) and in the presence of 5.0 nM miRNA-21 (1), 1-m (c) (2), 1-m (t) (3), NC1 (miRNA-205) (4), NC2 (miRNA-122) (5), and mixture solutions containing the target miRNA and the two 1-m (6) or the target miRNA and the two non-complementary (NC) sequences (7). Error bars estimated as three times the standard deviation of three replicates.
3.3. Determination of Mature miRNA-21 in RNAt Extracted from Cancer Cells
The clear association between the hyperexpression of miRNA-21 and the incidence and progression of breast cancer [37,38,39,40] led us to evaluate the applicability of the developed biosensor by determining the mature content of the target miRNA in breast cancer (MCF-7) and non-tumoral breast primary epithelial (MCF-10A) cells.
The amperometric signals provided by the biosensor in the analysis of different amounts of RNAt extracted from both types of cell are shown in Figure 5. As expected, substantially lower amperometric responses were measured for RNAt extracted from the MCF-7 when compared with those from MCF-10A cells according to the overexpression of the oncogenic target miRNA in the cancerous cells [7,26,27,32,41] as long as the amount of extracted RNAt from MCF-7 cells was at least 1 μg.
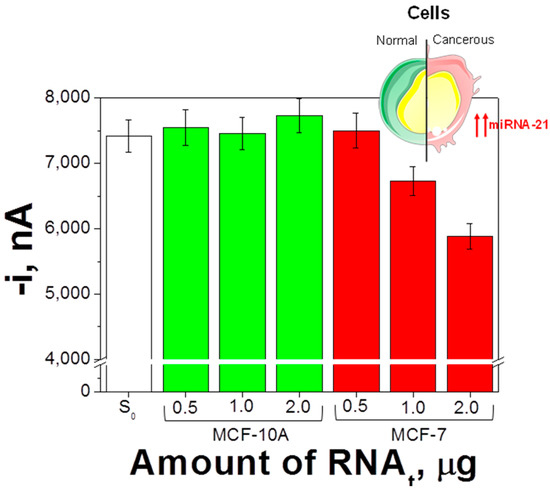
Figure 5.
Determination of the endogenous content of miRNA-21 in raw total RNA (RNAt) extracted from breast cells. Amperometric responses measured with the developed biosensor in the absence (S0) and in the presence of different amounts of RNAt extracted from MCF-10A and MCF-7 cells. Error bars estimated as three times the standard deviation of three replicates.
Moreover, the slope value of the linear calibration plot constructed for miRNA-21 in the presence of 1.0 μg of RNAt extracted from MCF-10A (−564 ± 87) nA·nM−1 was not significantly different than that measured for miRNA-21 in buffered solutions (−496 ± 12) nA·nM−1. Therefore, no apparent matrix effect occurred using this RNAt amount and the endogenous content of the target miRNA in the MCF-7 cells could be estimated by simple interpolation of the amperometric responses measured for these cells into the calibration graph constructed with miRNA-21 standards (Figure 4). Results obtained were (25 ± 4) amol miRNA-21 ng−1 RNAt (RSDn=5 = 12.1%), which were similar to those reported by other authors, (18 ± 2) [26], 21.7 [27], and (21 ± 3) amol miRNA-21 ng−1 of RNAt [32], using other electrochemical biosensing strategies. It is also useful to note that the reported levels of miRNA-21 in MCF-10A cells are on the order of 1.0 amol ng−1 RNAt, while they are in the range of 0.1−1.5 and 0.4−3.9 amol per ng RNAt in breast normal and tumor tissues, respectively [29,42].
Regarding the comparison with other electrochemical methods applied to the determination of the endogenous content of a target miRNA in cell lysates [26,27,29,30,31,32], as well as with other types of detection [43], the most relevant advantage of the methodology reported here is the simplicity of the determination in one single step, without long and expensive chemistry to label the target miRNA, and using just 1.0 μg of the extracted RNAt.
These results demonstrated an acceptable reproducibility of the whole method, including the RNAt-extraction protocol and the determination with the developed electrochemical biosensor, as well as the feasibility and potential applicability of the developed approach to perform a simple and accurate determination of the target miRNA in just one incubation step and directly in raw RNAt extracted from cancerous cells, without reverse transcription, amplification, preconcentration, or labeling steps. Moreover, the straightforward determination in this raw RNAt matrix, where many other non-target miRNAs are also present to a large extent, further highlights the excellent selectivity of the developed methodology.
4. Conclusions
This work describes an amperometric biosensor based on a competitive DNA/RNA hybridization format implemented on the surface of MBs and SPCEs for the determination of miRNA-21. This strategy exhibits an attractive analytical performance with a LOD of 0.2 nM (5.0 fmol in 25 μL of sample), a good reproducibility between the amperometric responses provided by different biosensors prepared in the same manner, a clear and acceptable discrimination towards non-complementary and 1-m sequences, respectively, and a 49-day storage stability of the magnetic bioconjugates.
It is worth mentioning also that the developed methodology allowed for the reliable and accurate determination of the target miRNA in cancer cells involving a simple and 1-step protocol (once the b-Cp-MBs were prepared) without requiring previous reverse transcription to cDNA, amplification, preconcentration, or purification steps. This performance makes it ideal for routine determinations in both clinical and research settings and as a basis for future diagnostic point-of-care (POC) devices. Moreover, although focused only on the determination of miRNA-21 as model, the same methodology can be applied to the determination of any miRNA in single or multiplexed formats simply by modifying the Strep-MBs with the appropriate complementary biotinylated DNA capture probes.
Acknowledgments
The financial support of the Spanish Ministerio de Economía y Competitividad, CTQ2015-64402-C2-1-R, and the NANOAVANSENS Program from the Comunidad de Madrid (S2013/MT−3029) and predoctoral contracts from the Spanish Ministerio de Economía y Competitividad (R.M.T.-R. and E.P.) and Universidad Complutense de Madrid (V.R.-V.M.) are gratefully acknowledged. E. Vargas acknowledges her contract to the Mirnax Biosens, S.L. The University of Tunis El Manar is acknowledged for the mobility grant (Bourse d’Alternance) awarded to M.Z. The Tunisian Ministry of Higher Education and Scientific Research is acknowledged for financial support to LR99ES15. The authors would also like to acknowledge J.M. Sánchez-Puelles and E. López-Hernández for kindly providing us with the RNAt samples extracted from cells.
Author Contributions
E.V., E.P., V.R.-V.M., S.C. and J.M.P. designed the experiments. E.V., E.P., V.R.-V.M., R.M.T.-R. and M.Z. performed the experiments. The manuscript was written through contributions of all authors. J.J.M., N.R., S.C. and J.M.P. wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hamam, R.; Hamam, D.; Alsaleh, K.A.; Kassem, M.; Zaher, W.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. Circulating microRNAs in breast cancer: Novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017, 8, e3045. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, S.; Pedrero, M.; Pingarrón, J.M. Non-Invasive Breast Cancer Diagnosis through Electrochemical Biosensing at Different Molecular Levels. Sensors 2017, 17, 1993. [Google Scholar] [CrossRef] [PubMed]
- Nicolini, A.; Ferrari, P.; Duffy, M.J. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin. Cancer Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-X.; Huang, K.-J.; Niu, K.-X. Recent advances in signal amplification strategy based on oligonucleotide and nanomaterials for microRNA detection-a review. Biosens. Bioelectron. 2018, 99, 612–624. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.W.; Chen, L.; Man, Y.-G. miRNA Biomarkers in Breast Cancer Detection and Management. J. Cancer 2011, 2, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Hasanzadeh, M.; Shadjou, N.; de la Guardia, M. Early stage screening of breast cancer using electrochemical biomarker detection. Trends Anal. Chem. 2017, 91, 67–76. [Google Scholar] [CrossRef]
- Cissell, K.A.; Rahimi, Y.; Shrestha, S.; Hunt, E.A.; Deo, S.K. Bioluminescence-Based Detection of MicroRNA, miR21 in Breast Cancer Cells. Anal. Chem. 2008, 80, 2319–2325. [Google Scholar] [CrossRef] [PubMed]
- Šípova, H.; Zhang, S.; Dudley, A.M.; Galas, D.; Wang, K.; Homola, J. Surface plasmon resonance biosensor for rapid label-free detection of microribonucleic acid at subfemtomole level. Anal. Chem. 2010, 82, 10110–10115. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, F. Label-free and enzyme-free homogeneous electrochemical biosensing strategy based on hybridization chain reaction: A facile, sensitive, and highly specific microRNA assay. Anal. Chem. 2015, 87, 11368–11374. [Google Scholar]
- Liao, Y.; Fu, Y.; Wu, Y.; Huang, R.; Zhou, X.; Xing, D. Ultrasensitive detection of microRNA in tumor cells and tissues via continuous assembly of DNA probe. Biomacromolecules 2015, 16, 3543–3551. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, D.-Z.; Cai, S.-X.; Chen, M.; Xia, Y.-K.; Wu, F.; Chen, J.-H. An immobilization-free electrochemical impedance biosensor based on duplex-specific nuclease assisted target recycling for amplified detection of microRNA. Biosens. Bioelectron. 2016, 75, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Kilic, T.; Erdem, A.; Ozsoz, M.; Carrara, S. MicroRNA biosensors: Opportunities and challenges among conventional and commercially available techniques. Biosens. Bioelectron. 2018, 99, 525–546. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.T.; Baldwin, D.A.; Scearce, L.M.; Oberholtzer, J.C.; Tobias, J.W.; Mourelatos, Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods 2004, 1, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.-Q.; Li, W.; Li, Y.; Tan, C.; Li, J.X.; Jin, Y.X.; Ruan, K.C. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Res. 2005, 33, e17. [Google Scholar] [CrossRef] [PubMed]
- Git, A.; Dvinge, H.; Salmon-Divon, M.; Osborne, M.; Kutter, C.; Hadfield, J.; Bertone, P.; Caldas, C. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 2010, 16, 991–1006. [Google Scholar] [CrossRef] [PubMed]
- Clancy, E.; Burke, M.; Arabkari, V.; Barry, T.; Kelly, H.; Dwyer, R.M.; Kerin, M.J.; Smith, T.J. Amplification-free detection of microRNAs via a rapid microarray-based sandwich assay. Anal. Bioanal. Chem. 2017, 409, 3497–3505. [Google Scholar] [CrossRef] [PubMed]
- Válóczi, A.; Hornyik, C.; Varga, N.; Burgyán, J.; Kauppinen, S.; Havelda, Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004, 32, e175. [Google Scholar] [CrossRef] [PubMed]
- Pall, G.S.; Codony-Servat, C.; Byrne, J.; Ritchie, L.; Hamilton, A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007, 35, e60. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yao, B.; Huang, H.; Wang, Z.; Sun, C.; Fan, Y.; Chang, Q.; Li, S.; Wang, X.; Xi, J. Real-time polymerase chain reaction microRNA detection based on enzymatic stem-loop probes ligation. Anal. Chem. 2009, 81, 5446–5451. [Google Scholar] [CrossRef] [PubMed]
- Kroh, E.M.; Parkin, R.K.; Mitchell, P.S.; Tewari, M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 2010, 50, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef] [PubMed]
- Centi, S.; Laschi, S.; Frànek, M.; Mascini, M. A disposable immunomagnetic electrochemical sensor based on functionalized magnetic beads and carbon-based screen-printed electrodes (SPCEs) for the detection of polychlorinated biphenyls (PCBs). Anal. Chim. Acta 2005, 538, 205–212. [Google Scholar] [CrossRef]
- Zacco, E.; Adrian, J.; Galve, R.; Marco, M.P.; Alegret, S.; Pividori, M.I. Electrochemical magneto immunosensing of antibiotic residues in milk. Biosens. Bioelectron. 2007, 22, 2184–2191. [Google Scholar] [CrossRef] [PubMed]
- Ricci, F.; Volpe, G.; Micheli, L.; Palleschi, G. A review on novel developments and applications of immunosensors in food analysis. Anal. Chim. Acta 2007, 605, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, E. Electrochemical biosensors based on magnetic micro/nano particles. Electrochim. Acta 2012, 84, 62–73. [Google Scholar] [CrossRef]
- Campuzano, S.; Torrente-Rodríguez, R.M.; López-Hernández, E.; Conzuelo, F.; Granados, R.; Sánchez-Puelles, J.M.; Pingarrón, J.M. Magnetobiosensors based on viral protein p19 for microRNA determination in cancer cells and tissues. Angew. Chem. Int. Ed. 2014, 53, 6168–6171. [Google Scholar] [CrossRef] [PubMed]
- Torrente-Rodríguez, R.M.; Campuzano, S.; López-Hernández, E.; Granados, R.; Sánchez-Puelles, J.M.; Pingarrón, J.M. Direct determination of miR-21 in total RNA extracted from breast cancer samples using magnetosensing platforms and the p19 viral protein as detector bioreceptor. Electroanalysis 2014, 26, 2080–2087. [Google Scholar] [CrossRef]
- Li, C.; Liu, Z.; Cai, S.; Wen, F.; Wu, D.; Liu, Y.; Wu, F.; Lan, J.; Han, Z.; Chen, J. An electrochemical microRNA biosensor based on protein p19 combining an acridone derivate as indicator and DNA concatamers for signal amplification. Electrochem. Commun. 2015, 60, 185–189. [Google Scholar] [CrossRef]
- Torrente-Rodríguez, R.M.; Ruiz-Valdepeñas Montiel, V.; Campuzano, S.; Fachardo-Dinia, M.; Barderas, R.; San Segundo-Acosta, P.; Montoya, J.J.; Pingarrón, J.M. Fast electrochemical miRNAs determination in cancer cells and tumor tissues with antibody-functionalized magnetic microcarriers. ACS Sens. 2016, 1, 896–903. [Google Scholar] [CrossRef]
- Torrente-Rodríguez, R.M.; Campuzano, S.; Ruiz-Valdepeñas Montiel, V.; Sagrera, A.; Domínguez-Cañete, J.J.; Vargas, E.; Montoya, J.J.; Granados, R.; Sánchez-Puelles, J.M.; Pingarrón, J.M. Electrochemical miRNAs Determination in Formalin-Fixed, Paraffin-Embedded Breast Tumor Tissues Association with HER2 Expression. JSM Biotechnol. Bioeng. 2016, 3, 1064. [Google Scholar]
- Vargas, E.; Torrente-Rodríguez, R.M.; Ruiz-Valdepeñas Montiel, V.; Povedano, E.; Pedrero, M.; Montoya, J.J.; Campuzano, S.; Pingarrón, J.M. Magnetic Beads-Based Sensor with tailored sensitivity for rapid and single-step amperometric determination of miRNAs. Int. J. Mol. Sci. 2017, 18, 2151. [Google Scholar] [CrossRef] [PubMed]
- Torrente-Rodríguez, R.M.; Campuzano, S.; Ruiz-Valdepeñas Montiel, V.; Montoya, J.J.; Pingarrón, J.M. Sensitive electrochemical determination of miRNAs based on a sandwich assay onto magnetic microcarriers and hybridization chain reaction amplification. Biosens. Bioelectron. 2016, 86, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Gangwar, R.S.; Rajagopalan, S.; Natarajan, R.; Deiuliis, J.A. Noncoding RNAs in Cardiovascular Disease: Pathological Relevance and Emerging Role as Biomarkers and Therapeutics. Am. J. Hypertens. 2018, 31, 150–165. [Google Scholar] [CrossRef] [PubMed]
- Gamella, M.; Campuzano, S.; Conzuelo, F.; Reviejo, A.J.; Pingarrón, J.M. Amperometric Magnetoimmunosensors for Direct Determination of D-Dimer in Human Serum. Electroanalysis 2012, 24, 2235–2243. [Google Scholar] [CrossRef]
- Wang, W.; Kong, T.; Zhang, D.; Zhang, J.; Cheng, G. Label-Free MicroRNA Detection Based on Fluorescence Quenching of Gold Nanoparticles with a Competitive Hybridization. Anal. Chem. 2015, 87, 10822–10829. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Peng, J.; Wang, J.; Tang, H.; Tan, L.; Xie, Q.; Yao, S. Carbon nanotube-based label-free electrochemical biosensor for sensitive detection of miRNA-24. Biosens. Bioelectron. 2014, 54, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Markou, A.; Yousef, G.M.; Stathopoulos, E.; Georgoulias, V.; Lianidou, E. Prognostic significance of metastasis-related microRNAs in early breast cancer patients with a long follow-up. Clin. Chem. 2014, 60, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, B.S.; Balslev, E.; Poulsen, T.S.; Nielsen, D.; Møller, T.; Mortensen, C.E.; Holmstrøm, K.; Høgdall, E. miR-21 expression in cancer cells may not predict resistance to adjuvant trastuzumab in primary breast cancer. Front. Oncol. 2014, 4, 207. [Google Scholar] [CrossRef] [PubMed]
- De Mattos-Arruda, L.; Bottai, G.; Nuciforo, P.G.; Di Tommaso, L.; Giovannetti, E.; Peg, V.; Losurdo, A.; Pérez-Garcia, J.; Masci, G.; Corsi, F.; et al. MicroRNA-21 links epithelial-to-mesenchymal transition and inflammatory signals to confer resistance to neoadjuvant trastuzumab and chemotherapy in HER2-positive breast cancer patients. Oncotarget 2015, 6, 37269–37280. [Google Scholar] [CrossRef] [PubMed]
- Kurozumi, S.; Yamaguchi, Y.; Kurosumi, M.; Ohira, M.; Matsumoto, H.; Horiguchi, J. Recent trends in microRNA research into breast cancer with particular focus on the associations between microRNAs and intrinsic subtypes. J. Hum. Genet. 2017, 62, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.X.; Wu, Q.N.; Zhang, Y.; Li, Y.Y.; Liao, D.Z.; Hou, J.H.; Fu, J.; Zeng, M.S.; Yun, J.P.; Wu, Q.L.; et al. Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, In Vitro migration and In Vivo tumor growth. Breast Cancer Res. 2011, 13, R2. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Yang, T.; Chen, Y. Quantification of microRNA by DNA−peptide probe and liquid chromatography−tandem mass spectrometry-based quasi-targeted proteomics. Anal. Chem. 2016, 88, 754–763. [Google Scholar] [CrossRef] [PubMed]
- Novara, C.; Chiadò, A.; Paccotti, N.; Catuogno, S.; Esposito, C.L.; Condorelli, G.; De Franciscis, V.; Geobaldo, F.; Rivolo, P.; Giorgis, F. SERS-active metal-dielectric nanostructures integrated in microfluidic devices for label-free quantitative detection of miRNA. Faraday Discuss. 2017, 205, 271–289. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).