Plant Temperature Sensors
Abstract
1. Temperature and Living Organisms
2. Effects of Temperature on Growth and Development
2.1. Tissue Elongation under High Temperature
2.2. Leaf Shape Modification
2.3. Thermotolerance
3. Temperature as a Seasonal Cue
3.1. Flowering Time
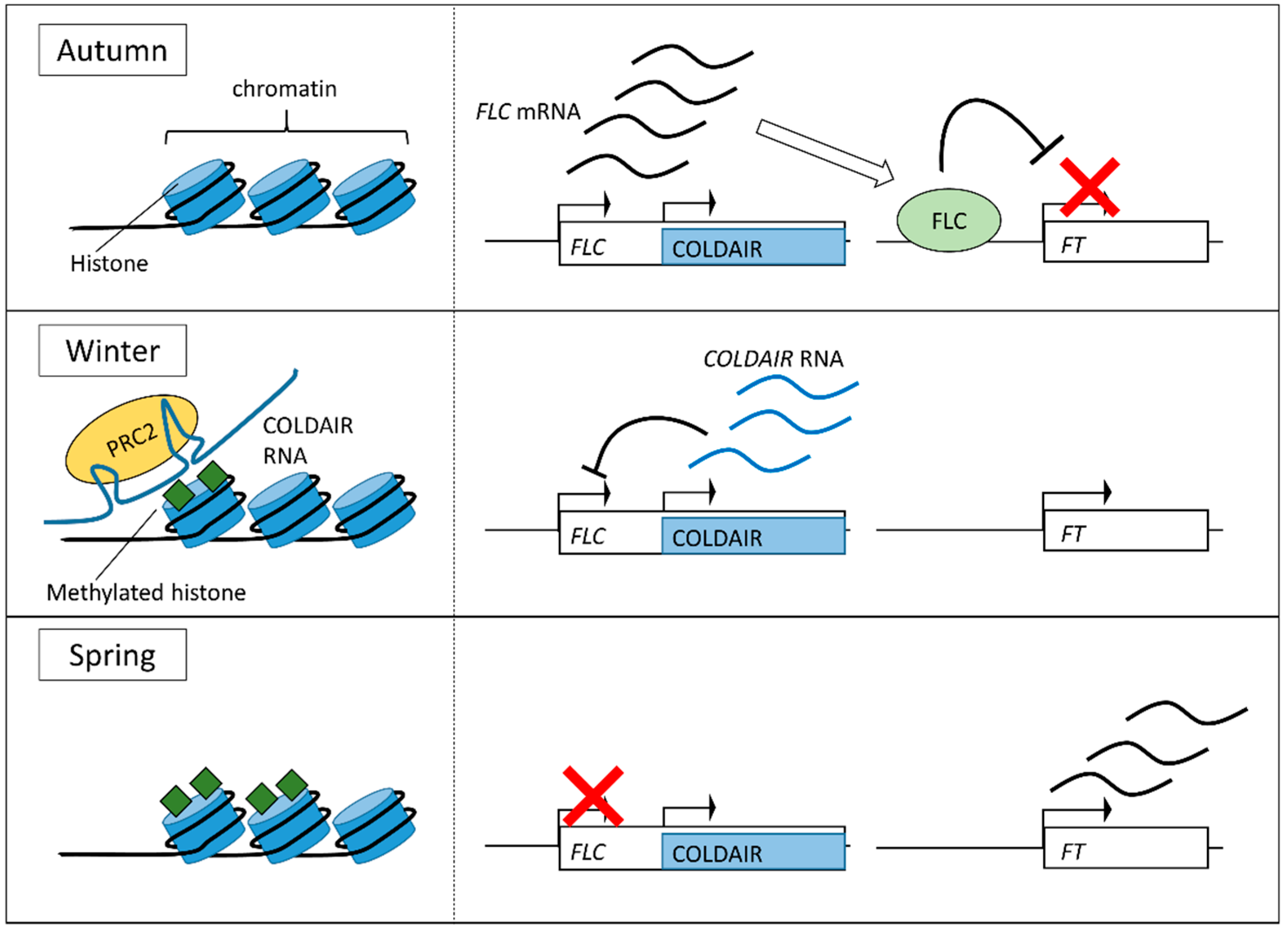
3.2. Vernalization
4. The Mechanisms Sensing Temperature
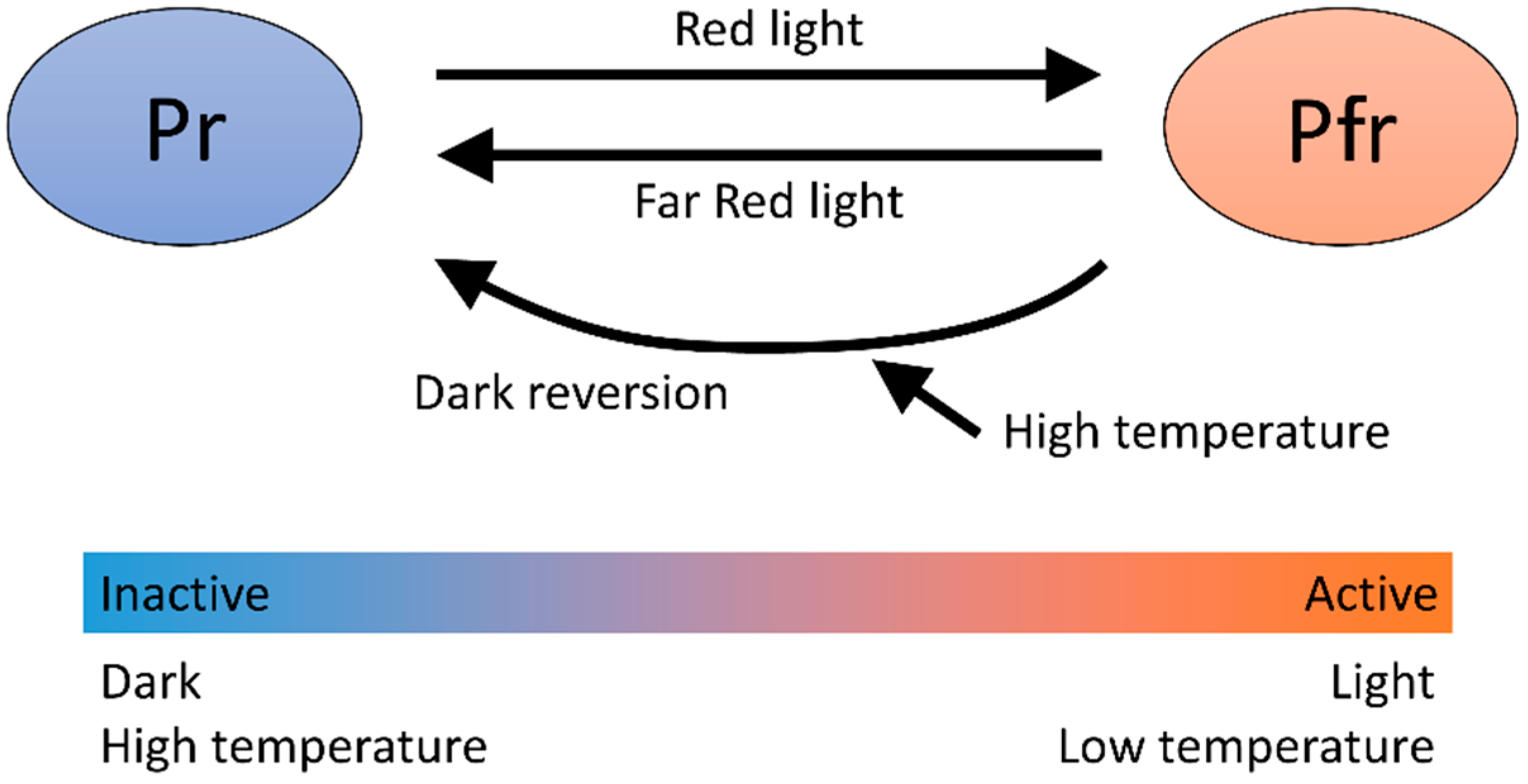
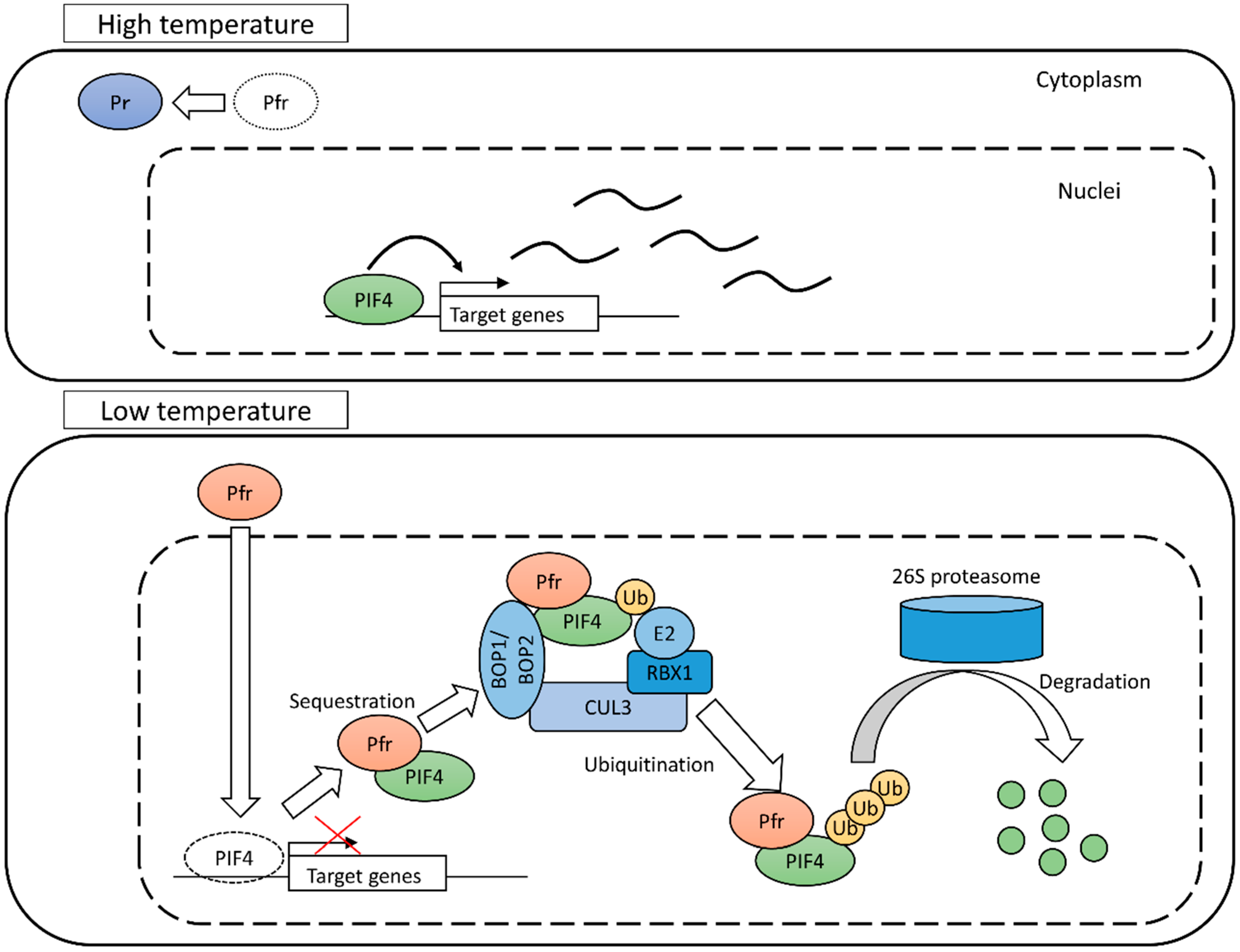
4.1. Phytochrome B
4.2. Temperature Sensing for Heat Shock Response
4.3. Other Temperature Sensing Systems
5. Conclusions
Funding
Conflicts of Interest
References
- Gray, W.M.; Ostin, A.; Sandberg, G.; Romano, C.P.; Estelle, M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 7197–7202. [Google Scholar] [CrossRef] [PubMed]
- Koini, M.A.; Alvey, L.; Allen, T.; Tilley, C.A.; Harberd, N.P.; Whitelam, G.C.; Franklin, K.A. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 2009, 19, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Nakayama, N.; Seiki, S.; Kojima, M.; Sakakibara, H.; Sinha, N.; Kimura, S. Regulation of the KNOX-GA gene module induces heterophyllic alteration in North American lake cress. Plant Cell 2014, 26, 4733–4748. [Google Scholar] [CrossRef] [PubMed]
- Royer, D.L. Leaf shape responds to temperature but not CO2 in Acer rubrum. PLoS ONE 2012, 7, e49559. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, K.; Fang, X.; Alvarez-Ponce, D.; Cai, H.; Carretero-Paulet, L.; Chen, C.; Chang, T.; Farr, K.; Fujita, T.; Hiwatashi, Y.; et al. Genome of the pitcher plant Cephalotus reveals genetic changes associated with carnivory. Nat. Ecol. Evol. 2017, 1, 59. [Google Scholar] [CrossRef]
- Larkindale, J.; Vierling, E. Core genome responses involved in acclimation to high temperature. Plant Physiol. 2008, 146, 748–761. [Google Scholar] [CrossRef]
- Hong, S.W.; Vierling, E. Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc. Natl. Acad. Sci. USA 2000, 97, 4392–4397. [Google Scholar] [CrossRef]
- Jagadish, S.V.K.; Bahuguna, R.N.; Djanaguiraman, M.; Gamuyao, R.; Prasad, P.V.V.; Craufurd, P.Q. Implications of High Temperature and Elevated CO2 on Flowering Time in Plants. Front. Plant Sci. 2016, 7, 913. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Sureshkumar, S.; Lempe, J.; Weigel, D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2006, 2, e106. [Google Scholar] [CrossRef]
- Luan, W.; Chen, H.; Fu, Y.; Si, H.; Peng, W.; Song, S.; Liu, W.; Hu, G.; Sun, Z.; Xie, D.; et al. The effect of the crosstalk between photoperiod and temperature on the heading-date in rice. PLoS ONE 2009, 4, e5891. [Google Scholar] [CrossRef]
- Nakano, Y.; Higuchi, Y.; Sumitomo, K.; Hisamatsu, T. Flowering retardation by high temperature in chrysanthemums: Involvement of FLOWERING LOCUS T-like 3 gene repression. J. Exp. Bot. 2013, 64, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Bouche, F.; Woods, D.P.; Amasino, R.M. Winter Memory throughout the Plant Kingdom: Different Paths to Flowering. Plant Physiol. 2017, 173, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Su, Y.; Lagarias, J.C. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 2006, 57, 837–858. [Google Scholar] [CrossRef]
- Burgie, E.S.; Bussell, A.N.; Walker, J.M.; Dubiel, K.; Vierstra, R.D. Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl. Acad. Sci. USA 2014, 111, 10179–10184. [Google Scholar] [CrossRef]
- Franklin, K.A.; Davis, S.J.; Stoddart, W.M.; Vierstra, R.D.; Whitelam, G.C. Mutant analyses define multiple roles for Phytochrome C in Arabidopsis photomorphogenesis. Plant Cell 2003, 15, 1981–1989. [Google Scholar] [CrossRef] [PubMed]
- Legris, M.; Klose, C.; Burgie, E.S.; Rojas, C.C.R.; Neme, M.; Hiltbrunner, A.; Wigge, P.A.; Schafer, E.; Vierstra, R.D.; Casal, J.J. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 2016, 354, 897–900. [Google Scholar] [CrossRef]
- Jung, J.; Domijan, M.; Klose, C.; Biswas, S.; Ezer, D.; Gao, M.; Khattak, A.K.; Box, M.; Charoensawan, V.; Cortijo, S.; et al. Phytochromes function as thermosensors in Arabidopsis. Science 2016, 354, 886–889. [Google Scholar] [CrossRef]
- Ni, M.; Tepperman, J.M.; Quail, P.H. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 1999, 400, 781–784. [Google Scholar] [CrossRef]
- Shimizu-Sato, S.; Huq, E.; Tepperman, J.M.; Quail, P.H. A light-switchable gene promoter system. Nat. Biotechnol. 2002, 20, 1041–1044. [Google Scholar] [CrossRef] [PubMed]
- Lorrain, S.; Allen, T.; Duek, P.D.; Whitelam, G.C.; Fankhauser, C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008, 53, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Foreman, J.; Johansson, H.; Hornitschek, P.; Josse, E.; Fankhauser, C.; Halliday, K.J. Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J. 2011, 65, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Legris, M.; Nieto, C.; Sellaro, R.; Prat, S.; Casal, J.J. Perception and signalling of light and temperature cues in plants. Plant J. 2017, 90, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Holmlund, M.; Lorrain, S.; Norberg, M.; Bako, L.; Fankhauser, C.; Nilsson, O. BLADE-ON-PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. eLife 2017, 6, e26759. [Google Scholar] [CrossRef] [PubMed]
- Genschik, P.; Sumara, I.; Lechner, E. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): Cellular functions and disease implications. EMBO J. 2013, 32, 2307–2320. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Kim, Y.; Choi, G. Phytochrome B Requires PIF Degradation and Sequestration to Induce Light Responses across a Wide Range of Light Conditions. Plant Cell 2018, 30, 1277–1292. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Lee, S.H.; Patel, D.; Kumar, S.V.; Spartz, A.K.; Gu, C.; Ye, S.; Yu, P.; Breen, G.; Cohen, J.D.; et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 2011, 108, 20231–20235. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Qi, L.; Li, Y.; Chu, J.; Li, C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 2012, 8, e1002594. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.K.; Tripp, J.; Winkelhaus, S.; Tschiersch, B.; Theres, K.; Nover, L.; Scharf, K.-D. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 2002, 16, 1555–1567. [Google Scholar] [CrossRef]
- Liu, H.-C.; Liao, H.-T.; Charng, Y.-Y. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011, 34, 738–751. [Google Scholar] [CrossRef]
- Liu, H.-T.; Gao, F.; Li, G.-L.; Han, J.-L.; Liu, D.-L.; Sun, D.-Y.; Zhou, R.-G. The calmodulin-binding protein kinase 3 is part of heat-shock signal transduction in Arabidopsis thaliana. Plant J. 2008, 55, 760–773. [Google Scholar] [CrossRef]
- Liu, H.-T.; Li, G.-L.; Chang, H.; Sun, D.-Y.; Zhou, R.-G.; Li, B. Calmodulin-binding protein phosphatase PP7 is involved in thermotolerance in Arabidopsis. Plant Cell Environ. 2007, 30, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Fukao, Y.; Hayashi, M.; Fukazawa, M.; Suzuki, I.; Nishimura, M. Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J. Biol. Chem. 2007, 282, 37794–37804. [Google Scholar] [CrossRef] [PubMed]
- Ohama, N.; Kusakabe, K.; Mizoi, J.; Zhao, H.; Kidokoro, S.; Koizumi, S.; Takahashi, F.; Ishida, T.; Yanagisawa, S.; Shinozaki, K.; et al. The Transcriptional Cascade in the Heat Stress Response of Arabidopsis Is Strictly Regulated at the Level of Transcription Factor Expression. Plant Cell 2016, 28, 181–201. [Google Scholar] [CrossRef] [PubMed]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, C.C.; Burn, J.E.; Perez, P.P.; Metzger, J.; Edwards, J.A.; Peacock, W.J.; Dennis, E.S. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 1999, 11, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Sung, S.; Amasino, R.M. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 2004, 427, 159–164. [Google Scholar] [CrossRef] [PubMed]
- De Lucia, F.; Crevillen, P.; Jones, A.M.E.; Greb, T.; Dean, C. A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. USA 2008, 105, 16831–16836. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Xi, Y.; Sung, S. Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 2017, 13, e1006939. [Google Scholar] [CrossRef]
- Kumar, S.V.; Wigge, P.A. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 2010, 140, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Lucyshyn, D.; Jaeger, K.E.; Alos, E.; Alvey, E.; Harberd, N.P.; Wigge, P.A. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 2012, 484, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Cortijo, S.; Charoensawan, V.; Brestovitsky, A.; Buning, R.; Ravarani, C.; Rhodes, D.; van Noort, J.; Jaeger, K.E.; Wigge, P.A. Transcriptional Regulation of the Ambient Temperature Response by H2A.Z Nucleosomes and HSF1 Transcription Factors in Arabidopsis. Mol. Plant 2017, 10, 1258–1273. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Li, X.; Guo, Y.; Chu, J.; Fang, S.; Yan, C.; Noel, J.P.; Liu, H. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA 2016, 113, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Hayes, S.; Sharma, A.; Fraser, D.P.; Trevisan, M.; Cragg-Barber, C.K.; Tavridou, E.; Fankhauser, C.; Jenkins, G.I.; Franklin, K.A. UV-B Perceived by the UVR8 Photoreceptor Inhibits Plant Thermomorphogenesis. Curr. Biol. 2017, 27, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Fujii, Y.; Tanaka, H.; Konno, N.; Ogasawara, Y.; Hamashima, N.; Tamura, S.; Hasegawa, S.; Hayasaki, Y.; Okajima, K.; Kodama, Y. Phototropin perceives temperature based on the lifetime of its photoactivated state. Proc. Natl. Acad. Sci. USA 2017, 114, 9206–9211. [Google Scholar] [CrossRef] [PubMed]
- Bechtold, U.; Albihlal, W.S.; Lawson, T.; Fryer, M.J.; Sparrow, P.A.C.; Richard, F.; Persad, R.; Bowden, L.; Hickman, R.; Martin, C.; et al. Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b overexpression enhances water productivity, resistance to drought, and infection. J. Exp. Bot. 2013, 64, 3467–3481. [Google Scholar] [CrossRef] [PubMed]
- Box, M.S.; Huang, B.E.; Domijan, M.; Jaeger, K.E.; Khattak, A.K.; Yoo, S.J.; Sedivy, E.L.; Jones, D.M.; Hearn, T.J.; Webb, A.A.R.; et al. ELF3 controls thermoresponsive growth in Arabidopsis. Curr. Biol. 2015, 25, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Ezer, D.; Jung, J.H.; Lan, H.; Biswas, S.; Gregoire, L.; Box, M.S.; Charoensawan, V.; Cortijo, S.; Lai, X.; Stockle, D.; et al. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants 2017, 3, 17087. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Alvarez, S.; Bindbeutel, R.; Shen, Z.; Naldrett, M.J.; Evans, B.S.; Briggs, S.P.; Hicks, L.M.; Kay, S.A.; Nusinow, D.A. Identification of Evening Complex Associated Proteins in Arabidopsis by Affinity Purification and Mass Spectrometry. Mol. Cell. Proteom. 2016, 15, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, P.J.; Kumar, M.; Martinho, C.; Yoo, S.J.; Lan, H.; Artavanis, G.; Charoensawan, V.; Schottler, M.A.; Bock, R.; Jaeger, K.E.; et al. Chloroplast Signaling Gates Thermotolerance in Arabidopsis. Cell Rep. 2018, 22, 1657–1665. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, C.; Delker, C.; Martinez, C.; Burstenbinder, K.; Janitza, P.; Lippmann, R.; Ludwig, W.; Sun, H.; James, G.V.; Klecker, M.; et al. Brassinosteroids Dominate Hormonal Regulation of Plant Thermomorphogenesis via BZR1. Curr. Biol. 2018, 28, 303–310. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakamoto, T.; Kimura, S. Plant Temperature Sensors. Sensors 2018, 18, 4365. https://doi.org/10.3390/s18124365
Sakamoto T, Kimura S. Plant Temperature Sensors. Sensors. 2018; 18(12):4365. https://doi.org/10.3390/s18124365
Chicago/Turabian StyleSakamoto, Tomoaki, and Seisuke Kimura. 2018. "Plant Temperature Sensors" Sensors 18, no. 12: 4365. https://doi.org/10.3390/s18124365
APA StyleSakamoto, T., & Kimura, S. (2018). Plant Temperature Sensors. Sensors, 18(12), 4365. https://doi.org/10.3390/s18124365




