Development and Testing of an LED-Based Near-Infrared Sensor for Human Kidney Tumor Diagnostics
Abstract
:1. Introduction
2. Materials and Methods
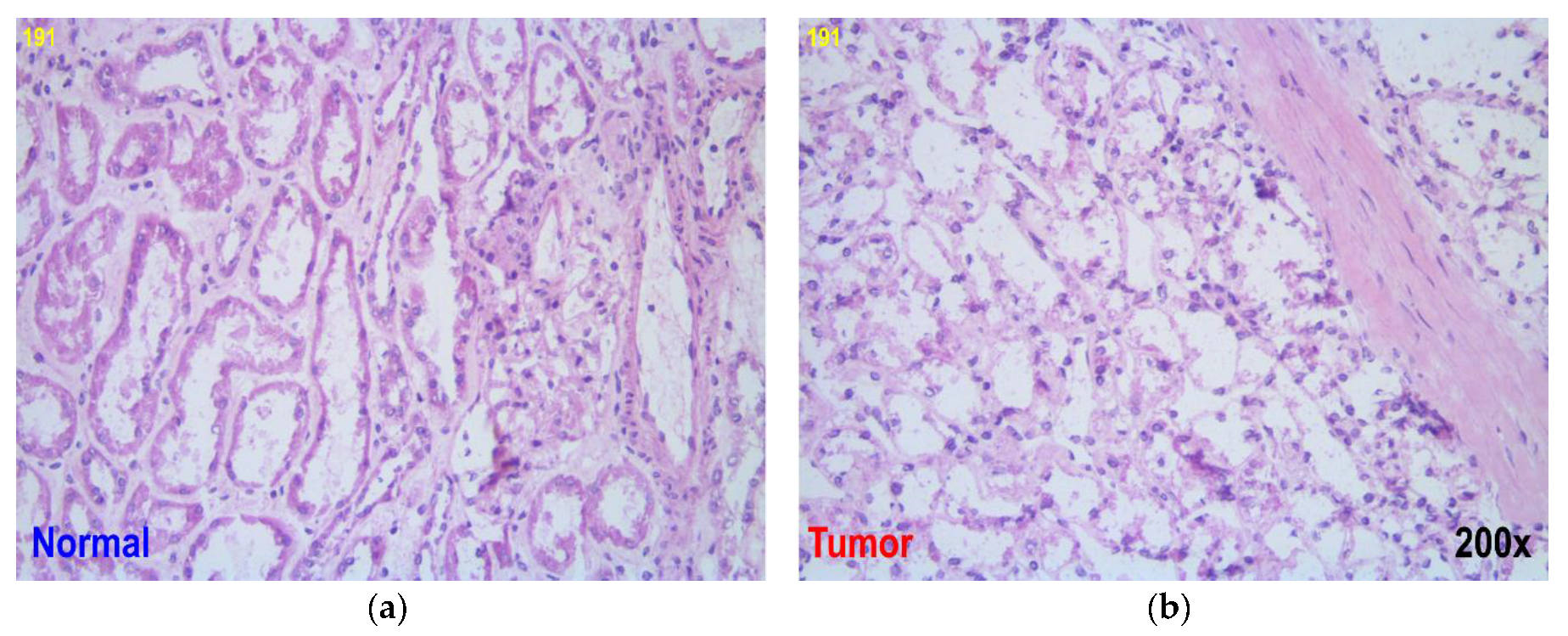
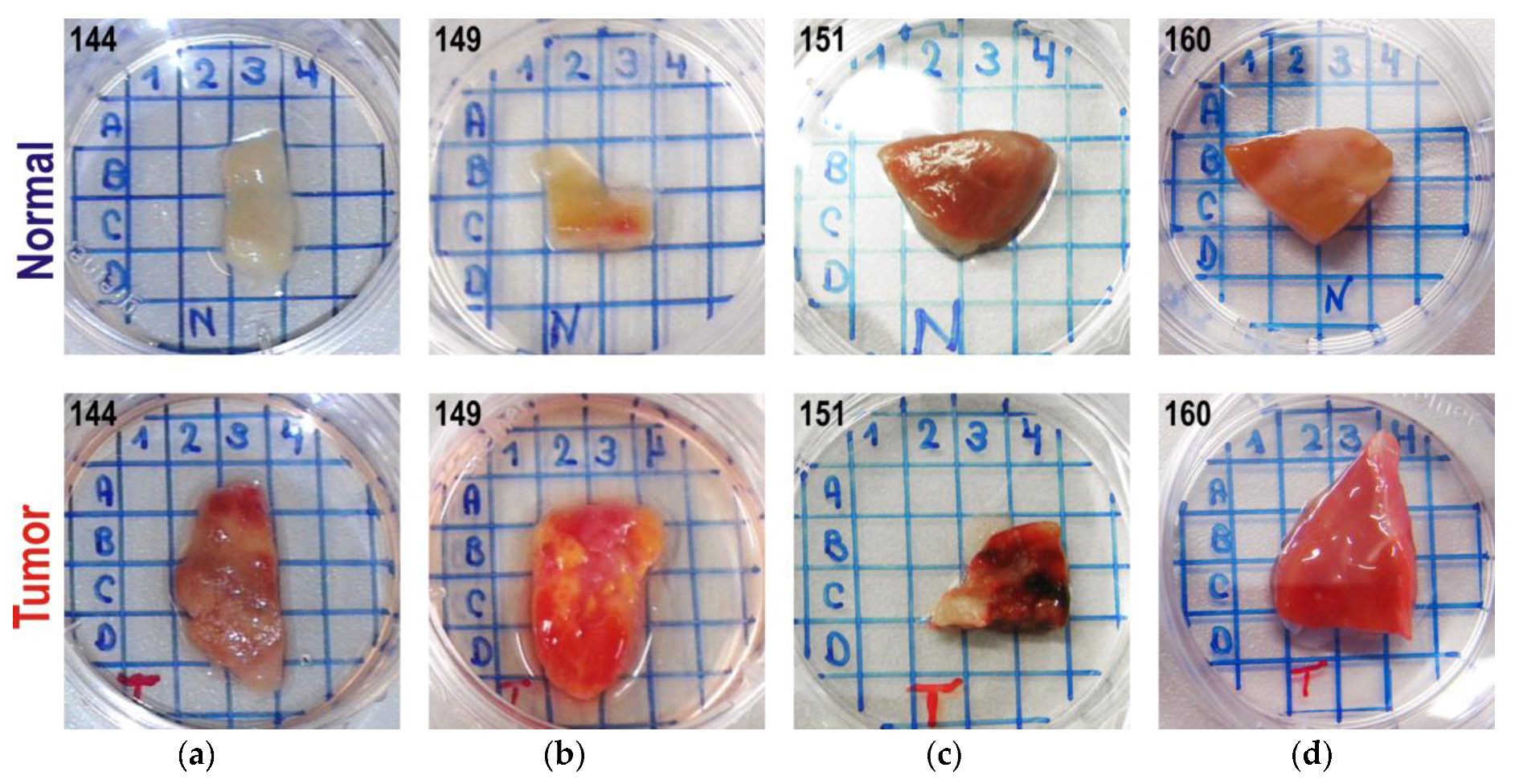
2.1. Samples
2.2. NIR Spectroscopy
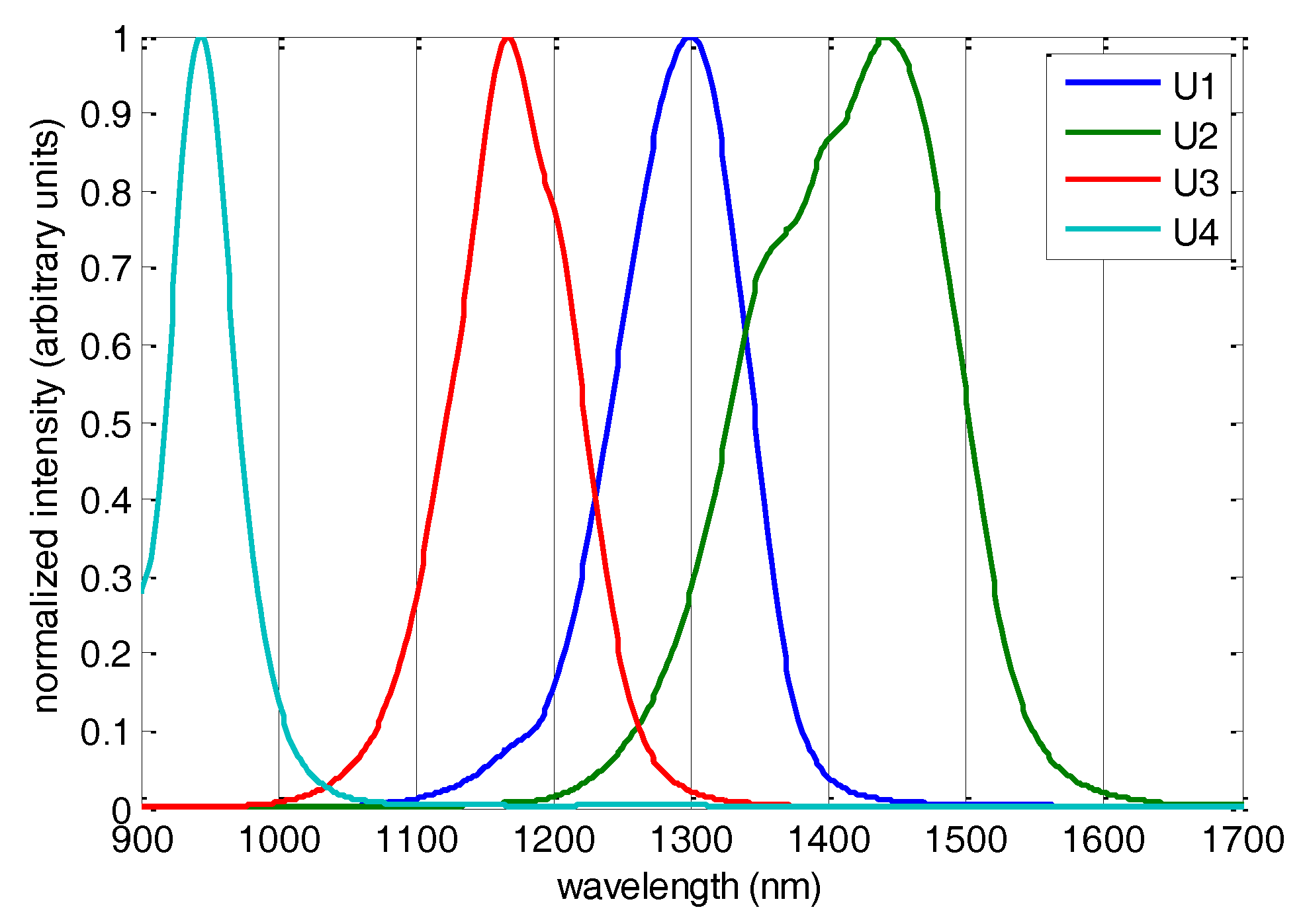
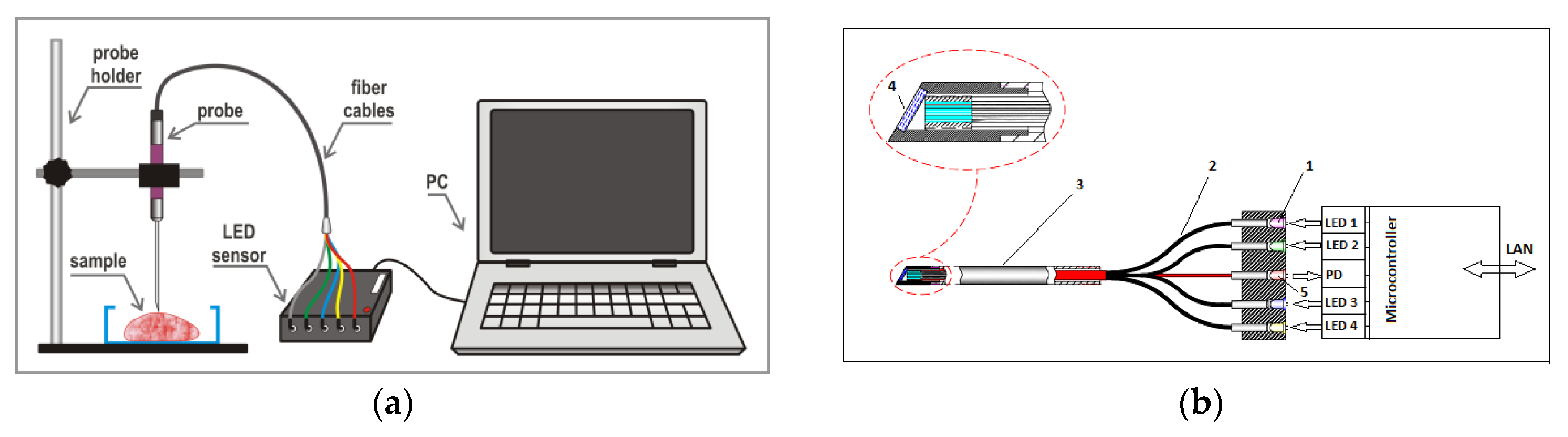
2.3. Sensor Design and Data Acquisition
2.4. Data Analysis Methods and Software
2.5. Model Validation
3. Results
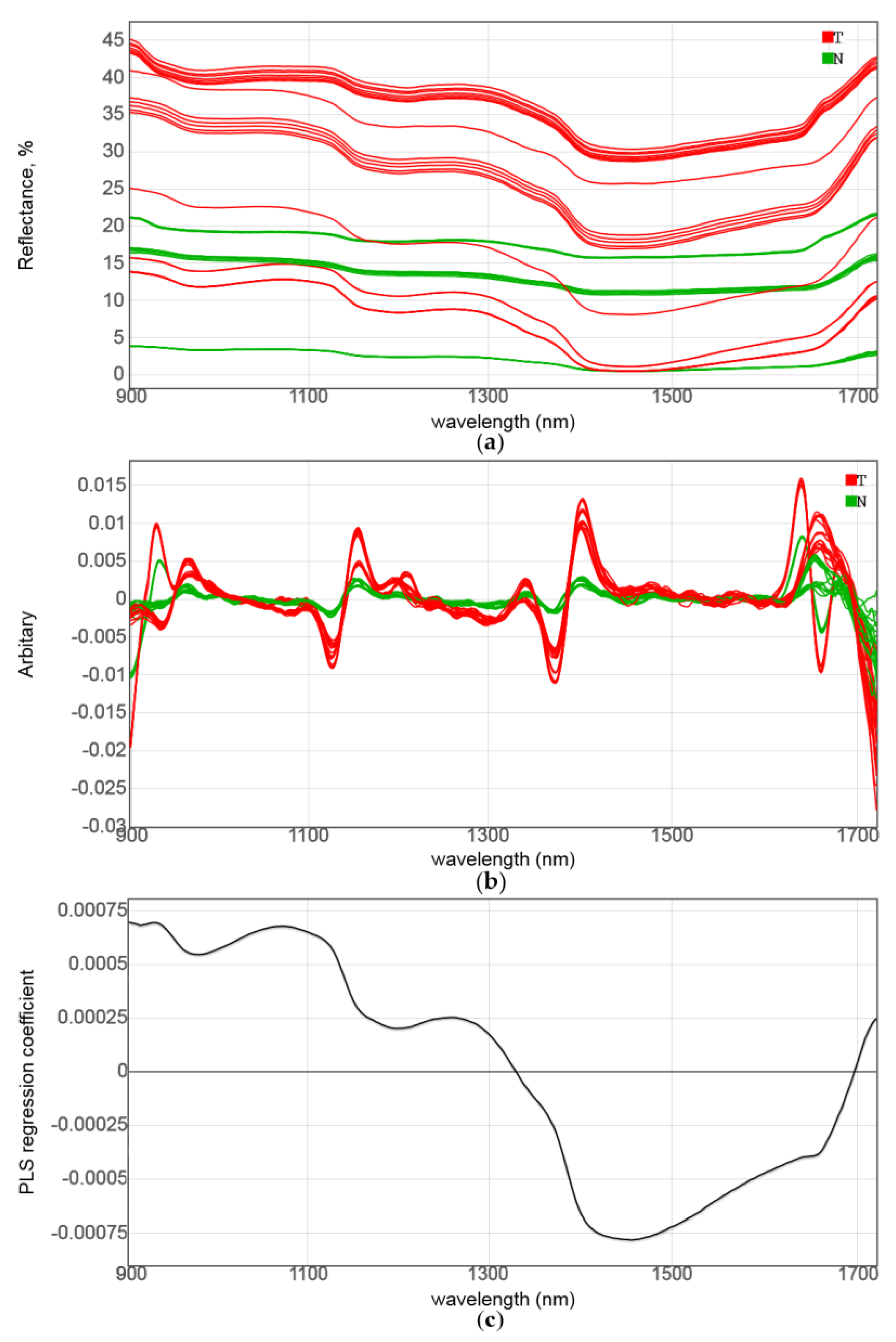
3.1. NIR Spectroscopic Analysis and Sensor Simulation
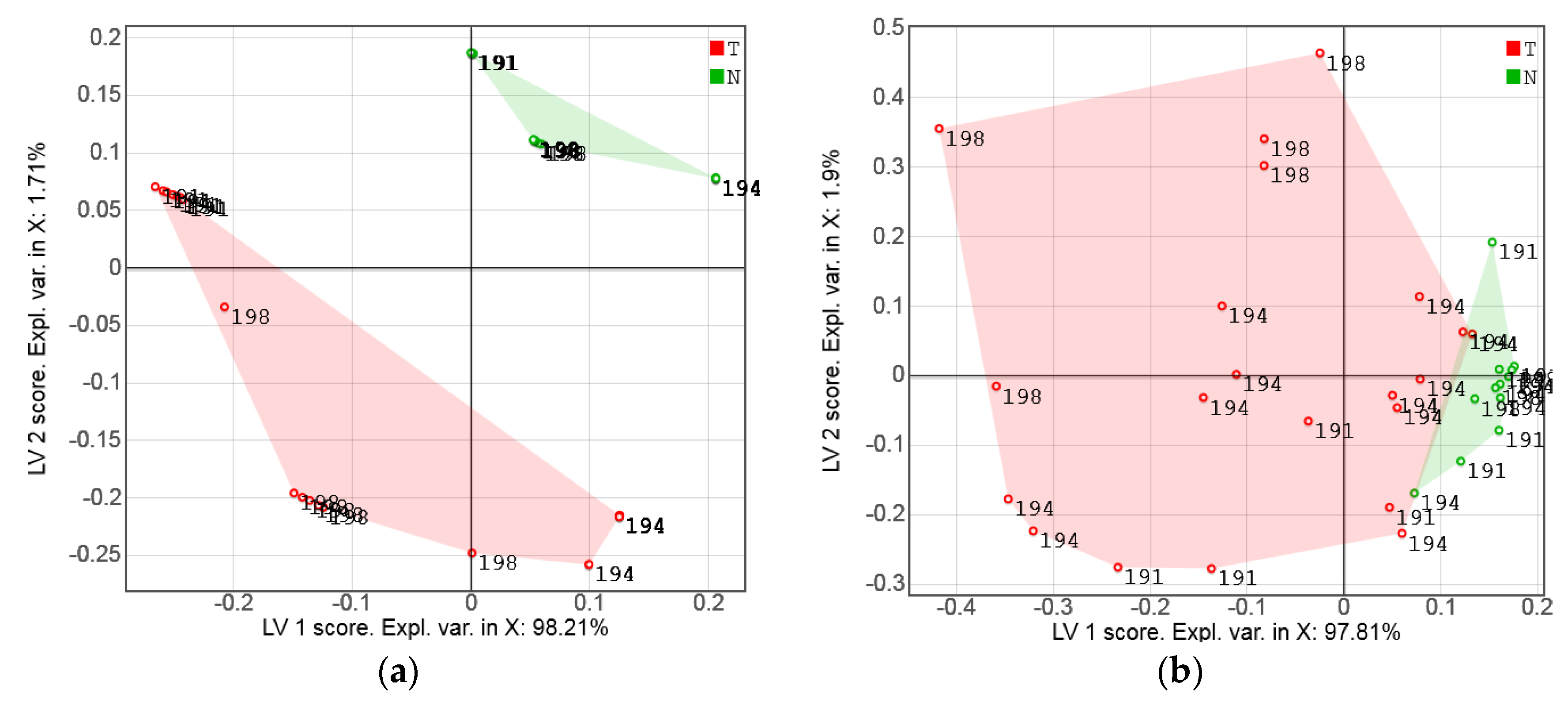
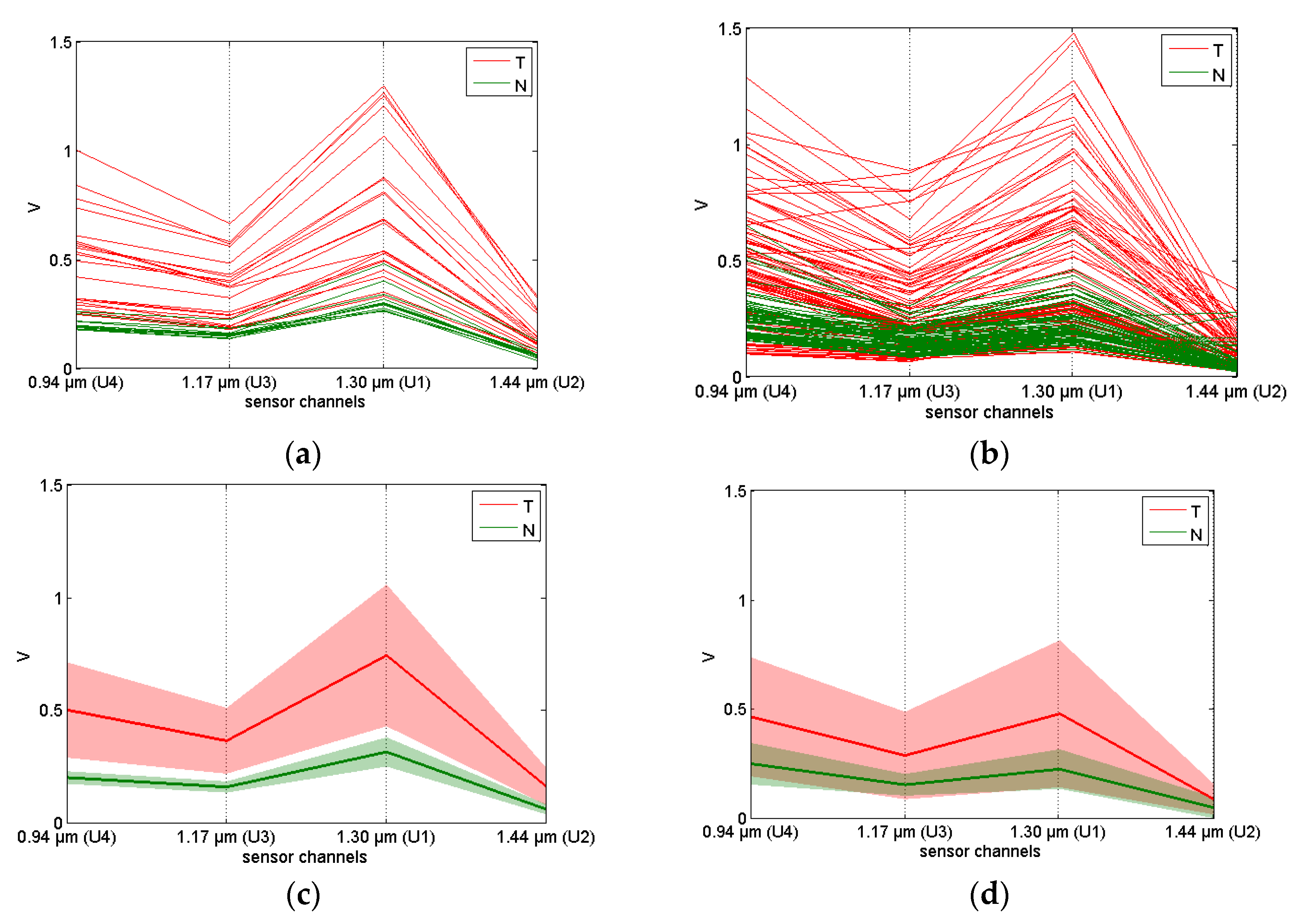
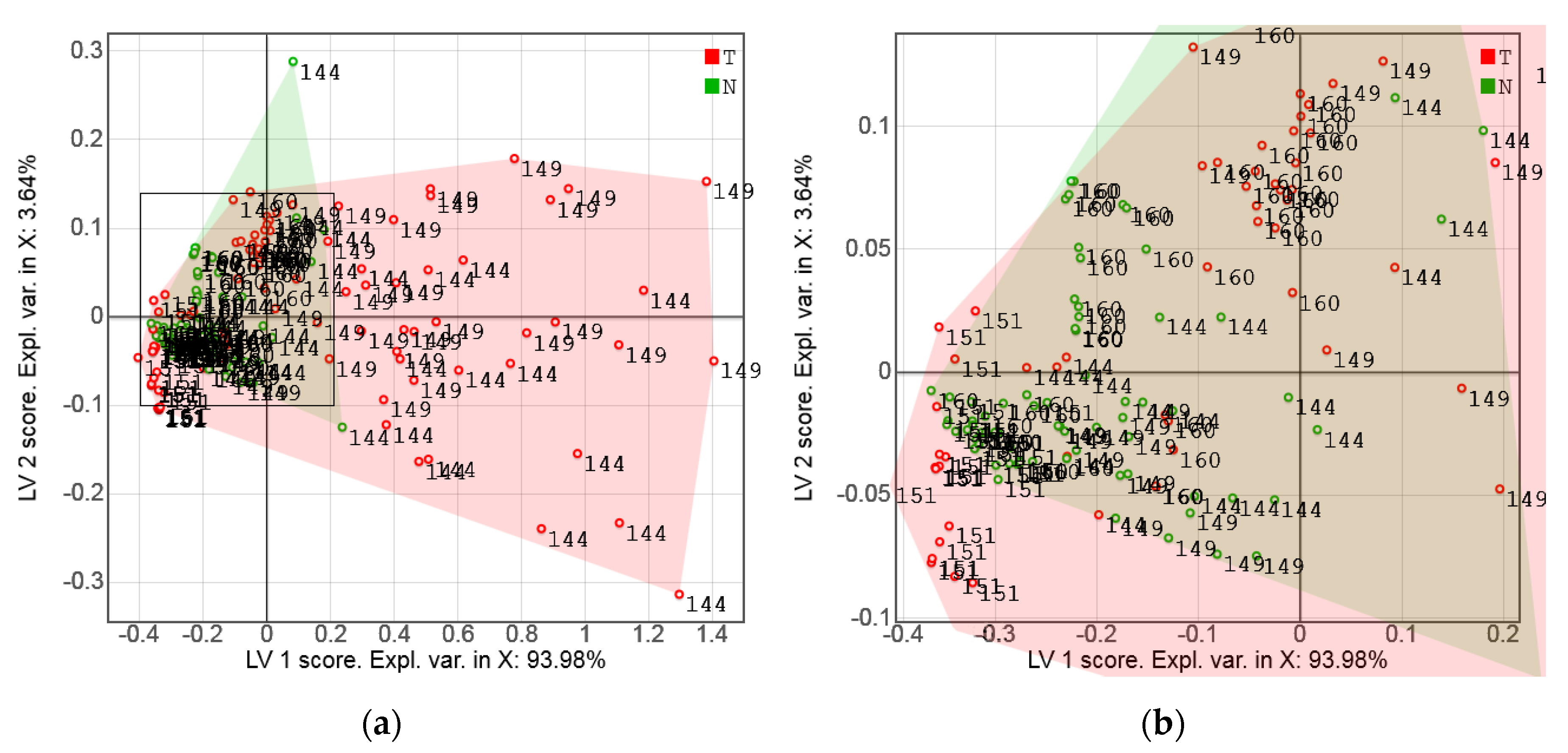
3.2. Exploratory Analysis of Sensor Data
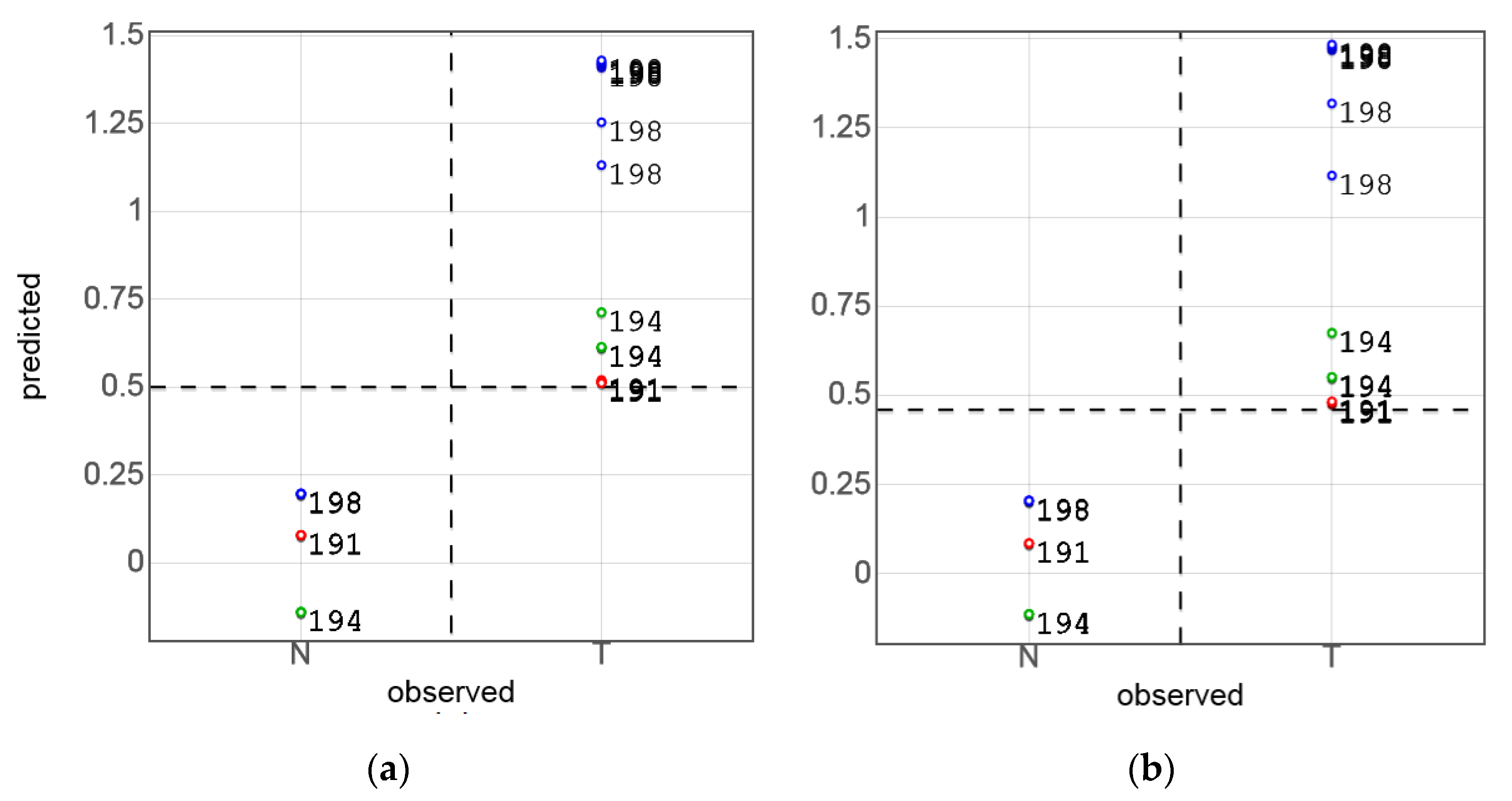
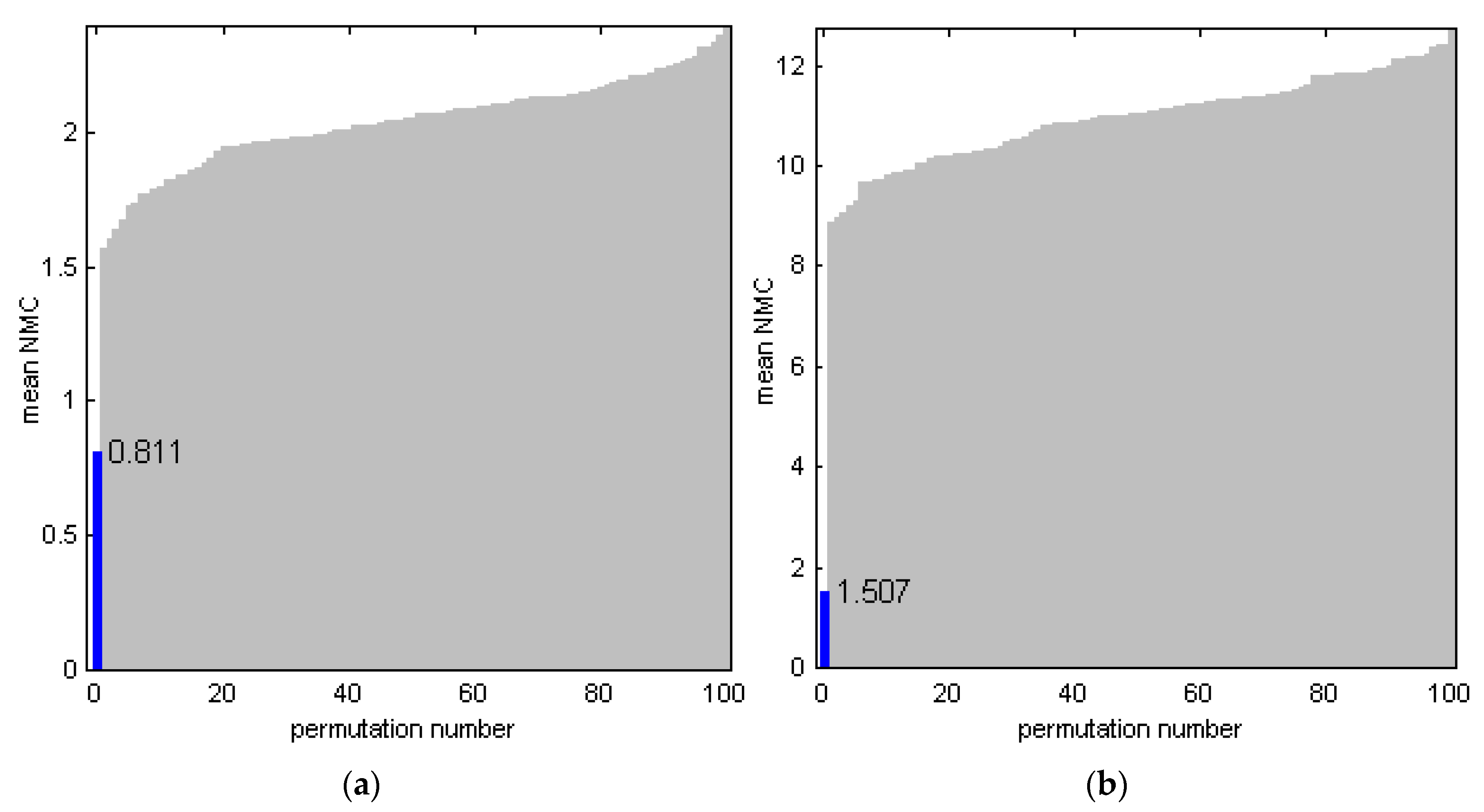
3.3. PLS-DA of Sensor Data and Model Validation
4. Discussion and Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Redaktion Barnes, B.; Kraywinkel, K.; Nowossadeck, E.; Schönfeld, I.; Starker, A.; Wienecke, A.; Wolf, U. Bericht Zum Krebsgeschehen in Deutschland 2016; Robert Koch-Institut (Hrsg): Berlin, Germany, 2016. (In German) [Google Scholar]
- Artyushenko, V.; Schulte, F.; Zabarylo, U.; Berlien, H.P.; Usenov, I.; Saeb Gilani, T.; Eichler, H.J.; Pieszczek, Ł.; Bogomolov, A.; Krause, H.; et al. Spectral Fiber Sensors for Cancer Diagnostics In Vitro. In Proceedings of the European Conference on Biomedical Optics, Munich, Germany, 21–25 June 2015. [Google Scholar]
- Kallenbach-Thieltges, A.; Großerüschkamp, F.; Mosig, A.; Diem, M.; Tannapfel, A.; Gerwert, K. Immunohistochemistry, histopathology and infrared spectral histopathology of colon cancer tissue sections. J. Biophotonics 2013, 6, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Dukor, R.K. Vibrational Spectroscopy in the Detection of Cancer. In Handbook of Vibrational Spectroscopy; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006. [Google Scholar]
- Nachabé, R.; Evers, D.J.; Hendriks, B.H.; Lucassen, G.W.; Van Der Voort, M.; Wesseling, J.; Ruers, T.J. Effect of bile absorption coefficients on the estimation of liver tissue optical properties and related implications in discriminating healthy and tumorous samples. Biomed. Opt. Express 2011, 2, 600–614. [Google Scholar] [CrossRef] [PubMed]
- Volynskaya, Z.; Haka, A.S.; Bechtel, K.L.; Fitzmaurice, M.; Shenk, R.; Wang, N.; Nazemi, J.; Dasari, R.R.; Feld, M.S. Diagnosing breast cancer using diffuse reflectance spectroscopy and intrinsic fluorescence spectroscopy. J. Biomed. Opt. 2008, 13, 024012. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Breslin, T.M.; Harter, J.; Ramanujam, N. Model based and empirical spectral analysis for the diagnosis of breast cancer. Opt. Express 2008, 16, 14961–14978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Y.; Yu, Y.; Xue, X.; Tuchin, V.V.; Zhu, D. Visible and near-infrared spectroscopy for distinguishing malignant tumor tissue from benign tumor and normal breast tissues in vitro. J. Biomed. Opt. 2013, 18, 077003. [Google Scholar] [CrossRef] [PubMed]
- Cerussi, A.; Shah, N.; Hsiang, D.; Durkin, A.; Butler, J.; Tromberg, B.J. In vivo absorption, scattering, and physiologic properties of 58 malignant breast tumors determined by broadband diffuse optical spectroscopy. J. Biomed. Opt. 2006, 11, 044005. [Google Scholar] [CrossRef] [PubMed]
- Chang, V.T.C.; Bean, S.M.; Cartwright, P.S.; Ramanujam, N. Visible light optical spectroscopy is sensitive to neovascularization in the dysplastic cervix. J. Biomed. Opt. 2010, 15, 057006. [Google Scholar] [CrossRef] [PubMed]
- Subhash, N.; Mallia, J.R.; Thomas, S.S.; Mathews, A.; Sebastian, P.; Madhavan, J. Oral cancer detection using diffuse reflectance spectral ratio R540/R575 of oxygenated hemoglobin bands. J. Biomed. Opt. 2006, 11, 014018. [Google Scholar] [CrossRef] [PubMed]
- Mallia, R.J.; Narayanan, S.; Madhavan, J.; Sebastian, P.; Kumar, R.; Mathews, A.; Thomas, G.; Radhakrishnan, J. Diffuse reflection spectroscopy: An alternative to autofluorescence spectroscopy in tongue cancer detection. Appl. Spectrosc. 2010, 64, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Amelink, A.; Kaspers, O.P.; Sterenborg, H.J.; Van Der Wal, J.E.; Roodenburg, J.L.; Witjes, M.J. Non-invasive measurement of the morphology and physiology of oral mucosa by use of optical spectroscopy. Oral Oncol. 2008, 44, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.B.; Temiyasathit, C.; Bensalah, K.; Tuncel, A.; Cadeddu, J.; Kabbani, W.; Liu, H. An effective classification procedure for diagnosis of prostate cancer in near infrared spectra. Expert Syst. Appl. 2010, 37, 3863–3869. [Google Scholar] [CrossRef]
- Sharma, V.; Olweny, E.; Kapur, P.; Cadeddu, J.; Roehrborn, C.; Liu, H. Prostate cancer detection using combined auto-fluorescence and light reflectance spectroscopy: Ex vivo study of human prostates. Biomed. Opt. Express 2014, 5, 1512–1529. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Xu, Y.; Wu, J.; Zhang, Y.; Sun, K. Detection of lung cancer tissue by attenuated total reflection-Fourier transform infrared spectroscopy—A pilot study of 60 samples. J. Surg. Res. 2013, 179, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Yi, W.S.; Cui, D.S.; Li, Z.; Wu, L.L.; Shen, A.G.; Hu, J.M. Gastric cancer differentiation using Fourier transform near-infrared spectroscopy with unsupervised pattern recognition. Spectrochim. Acta Mol. Biomol. Spectrosc. 2012, 101, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Maziak, D.E.; Do, M.T.; Shamji, F.M.; Sundaresan, S.R.; Perkins, D.G.; Wong, P.T.T. Fourier-transform infrared spectroscopic study of characteristic molecular structure in cancer cells of esophagus: An exploratory study. Cancer Detect. Prev. 2007, 31, 244–253. [Google Scholar] [CrossRef] [PubMed]
- De Boer, L.L.; Molenkamp, B.G.; Bydlon, T.M.; Hendriks, B.H.; Wesseling, J.; Sterenborg, H.J.; Ruers, T.J.M. Fat/water ratios measured with diffuse reflectance spectroscopy to detect breast tumor boundaries. Breast Cancer Res. Treat. 2015, 152, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Eichler, J.; Knot, J.; Lenz, H. Measurements on the depth of penetration of light (0.35–1.0 µm) in tissue. Radiat. Environ. Biophys. 1977, 14, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Svaasand, L. On the Physical Rationale of Photodynamic Therapy. In Proceedings of the Future Directions and Application in Photodynamic Therapy, San Diego, CA, USA, 19–21 January 1990. [Google Scholar]
- Galyanin, V.; Melenteva, A.; Bogomolov, A. Selecting optimal wavelength intervals for an optical sensor: A case study of milk fat and total protein analysis in the region 400–1100 nm. Sens. Actuators B Chem. 2015, 218, 97–104. [Google Scholar] [CrossRef]
- Melenteva, A.; Galyanin, V.; Savenkova, E.; Bogomolov, A. Building global models for fat and total protein content in raw milk based on historical spectroscopic data in the visible and short-wave near infrared range. Food Chem. 2016, 203, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Ouyang, Q.; Chen, Q.; Zhao, J. Real-time monitoring of total polyphenols content in tea using a developed optical sensors system. J. Pharm. Biomed. Anal. 2014, 97, 116–122. [Google Scholar] [CrossRef] [PubMed]
- De Lima, K.M.G. A portable photometer based on LED for the determination of aromatic hydrocarbons in water. Microchem. J. 2012, 103, 62–67. [Google Scholar] [CrossRef]
- Fuhrman, S.A.; Lasky, L.C.; Limas, C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am. J. Surg. Pathol. 1982, 6, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Moch, H.; Artibani, W.; Delahunt, B.; Ficarra, V.; Knuechel, R.; Montorsi, F.; Patard, J.J.; Stief, C.G.; Sulser, T.; Wild, P.J. Reassessing the current UICC/AJCC TNM staging for renal cell carcinoma. Eur. Urol. 2009, 56, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Wills, H.; Kast, R.; Stewart, C.; Rabah, R.; Pandya, A.; Poulik, J.; Auner, G.; Klein, M.D. Raman spectroscopy detects and distinguishes neuroblastoma and related tissues in fresh and (banked) frozen specimens. J. Pediatr. Surg. 2009, 44, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Bogomolov, A.; Ageev, V.; Zabarylo, U.; Usenov, I.; Schulte, F.; Kirsanov, D.; Belikova, V.; Minet, O.; Feliksberger, E.; Meshkovsky, I.; et al. LED-based near infrared sensor for cancer diagnostics. Proc. SPIE 2016, 9715. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Brereton, R.G. Chemometrics for Pattern Recognition; John Wiley & Sons, Ltd.: Bristol, UK, 2009; pp. 199–200. [Google Scholar]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Cruciani, G.; Baroni, M.; Clementi, S. Predictive ability of regression-Models.1. Standard-deviation of prediction errors (SDEP). J. Chemom. 1992, 6, 335–346. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Velzen, E.J.J.; Hoefsloot, H.C.J.; Smilde, A.K. Discriminant Q2 (DQ2) for improved discrimination in PLSDA models. Metabolomics 2008, 4, 293–296. [Google Scholar] [CrossRef]
- Van der Voet, H. Comparing the predictive accuracy of models using a simple randomization test. Chemom. Intell. Lab. Syst. 1994, 25, 313–323. [Google Scholar] [CrossRef]
- Workman, J., Jr.; Weyer, L. Practical Guide to Interpretive Near-Infrared Spectroscopy; Taylor and Francis Group, CRC Press: Boca Raton, FL, USA, 2008; pp. 239–286. [Google Scholar]
- Nachabé, R.; Evers, D.J.; Hendriks, B.H.W.; Lucassen, G.W.; Van Der Voort, M.; Rutgers, E.J.; Vrancken Peeters, M.J.; Van Der Hage, J.A.; Oldenburg, H.S.; Wesseling, J.; et al. Diagnosis of breast cancer using diffuse optical spectroscopy from 500 to 1600 nm: comparison of classification methods. J. Biomed. Opt. 2011, 16, 087010. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Cerussi, A.E.; Jakubowski, D.; Hsiang, D.; Butler, J.; Tromberg, B.J. The role of diffuse optical spectroscopy in the clinical management of breast cancer. Dis. Markers 2003–2004, 19, 95–105. [Google Scholar] [CrossRef]
- Drezek, R.; Dunn, A.; Richards-Kortum, R. A pulsed finite-difference time-domain (FDTD) method for calculating light scattering from biological cells over broad wavelength ranges. Opt. Express 2000, 6, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Guo, X.; Fang, B.; Song, C. Study of back-scattering microspectrum for stomach cells at single-cell scale. J. Biomed. Opt. 2010, 15, 040505. [Google Scholar] [CrossRef] [PubMed]
- Roth, W.; Macher-Goeppinger, S.; Schirmacher, P. Pathology and molecular pathogenesis of renal cell carcinoma. Nephrologe 2011, 6, 315–322. [Google Scholar] [CrossRef]
- Rücker, C.; Rücker, G.; Meringer, M. y-Randomization and Its Variants in QSPR/QSAR. J. Chem. Inf. Model. 2007, 47, 2345–2357. [Google Scholar] [CrossRef] [PubMed]
- Reddy, G.S.; Rao, K.E.; Kumar, K.K.; Sekhar, P.C.; Prakash Chandra, K.L.; Ramana Reddy, B.V. Diagnosis of oral cancer: The past and present. J. Orofac. Sci. 2014, 6, 10–16. [Google Scholar] [CrossRef]
| Data | DQ2 | TP | FP | TN | FN | %Sn | %Sp | %Ac |
|---|---|---|---|---|---|---|---|---|
| Calibration 1 | ||||||||
| An41 2 | 0.932 | 21 | 0 | 20 | 0 | 100.0 | 100.0 | 100.0 |
| As41 3 | 0.917 | 21 | 0 | 20 | 0 | 100.0 | 100.0 | 100.0 |
| As33 4 | 0.437 | 19 | 1 | 11 | 2 | 90.5 | 91.7 | 90.9 |
| Bs170 5 | 0.181 | 62 | 5 | 70 | 33 | 65.3 | 93.3 | 77.6 |
| Bs140 6 | 0.500 | 64 | 3 | 67 | 6 | 91.4 | 95.7 | 93.6 |
| Full (leave-one-out) cross-validation | ||||||||
| An41 | 0.920 | 21 | 0 | 20 | 0 | 100.0 | 100.0 | 100.0 |
| As41 | 0.902 | 21 | 0 | 20 | 0 | 100.0 | 100.0 | 100.0 |
| As33 | 0.358 | 18 | 1 | 11 | 3 | 85.7 | 91.7 | 87.9 |
| Bs170 | 0.153 | 59 | 7 | 68 | 36 | 62.1 | 90.7 | 74.7 |
| Bs140 | 0.478 | 63 | 3 | 67 | 7 | 90.0 | 95.7 | 92.9 |
| Segmented cross-validation 7 | ||||||||
| An41 | 0.574 | 21 | 0 | 20 | 0 | 100.0 | 100.0 | 100.0 |
| As41 | 0.488 | 14 | 0 | 20 | 7 | 66.7 | 100.0 | 82.9 |
| As33 | 0.051 | 13 | 3 | 9 | 8 | 61.9 | 75.0 | 66.7 |
| Bs170 | 0.068 | 51 | 7 | 68 | 44 | 53.7 | 90.7 | 70.0 |
| Bs140 | 0.416 | 61 | 3 | 67 | 9 | 87.1 | 95.7 | 91.4 |
| Random-subset validation 8 | ||||||||
| An41 | 0.916 | 100.0 | 100.0 | 100.0 | ||||
| As41 | 0.899 | 100.0 | 100.0 | 100.0 | ||||
| As33 | 0.345 | 82.4 | 86.1 | 83.8 | ||||
| Bs170 | 0.149 | 60.2 | 91.0 | 74.8 | ||||
| Bs140 | 0.480 | 89.9 | 95.7 | 92.8 | ||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bogomolov, A.; Zabarylo, U.; Kirsanov, D.; Belikova, V.; Ageev, V.; Usenov, I.; Galyanin, V.; Minet, O.; Sakharova, T.; Danielyan, G.; et al. Development and Testing of an LED-Based Near-Infrared Sensor for Human Kidney Tumor Diagnostics. Sensors 2017, 17, 1914. https://doi.org/10.3390/s17081914
Bogomolov A, Zabarylo U, Kirsanov D, Belikova V, Ageev V, Usenov I, Galyanin V, Minet O, Sakharova T, Danielyan G, et al. Development and Testing of an LED-Based Near-Infrared Sensor for Human Kidney Tumor Diagnostics. Sensors. 2017; 17(8):1914. https://doi.org/10.3390/s17081914
Chicago/Turabian StyleBogomolov, Andrey, Urszula Zabarylo, Dmitry Kirsanov, Valeria Belikova, Vladimir Ageev, Iskander Usenov, Vladislav Galyanin, Olaf Minet, Tatiana Sakharova, Georgy Danielyan, and et al. 2017. "Development and Testing of an LED-Based Near-Infrared Sensor for Human Kidney Tumor Diagnostics" Sensors 17, no. 8: 1914. https://doi.org/10.3390/s17081914
APA StyleBogomolov, A., Zabarylo, U., Kirsanov, D., Belikova, V., Ageev, V., Usenov, I., Galyanin, V., Minet, O., Sakharova, T., Danielyan, G., Feliksberger, E., & Artyushenko, V. (2017). Development and Testing of an LED-Based Near-Infrared Sensor for Human Kidney Tumor Diagnostics. Sensors, 17(8), 1914. https://doi.org/10.3390/s17081914








