Early Diagnosis of Breast Cancer
Abstract
:1. Introduction
2. Clinical Breast Imaging Techniques
2.1. Mammography
2.2. Ultrasound
2.3. MRI
3. Microwave Breast Imaging Methods and Measurement Systems
3.1. Microwave Tomography
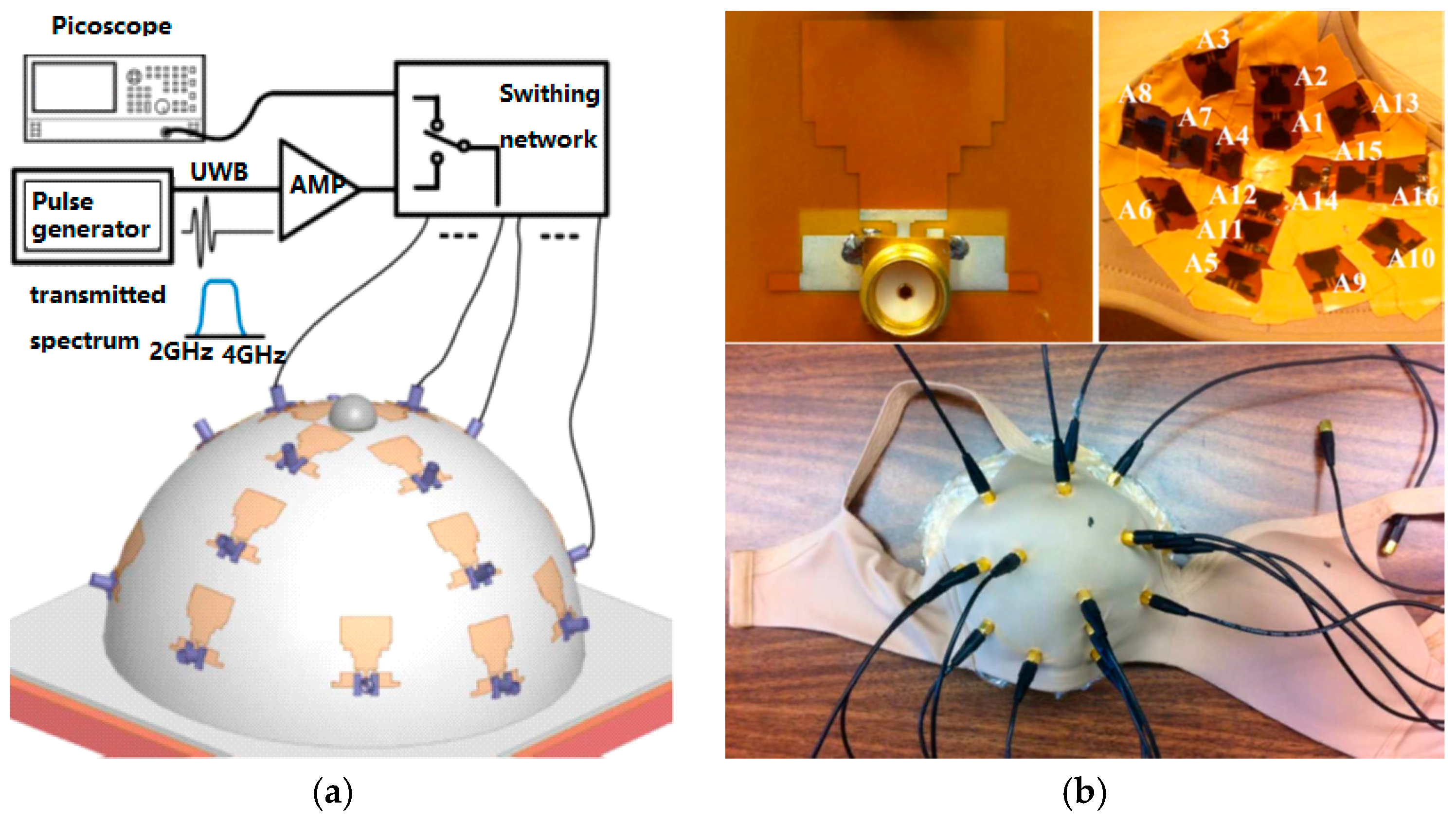
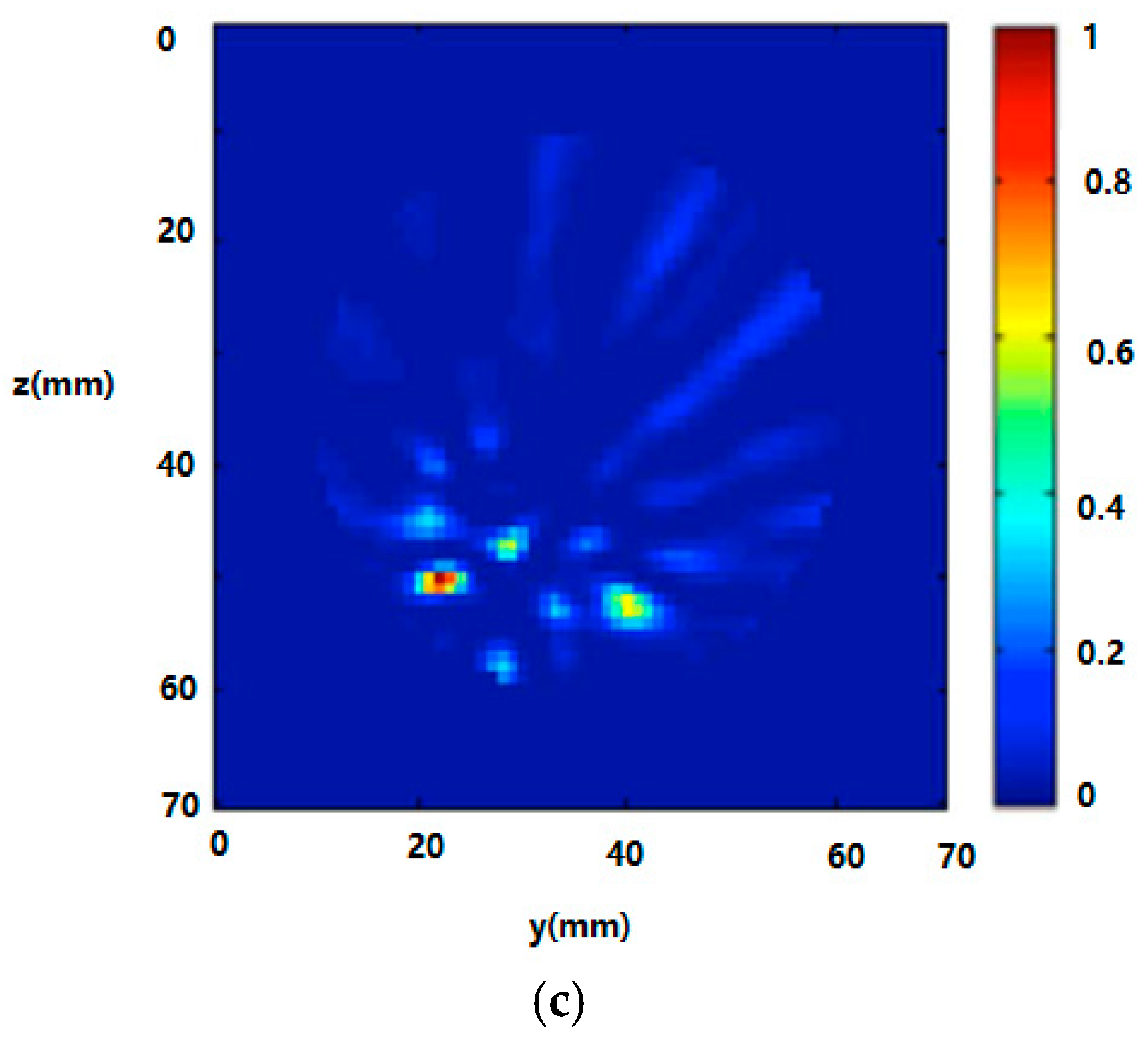
3.2. Radar-Based Microwave Imaging
3.3. RF Sensors and Sensor Arrays
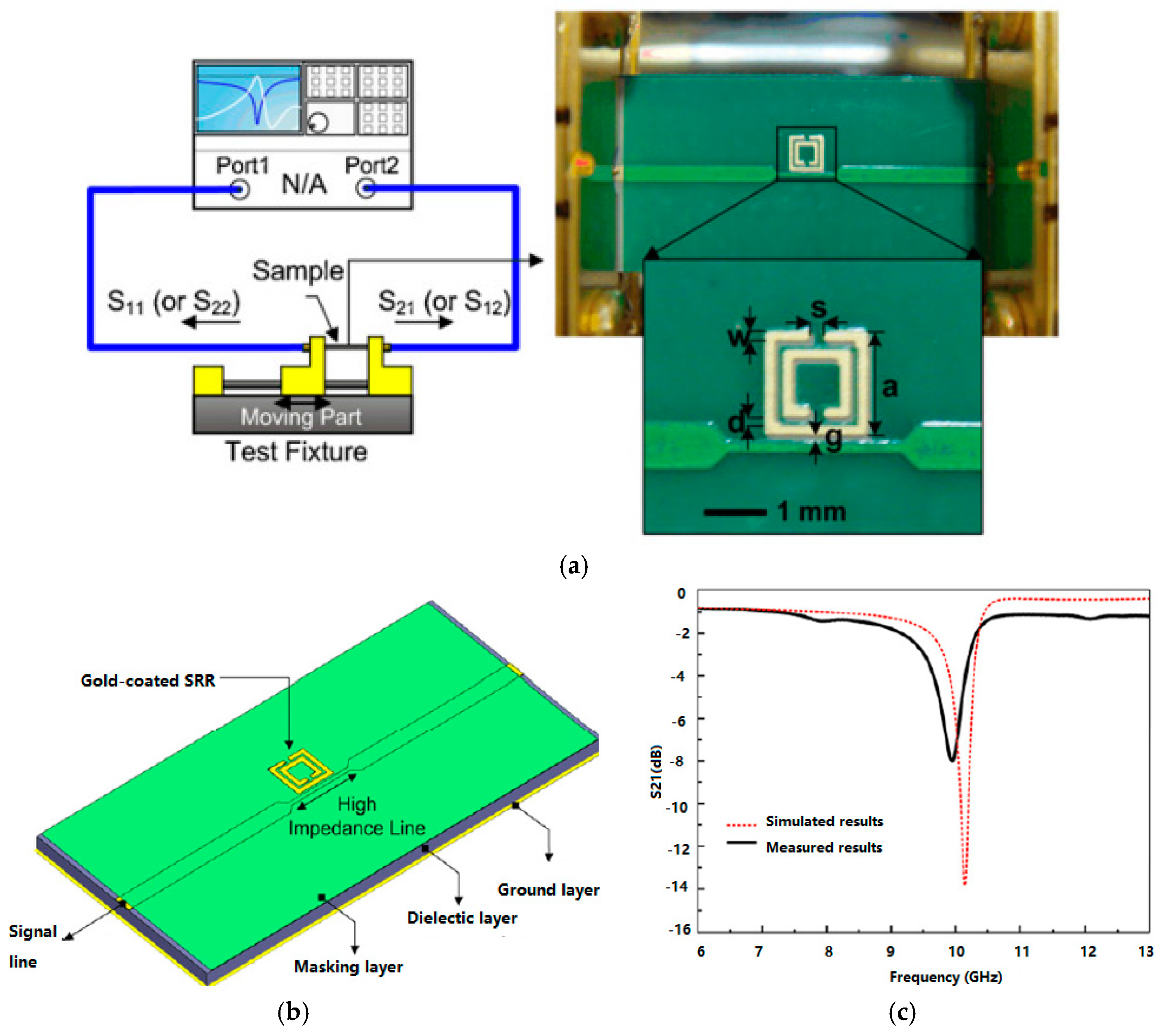
3.3.1. RF Sensors for MI Systems
3.3.2. RF Biosensors
3.3.3. RF Sensor Arrays
4. Biomarkers for Breast Cancer Detection
4.1. Proteomic Biomarkers
4.2. Gene Biomarkers
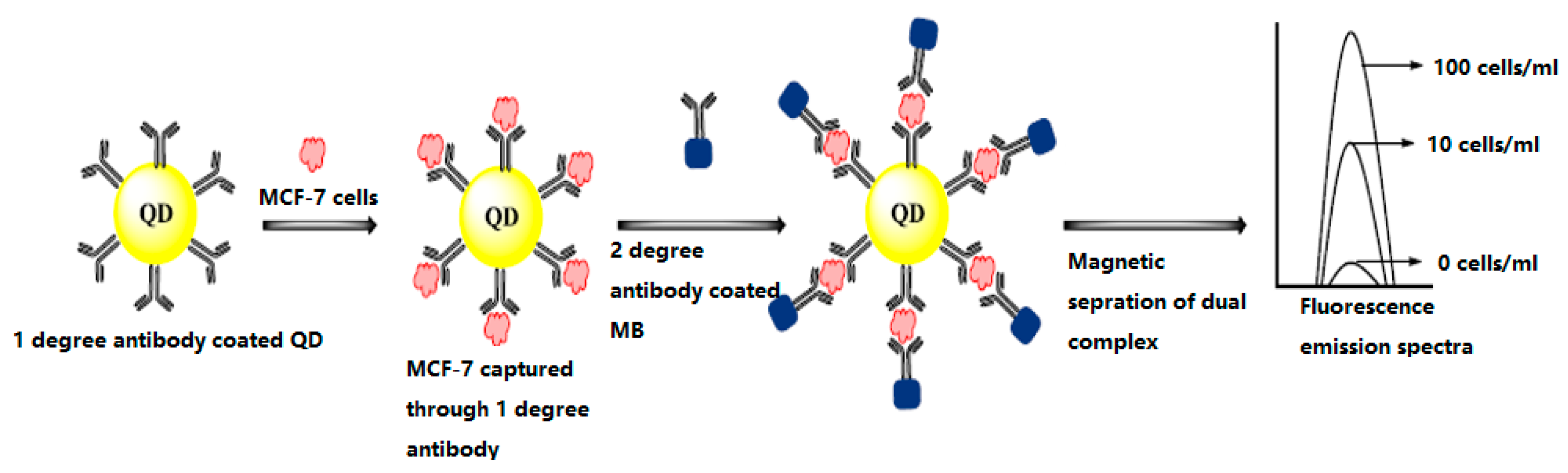
4.3. Biosensors for Cancer Markers Detection
4.3.1. Optical Biosensors
4.3.2. Piezoelectric Biosensors
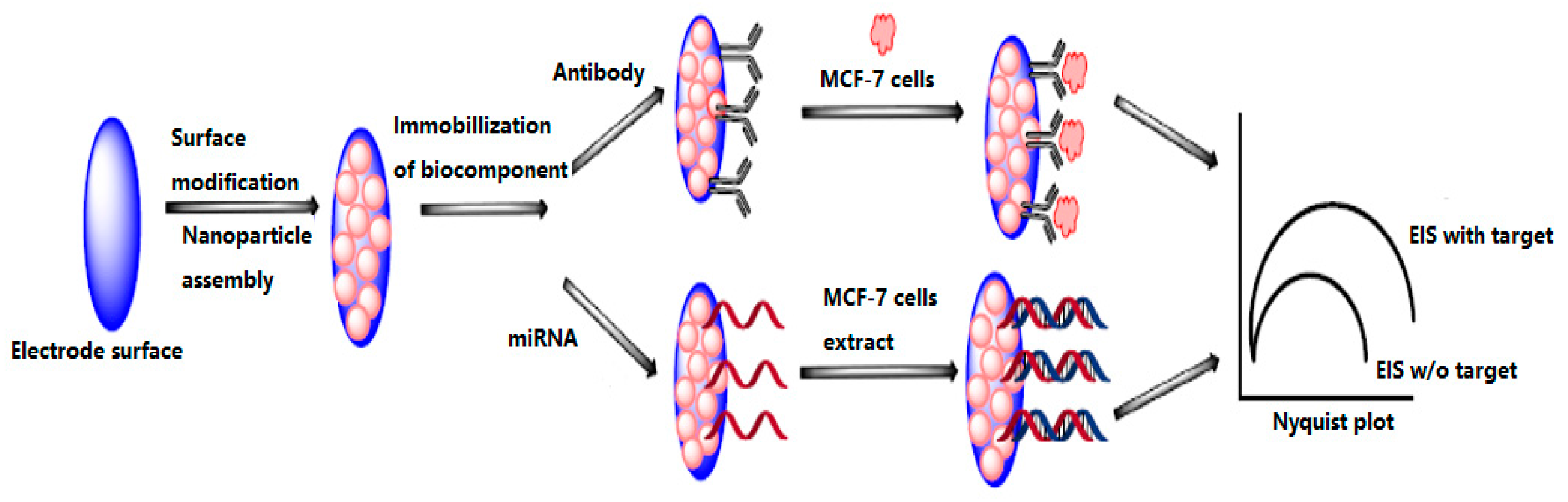
4.3.3. Electrochemical Biosensors
5. Current Trends and Future Perspectives
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Mohebian, M.R.; Marateb, H.R.; Mansourian, M.; Mañanas, M.A.; Mokarian, F. A hybrid computer-aided-diagnosis system for prediction of breast cancer recurrence (HPBCR) using optimized ensemble learning. Comput. Struct. Biotechnol. J. 2017, 15, 75–85. [Google Scholar] [CrossRef] [PubMed]
- U.S. Breast Cancer Statistics 2017. Available online: http://www.breastcancer.org/symptoms/understand_bc/statistics (accessed on 2 July 2017).
- Migowski, A. Early detection of breast cancer and the interpretation of results of survival studies. Cienc. Saude Coletiva 2015, 20, 1309. [Google Scholar] [CrossRef] [PubMed]
- Hellquist, B.N.; Czene, K.; Hjälm, A.; Nyström, L.; Jonsson, H. Effectiveness of population-based service screening with mammography for women ages 40 to 49 years with a high or low risk of breast cancer: Socioeconomic status, parity, and age at birth of first child. Cancer 2012, 118, 1170–1171. [Google Scholar] [CrossRef]
- Onega, T.; Goldman, L.E.; Walker, R.L.; Miglioretti, D.L.; Buist, D.S.; Taplin, S.; Geller, B.M.; Hill, D.A.; Smith-Bindman, R. Facility mammography volume in relation to breast cancer screening outcomes. J. Med. Screen. 2016, 23, 31. [Google Scholar] [CrossRef] [PubMed]
- Lewis, T.C.; Pizzitola, V.J.; Giurescu, M.E.; Eversman, W.G.; Lorans, R.; Robinson, K.A.; Patel, B.K. Contrast-enhanced digital mammography: A single-institution experience of the first 208 cases. Breast J. 2017, 23, 67. [Google Scholar] [CrossRef] [PubMed]
- Ozmen, N.; Dapp, R.; Zapf, M.; Gemmeke, H.; van Ruiter, N.V.; Dongen, K.W. Comparing different ultrasound imaging methods for breast cancer detection. IEEE Tran. Ultrason. Ferroelectr. Freq. Control. 2015, 62, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Roganovic, D.; Djilas, D.; Vujnovic, S.; Pavic, D.; Stojanov, D. Breast MRI, digital mammography and breast tomosynthesis: Comparison of three methods for early detection of breast cancer. Bosn. J. Basic Med. Sci. 2015, 15, 64. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.M.; El-Shenawee, M. Review of electromagnetic techniques for breast cancer detection. IEEE Rev. Biomed. Eng. 2011, 4, 103. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Peng, Y.; Sun, M.; Yang, X. SU-E-I-81: Targeting of HER2-Expressing Tumors with Dual PET-MR Imaging Probes. Med. Phys. 2015, 42, 3260. [Google Scholar] [CrossRef]
- Chen, T.; Artis, F.; Dubuc, D.; Fournie, J.J.; Poupot, M.; Grenier, K. Microwave biosensor dedicated to the dielectric spectroscopy of a single alive biological cell in its culture medium. In Proceedings of the 2013 IEEE MTT-S International Microwave Symposium Digest (IMS), Seattle, WA, USA, 2–7 June 2013. [Google Scholar]
- Löhndorf, M.; Schlecht, U.; Gronewold, T.M.A.; Malavé, A.; Tewes, M. Microfabricated high-performance microwave impedance biosensors for detection of aptamer-protein interactions. Appl. Phys. Lett. 2015, 87, 1289. [Google Scholar] [CrossRef]
- Kim, N.Y.; Dhakal, R.; Adhikari, K.K. Practical RF Biosensor Technology; LAP Lambert Academic Publishing: Saarbrücken, Germany, 2017. [Google Scholar]
- Xu, X.; Gifford-Hollingsworth, C.; Sensenig, R.; Shih, W.H.; Shih, W.Y.; Brooks, A.D. Breast tumor detection using piezoelectric fingers: First clinical report. J. Am. Coll. Surg. 2013, 216, 1168–1173. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chung, Y.; Brooks, A.D.; Shih, W.H.; Shih, W.Y. Development of array piezoelectric fingers towards in vivo breast tumor detection. Rev. Sci. Instrum. 2016, 87, 124301. [Google Scholar] [CrossRef] [PubMed]
- Nassar, F.J.; Nasr, R.; Talhouk, R. Micrornas as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol. Ther. 2017, 172, 34–49. [Google Scholar] [CrossRef] [PubMed]
- Giovannetti, G.; Tiberi, G.; Tosetti, M. Finite element method-based approach for radiofrequency magnetic resonance coil losses estimation. Concepts Magn. Reson. Part B 2017, 46, 186–190. [Google Scholar] [CrossRef]
- Kim, N.Y.; Dhakal, R.; Adhikari, K.K.; Kim, E.S.; Wang, C. A reusable robust radio frequency biosensor using microwave resonator by integrated passive device technology for quantitative detection of glucose level. Biosens. Bioelectron. 2014, 67, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Lee, H.J.; Choi, H.H.; Yook, J.G.; Yoo, K.H. Carbon-nanotube-resonator-based biosensors. Small 2008, 4, 1723–1727. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Porter, E.; Santorelli, A.; Popović, M.; Coates, M. Microwave breast cancer detection via cost-sensitive ensemble classifiers: Phantom and patient investigation. Biomed. Signal Process. Control 2017, 31, 366–376. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, J.H.; Moon, H.S.; Jang, I.S.; Choi, J.S.; Yook, J.G.; Jung, H. A planar split-ring resonator-based microwave biosensor for label-free detection of biomolecules. Sens. Actuators B Chem. 2012, 169, 26–31. [Google Scholar] [CrossRef]
- Yen, T.W.F.; Li, J.; Sparapani, R.A.; Laud, P.W.; Nattinger, A.B. The interplay between hospital and surgeon factors and the use of sentinel lymph node biopsy for breast cancer. Medicine 2016, 95, e4392. [Google Scholar] [CrossRef] [PubMed]
- Abel, E.J.; Carrasco, A.; Culp, S.H.; Matin, S.F.; Tamboli, P.; Tannir, N.M.; Wood, C.G. Limitations of preoperative biopsy in patients with metastatic renal cell carcinoma: Comparison to surgical pathology in 405 cases. BJU Int. 2013, 189, 1692. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.Y. Development of a chemiluminescent immunoassay for cancer antigen 15–3. Labeled Immunoass. Clin. Med. 2016, 23, 1348–1351. [Google Scholar]
- Li, L.; Feng, D.; Zhao, J.; Guo, Z.; Zhang, Y. Simultaneous fluoroimmunoassay of two tumor markers based on CDTE quantum dots and gold nanocluster coated-silica nanospheres as labels. RSC Adv. 2015, 5, 105992–105998. [Google Scholar] [CrossRef]
- Mittal, S.; Kaur, H.; Gautam, N.; Mantha, A.K. Biosensors for breast cancer diagnosis: A review of bioreceptors, biotransducers and signal amplification strategies. Biosens. Bioelectron. 2017, 88, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Ji, Z.; Ding, W.; Lou, C.; Yang, S.; Xing, D. Ultrashort microwave-pumped real-time thermoacoustic breast tumor imaging system. IEEE Trans. Med. Imaging 2015, 35, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Nelson, H.D.; Tyne, K.; Naik, A.; Bougatsos, C.; Chan, B.K.; Humphrey, L.; U.S. Preventive Services Task Force. Screening for breast cancer: An update for the U.S. Preventive Services Task Force. Ann. Intern. Med. 2009, 151, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Pisano, E.D.; Gatsonis, C.; Hendrick, E.; Yaffe, M.; Baum, J.K.; Acharyya, S.; Conant, E.F.; Fajardo, L.L.; Bassett, L.; D’Orsi, C.; et al. Diagnostic performance of digital versus film mammography for breast-cancer screening. N. Engl. J. Med. 2005, 353, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Smart, C.R. Limitations of the randomized trial for the early detection of cancer. Cancer 1997, 79, 1740–1746. [Google Scholar] [CrossRef]
- Lobbes, M.B.; Smidt, M.L.; Houwers, J.; Tjan-Heijnen, V.C.; Wildberger, J.E. Contrast enhanced mammography: Techniques, current results, and potential indications. Clin. Radiol. 2013, 68, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Hooley, R.J.; Scoutt, L.M.; Philpotts, L.E. Breast ultrasonography: State of the art. Radiology 2013, 268, 642–659. [Google Scholar] [CrossRef] [PubMed]
- Schneble, E.J.; Graham, L.J.; Shupe, M.P.; Flynt, F.L.; Banks, K.P.; Kirkpatrick, A.D.; Nissan, A.; Henry, L.; Stojadinovic, A.; Shumway, N.M.; et al. Future directions for the early detection of recurrent breast cancer. J. Cancer 2014, 5, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.M.; Abbosh, S.; Mustafa, D.; Ireland, D. Microwave system for head imaging. IEEE Trans. Instrum. Meas. 2014, 63, 117–123. [Google Scholar] [CrossRef]
- Meaney, P.M.; Fanning, M.W.; Di, F.A.R.; Kaufman, P.A.; Geimer, S.D.; Zhou, T.; Paulsen, K.D. Microwave tomography in the context of complex breast cancer imaging. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Buenos Aires, Argentina, 31 August–4 September 2010; pp. 3398–3401. [Google Scholar]
- Zhu, X.; Zhao, Z.; Wang, J.; Song, J.; Liu, Q.H. Microwave-induced thermal acoustic tomography for breast tumor based on compressive sensing. IEEE Trans. Biomed. Eng. 2013, 60, 1298–1307. [Google Scholar] [PubMed]
- Bevacqua, M.T.; Scapaticci, R. A compressive sensing approach for 3D breast cancer microwave imaging with magnetic nanoparticles as contrast agent. IEEE Trans. Med. Imaging 2016, 35, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Hagness, S.C.; Taflove, A.; Bridges, J.E. Three-dimensional FDTD analysis of an ultrawideband antenna-array element for confocal microwave imaging of nonpalpable breast tumors. Antennas Propag. Soc. Int. Sympos. 1999, 3, 1886–1889. [Google Scholar]
- Fear, E.C.; Sill, J.M. Preliminary investigations of tissue sensing adaptive radar for breast tumor detection. In Proceedings of the 25th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Cancun, Mexico, 17–21 September 2003; Volume 4, pp. 3787–3790. [Google Scholar]
- Khosrowshahli, E.; Jeremić, A. Bayesian estimation of tumours in breasts using microwave imaging. Antimicrob. Agents Chemother. 2013, 52, 1670–1676. [Google Scholar]
- Bond, E.J.; Li, X.; Hagness, S.C.; Van Veen, B.D. Microwave imaging via space-time beamforming for early detection of breast cancer. IEEE Trans. Antennas Propag. 2003, 51, 1690–1705. [Google Scholar] [CrossRef]
- Smith, D.; Yurduseven, O.; Livingstone, B.; Schejbal, V. Microwave imaging using indirect holographic techniques. IEEE Antennas Propag. 2014, 56, 104–117. [Google Scholar] [CrossRef]
- Wang, L.; Al-Jumaily, A.M.; Simpkin, R. Imaging of 3-D dielectric objects using far-field holographic microwave imaging technique. Prog. Electromagn. Res. B 2014, 61, 135–147. [Google Scholar] [CrossRef]
- Wang, L.; Simpkin, R.; Al-Jumaily, A.M. Three-dimensional far-field holographic microwave imaging: An experimental investigation of dielectric object. Prog. Electromagn. Res. B 2014, 61, 169–184. [Google Scholar] [CrossRef]
- Fear, E.C.; Meaney, P.M.; Stuchly, M.A. Microwaves for breast cancer detection? IEEE Potentials 2003, 22, 12–18. [Google Scholar] [CrossRef]
- Meaney, P.M.; Fanning, M.W.; Li, D.; Poplack, S.P. A clinical prototype for active microwave imaging of the breast. IEEE Trans. Microw. Theory Tech. 2002, 48, 1841–1853. [Google Scholar]
- Meaney, P.M.; Fanning, M.W.; Zhou, T.; Golnabi, A. Clinical microwave breast imaging—2D results and the evolution to 3D. In Proceedings of the IEEE International Conference on Electromagnetics in Advanced Applications, Torino, Italy, 14–18 September 2009; pp. 881–884. [Google Scholar]
- Bourqui, J.; Sill, J.M.; Fear, E.C. A prototype system for measuring microwave frequency reflections from the breast. Int. J. Biomed. Imaging 2012, 2012, 851234. [Google Scholar] [CrossRef] [PubMed]
- Craddock, I.J.; Klemm, M.; Leendertz, J.; Preece, A.W.; Benjamin, R. Development and application of a UWB radar system for breast imaging. J. Pure Appl. Algebra 2008, 216, 314–322. [Google Scholar]
- Klemm, M.; Craddock, I.; Leendertz, J.; Preece, A. Experimental and clinical results of breast cancer detection using UWB microwave radar. In Proceedings of the IEEE Antennas and Propagation Society International Symposium, San Diego, CA, USA, 5–11 July 2008; pp. 1–4. [Google Scholar]
- Shahzad, A.; O’Halloran, M.; Jones, E.; Glavin, M. A preprocessing filter for multistatic microwave breast imaging for enhanced tumour detection. Prog. Electromagn. Res. B 2014, 57, 115–126. [Google Scholar] [CrossRef]
- Porter, E.; Coates, M.; Popović, M. An early clinical study of time-domain microwave radar for breast health monitoring. IEEE Trans. Biomed. Eng. 2016, 63, 530–539. [Google Scholar] [CrossRef] [PubMed]
- Porter, E.; Bahrami, H.; Santorelli, A.; Gosselin, B.; Rusch, L.; Popovich, M. A wearable microwave antenna array for time-domain breast tumor screening. IEEE Trans. Med. Imaging 2016, 35, 1501–1509. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Simpkin, R.; Al-Jumaily, A.M. Open-ended waveguide antenna for microwave breast cancer detection. Proceeedings of the 2013 IEEE International Workshop on Electromagnetics, Applications and Student Innovation Competition, Kowloon, Hongkong, China, 1–3 August 2013; pp. 65–68. [Google Scholar]
- Wang, Y.; Bakar, A.A.; Bialkowski, M.E. Compact Tapered Slot Antennas for UWB microwave imaging applications. In Proceedings of the IEEE International Conference on Microwave Radar and Wireless Communications, Vilnius, Lithuania, 14–16 June 2010; pp. 1–4. [Google Scholar]
- Azim, R.; Islam, M.T.; Misran, N. Compact tapered-shape slot antenna for UWB applications. IEEE Antennas Wirel. Propag. Lett. 2011, 10, 1190–1193. [Google Scholar] [CrossRef]
- Pallone, M.J.; Meaney, P.M.; Paulsen, K.D. Surface scanning through a cylindrical tank of coupling fluid for clinical microwave breast imaging exams. Med. Phys. 2012, 39, 3102. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Yook, J.G. Recent research trends of radio-frequency biosensors for biomolecular detection. Biosens. Bioelectron. 2014, 61, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.H.; Kuo, L.S.; Chen, P.H.; Yang, C.R.; Tsai, Z.M. Development of a multilayered polymeric DNA biosensor using radio frequency technology with gold and magnetic nanoparticles. Biosens. Bioelectron. 2012, 31, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Yoon, T.J.; Lee, H.; Andress, W.; Weissleder, R.; Ham, D. Palm NMR and 1-chip NMR. IEEE J. Solid-State Circuits 2011, 46, 342–352. [Google Scholar] [CrossRef]
- Kim, S.G.; Lee, H.J.; Lee, J.H.; Jung, H.I.; Yook, J.G. A highly sensitive and label free biosensing platform for wireless sensor node system. Biosens. Bioelectron. 2013, 50, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.P.; Yuan, R.; Chai, Y.Q. Novel immunoassay for carcinoembryonic antigen based on protein a-conjugated immunosensor chip by surface plasmon resonance and cyclic voltammetry. Bioprocess Biosyst. Eng. 2006, 28, 315. [Google Scholar] [CrossRef] [PubMed]
- Northardt, T.; Kasilingam, D. Spectral extrapolation for ultra-wide band radio frequency super-resolution tumor localization in the breast. Biomed. Eng. Lett. 2017, 7, 1–6. [Google Scholar] [CrossRef]
- Misek, D.E.; Kim, E.H. Protein biomarkers for the early detection of breast cancer. Int. J. Proteom. 2011, 2090–2166, 343582. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.K.; Zetter, B.R. Cancer biomarkers: Knowing the present and predicting the future. Future Oncol. 2017, 1, 37. [Google Scholar] [CrossRef] [PubMed]
- Porto-Mascarenhas, E.C.; Assad, D.X.; Chardin, H.; Gozal, D.; De Luca Canto, G.; Acevedo, A.C.; Guerra, E.N. Salivary biomarkers in the diagnosis of breast cancer: A review. Crit. Rev. Oncol. Hematol. 2017, 110, 62. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, A.; Koch, A.; Krockenberger, K.; Grosshennig, A. Personalized medicine using DNA biomarkers: A review. Hum. Genet. 2012, 131, 1627–1638. [Google Scholar] [CrossRef] [PubMed]
- Opstal-van Winden, A.W.; Vermeulen, R.C.; Peeters, P.H.; Beijnen, J.H.; van Gils, C.H. Early diagnostic protein biomarkers for breast cancer: How far have we come? Breast Cancer Res. Treat. 2012, 134, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Le, N.F.; Misek, D.E.; Krause, M.C.; Deneux, L.; Giordano, T.J.; Scholl, S.; Hanash, S.M. Proteomics-based identification of RS/DJ-1 as a novel circulating tumor antigen in breast cancer. Clin. Cancer Res. 2001, 7, 3328–3335. [Google Scholar]
- Su, M.; Wheeler, T.K.; Picken, S.; Negus, S.; Milner, A.J. P53 autoantibodies in 1006 patients followed up for breast cancer. Breast Cancer Res. 2000, 2, 438–443. [Google Scholar]
- Kulić, A.; Sirotković-Skerlev, M.; Jelisavac-Cosić, S.; Herceg, D.; Kovac, Z.; Vrbanec, D. Anti-p53 antibodies in serum: Relationship to tumor biology and prognosis of breast cancer patients. Med. Oncol. 2010, 27, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Park, S.; Ji, M.P.; Cho, J.H.; Kim, S.I.; Park, B.W. Elevated levels of serum tumor markers CA 15–3 and CEA are prognostic factors for diagnosis of metastatic breast cancers. Breast Cancer Res. Treat. 2013, 141, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.J.; Park, H.Y.; Lee, Y.H. Clinical value of CEA, CEA15-3 and TPS in breast cancer. J. Korean Breast Cancer Soc. 2001, 4, 136–143. [Google Scholar]
- Duffy, M.J. CA 15–3 and related mucins as circulating markers in breast cancer. Ann. Clin. Biochem. 1999, 36, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Orlandi, A.; Dio, C.D.; Calegari, M.A.; Barone, C. Paradox, CA 15–3 increase in metastatic breast cancer patients treated with everolimus: A change of paradigm in a case series. Biomark. Med. 2017, 10, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.W.; Li, W.H.; Wang, J.; Li, A.L.; Li, H.Y.; Wang, H.X.; Li, W.; Kang, L.H.; Yu, M.; Shen, B.F.; et al. Proteomics-based identification of human acute leukemia antigens that induce humoral immune response. Mol. Cell. Proteom. 2005, 4, 1718. [Google Scholar] [CrossRef] [PubMed]
- Asif, H.M.; Sultana, S.; Ahmed, S.; Akhtar, N.; Tariq, M. HER-2 positive breast cancer—A mini-review. Asian Pac. J. Cancer Prev. APJCP 2016, 17, 1609. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, P.D.; Jakobsen, E.H.; Madsen, J.S.; Petersen, E.B.; Andersen, R.F.; Østergaard, B.; Brandslund, I. Serum HER-2: Sensitivity, specificity, and predictive values for detecting metastatic recurrence in breast cancer patients. J. Cancer Res. Clin. Oncol. 2013, 139, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Molina, R.; Escudero, J.M.; Muñoz, M.; Augé, J.M.; Filella, X. Circulating levels of HER-2/neu oncoprotein in breast cancer. Clin. Chem. Lab. Med. 2012, 50, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Mattos-Arruda, L.D.; Cortes, J.; Santarpia, L.; Vivancos, A.; Tabernero, J.; Reis-Filho, J.S.; Seoane, J. Circulating tumour cells and cell-free DNA as tools for managing breast cancer. Nat. Rev. Clin. Oncol. 2013, 10, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Gracia-Aznarez, F.J.; Fernandez, V.; Pita, G.; Peterlongo, P.; Dominguez, O.; de la Hoya, M.D.L.; Duran, M.; Osorio, A.; Moreno, L.; Gonzalez-Neira, A.; et al. Whole exome sequencing suggests much of non-BRCA1/BRCA2 familial breast cancer is due to moderate and low penetrance susceptibility alleles. PLoS ONE 2013, 8, e55681. [Google Scholar] [CrossRef] [PubMed]
- Konishi, H.; Mohseni, M.; Tamaki, A.; Gary, J.P.; Croessmann, S.; Karnan, S.; Ota, A.; Wong, H.Y.; Konishi, Y.; Karakas, B.; et al. Mutation of a single allele of the cancer susceptibility gene brca1 leads to genomic instability in human breast epithelial cells. Proc. Natl. Acad. Sci. USA 2011, 108, 17773. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, P.A.; Sandhyarani, N. Graphene-DNA electrochemical sensor for the sensitive detection of BRCA1 gene. Sens. Actuators B Chem. 2014, 204, 777–782. [Google Scholar] [CrossRef]
- Kabat, G.C.; Kandel, R.A.; Glass, A.G.; Jones, J.G.; Olson, N.; Duggan, C.; Ginsberg, M.; Negassa, A.; Rohan, T. A cohort study of p53 mutations and protein accumulation in benign breast tissue and subsequent breast cancer risk. J. Oncol. 2011, 2011, 970804. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Tao, Y.; Yue, G.; Li, R.; Qiu, B.; Guo, L.; Lin, Z.; Yang, H.H. Highly selective and sensitive electrochemiluminescence biosensor for p53 DNA sequence based on nicking endonuclease assisted target recycling and hyperbranched rolling circle amplification. Anal. Chem. 2016, 88, 5097. [Google Scholar] [CrossRef] [PubMed]
- Chase, J.W.; L’Italien, J.J.; Murphy, J.B.; Spicer, E.K.; Williams, K.R. Characterization of the escherichia coli ssb-113 mutant single-stranded DNA-binding protein. cloning of the gene, DNA and protein sequence analysis, high pressure liquid chromatography peptide mapping, and DNA-binding studies. J. Biol. Chem. 1984, 259, 805–814. [Google Scholar] [PubMed]
- Singh, B.; Chatterjee, A.; Ronghe, A.M.; Bhat, N.K.; Bhat, H.K. Antioxidant-mediated up-regulation of OGG1 via NRF2 induction is associated with inhibition of oxidative DNA damage in estrogen-induced breast cancer. BMC Cancer 2013, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Zhang, A.; Powell, M.J. Genetic alteration and mutation profiling of circulating cell-free tumor DNA (cfDNA) for diagnosis and targeted therapy of gastrointestinal stromal tumors. Chin. J. Cancer 2016, 35, 68. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Pantel, K. Circulating DNA as biomarker in breast cancer. Breast Cancer Res. 2015, 17, 136. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, G.; Cava, C.; Castiglioni, I. Micrornas: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 2014, 5, 1122–1143. [Google Scholar] [CrossRef] [PubMed]
- Matamala, N.; Vargas, M.T.; González-Cámpora, R.; Miñambres, R.; Arias, J.I.; Menéndez, P.; Andrés-León, E.; Gómez-López, G.; Yanowsky, K.; Calvete-Candenas, J.; et al. Tumor microrna expression profiling identifies circulating micrornas for early breast cancer detection. Clin. Chem. 2015, 61, 1098–1106. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, X.; Yang, J.; Zhen, J.; Zhang, D. Serum microRNA-21 as a potential diagnostic biomarker for breast cancer: A systematic review and meta-analysis. Clin. Exp. Med. 2016, 16, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.W.; Wu, J.; Lu, J.S.; Han, Q.X.; Shen, Z.Z.; Nguyen, M.; Barsky, S.H.; Shao, Z.M. Fiberoptic ductoscopy for breast cancer patients with nipple discharge. Surg. Endosc. 2011, 15, 1340–1345. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Bai, H.; Luo, J.; Yang, P.; Cai, J. A recyclable chitosan-based QCM biosensor for sensitive and selective detection of breast cancer cells in real time. Analyst 2014, 139, 6259. [Google Scholar] [CrossRef] [PubMed]
- Daghestani, H.N.; Day, B.W. Theory and applications of surface plasmon resonance, resonant mirror, resonant waveguide grating, and dual polarization interferometry biosensors. Sensors 2010, 10, 9630–9646. [Google Scholar] [CrossRef] [PubMed]
- Lepage, D.; Carrier, D.; Jiménez, A.; Beauvais, J.; Dubowski, J.J. Plasmonic propagations distances for interferometric surface plasmon resonance biosensing. Nanoscale Res. Lett. 2011, 6, 388. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Mishra, S.K.; Gupta, B.D. SPR based fibre optic biosensor for phenolic compounds using immobilization of tyrosinase in polyacrylamide gel. Sens. Actuators B Chem. 2013, 186, 388–395. [Google Scholar] [CrossRef]
- Dey, D. Optical biosensors: A revolution towards quantum nanoscale electronics device fabrication. Biomed. Res. Int. 2011, 1, 348218. [Google Scholar] [CrossRef] [PubMed]
- Becker, B.; Cooper, M.A. A survey of the 2006–2009 quartz crystal microbalance biosensor literature. J. Mol. Recognit. 2011, 24, 754. [Google Scholar] [CrossRef] [PubMed]
- Arif, S.; Qudsia, S.; Urooj, S.; Chaudry, N.; Arshad, A.; Andleeb, S. Blueprint of quartz crystal microbalance biosensor for early detection of breast cancer through salivary autoantibodies against ATP6AP1. Biosens. Bioelectron. 2015, 65, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Chatterjee, S. Nanomaterials based electrochemical sensors for biomedical applications. Chem. Soc. Rev. 2013, 42, 5425. [Google Scholar] [CrossRef] [PubMed]
- Arya, S.K.; Wang, K.Y.; Wong, C.C.; Rahman, A.R. Anti-EpCAM modified LC-SPDP monolayer on gold microelectrode based electrochemical biosensor for MCF-7 cells detection. Biosens. Bioelectron. 2013, 41, 446. [Google Scholar] [CrossRef] [PubMed]
- Selwyna, P.G.C.; Loganathan, P.R.; Begam, K.H. Development of electrochemical biosensor for breast cancer detection using gold nanoparticle doped CA 15–3 antibody and antigen interaction. In Proceedings of the 2013 International Conference on Signal Processing, Image Processing & Pattern Recognition, Coimbatore, India, 7–8 February 2013; Volume 18, pp. 75–81. [Google Scholar]
- Zhou, T.; Zhang, B.; Wei, P.; Du, Y.; Zhou, H.; Yu, M.; Yan, L.; Zhang, W.; Nie, G.; Chen, C.; et al. Energy metabolism analysis reveals the mechanism of inhibition of breast cancer cell metastasis by peg-modified graphene oxide nanosheets. Biomaterials 2014, 35, 9833. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Kumar, R.; Kaur, H.; Rayat, C.S.; Kaur, I.; Arora, S.K.; Srivastava, J.; Bharadwaj, L.M. Translocation and toxicity of docetaxel multi-walled carbon nanotube conjugates in mammalian breast cancer cells. J. Biomed. Nanotechnol. 2014, 10, 3601–3609. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xie, L.; Li, H. Electrochemical biosensor based on reduced graphene oxide and Au nanoparticles entrapped in chitosan/silica sol–gel hybrid membranes for determination of dopamine and uric acid. J. Electroanal. Chem. 2012, 682, 158–163. [Google Scholar] [CrossRef]
- Abruzzi, A. Combination of single walled carbon nanotubes/graphene oxide with paclitaxel: A reactive oxygen species mediated synergism for treatment of lung cancer. Nanoscale 2013, 5, 2818–2829. [Google Scholar]
- Zhao, H.; Liang, H.; Vidal, F.; Rosei, F.; Vomiero, A.; Ma, D. Size dependence of temperature-related optical properties of PbS and PbS/CdS core/shell quantum dots. J. Phys. Chem. C 2014, 118, 20585–20593. [Google Scholar] [CrossRef]
- Peng, Y.; Yi, G.; Gao, Z. A highly sensitive microrna biosensor based on ruthenium oxide nanoparticle-initiated polymerization of aniline. Chem. Commun. 2010, 46, 9131–9133. [Google Scholar] [CrossRef] [PubMed]
- Pérez, W.I.; Soto, Y.; Ramirez-Vick, J.E.; Meléndez, E. Nanostructured gold dsDNA sensor for early detection of breast cancer by beta protein 1 (BP1). J. Electroanal. Chem. 2015, 751, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, A.; Mahdi, R.; Navid, N.; Khadijeh, A.; Hossein, N.M. An electrochemical nanobiosensor for plasma miRNA-155, based on graphene oxide and gold nanorod, for early detection of breast cancer. Biosens. Bioelectron. 2016, 77, 99–106. [Google Scholar]
- Wang, K.; He, M.Q.; Zhai, F.H.; He, R.H.; Yu, Y.L. A novel electrochemical biosensor based on polyadenine modified aptamer for label-free and ultrasensitive detection of human breast cancer cells. Talanta 2017, 166, 87–92. [Google Scholar] [CrossRef] [PubMed]
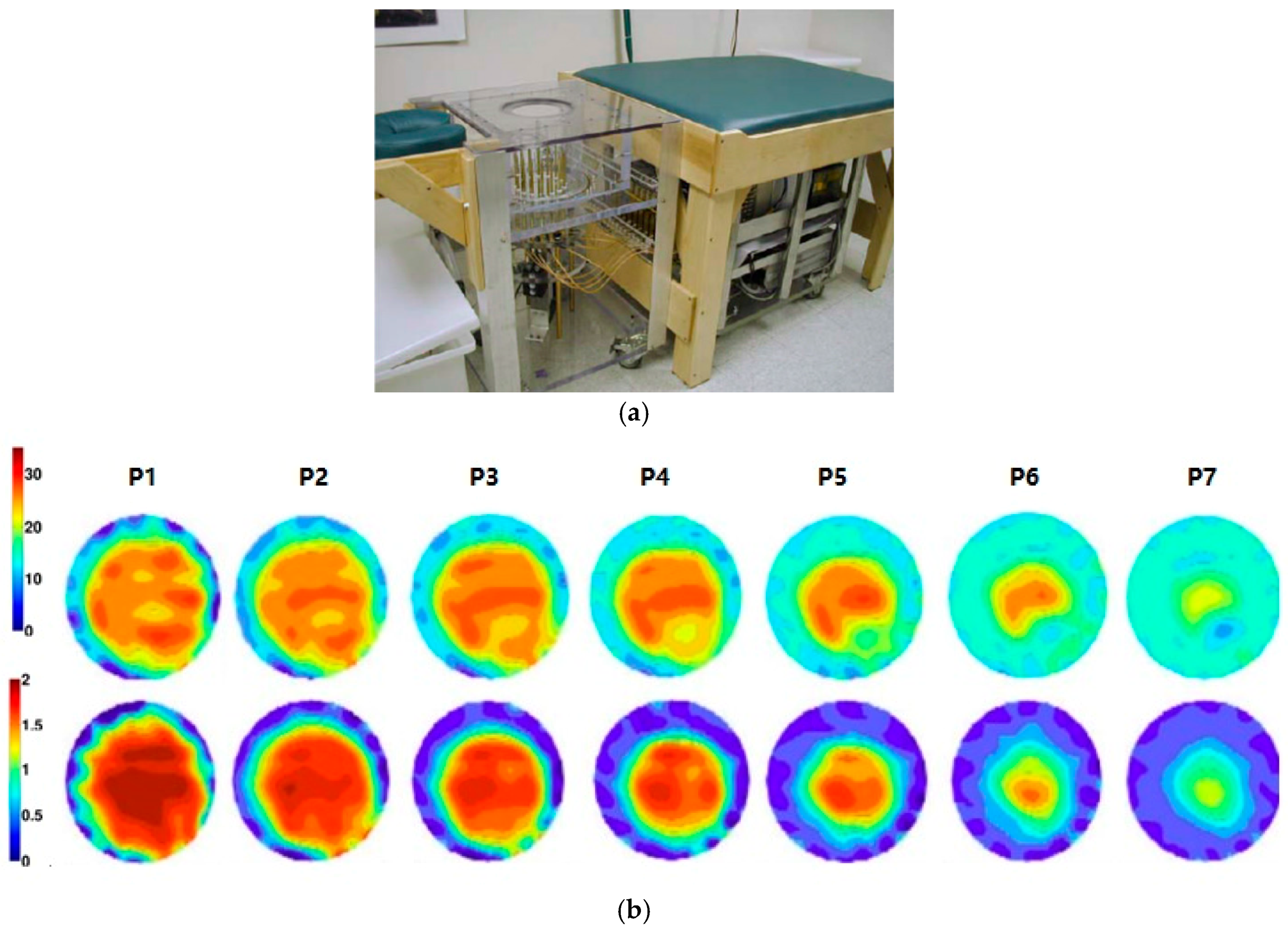
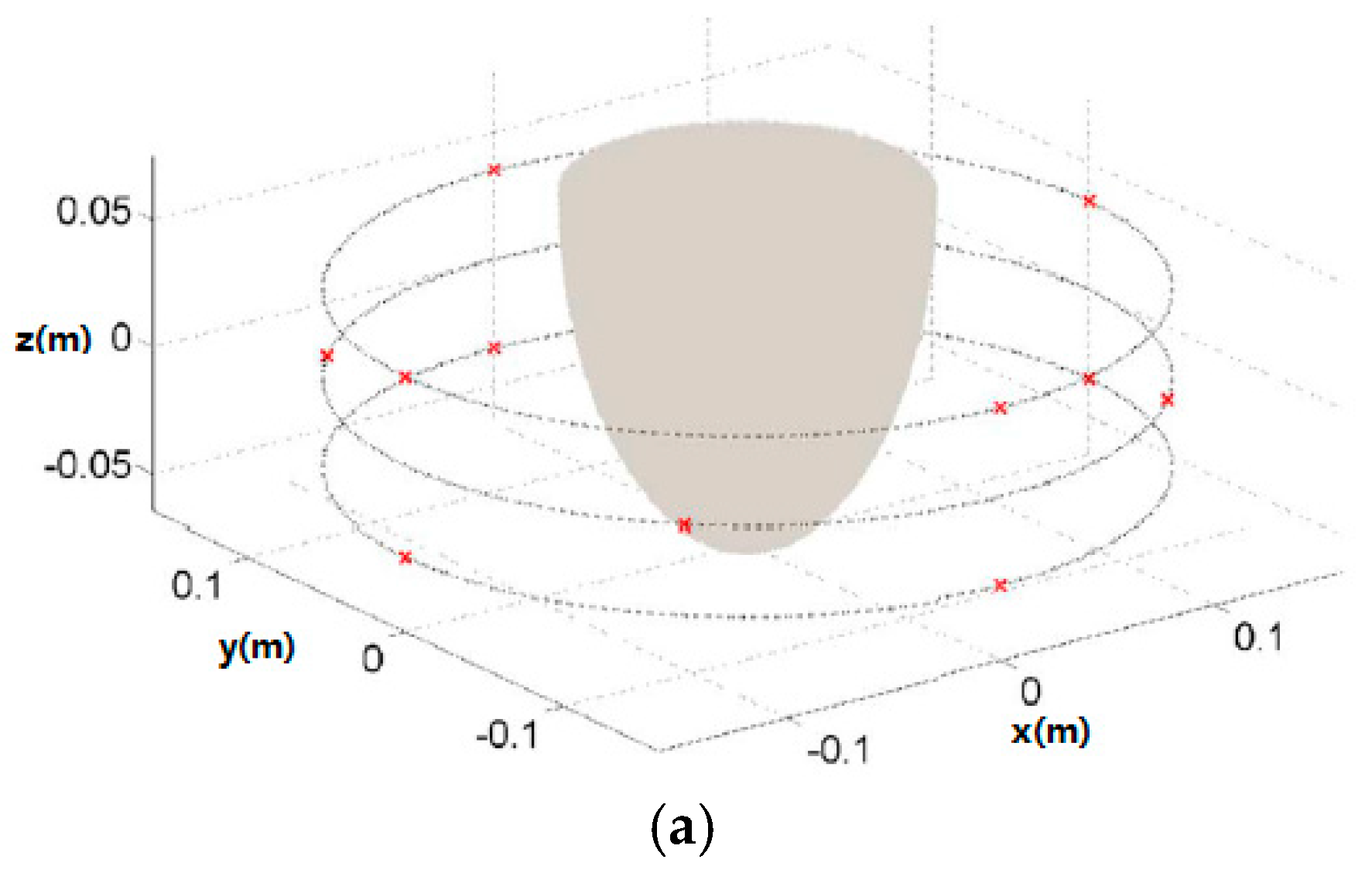
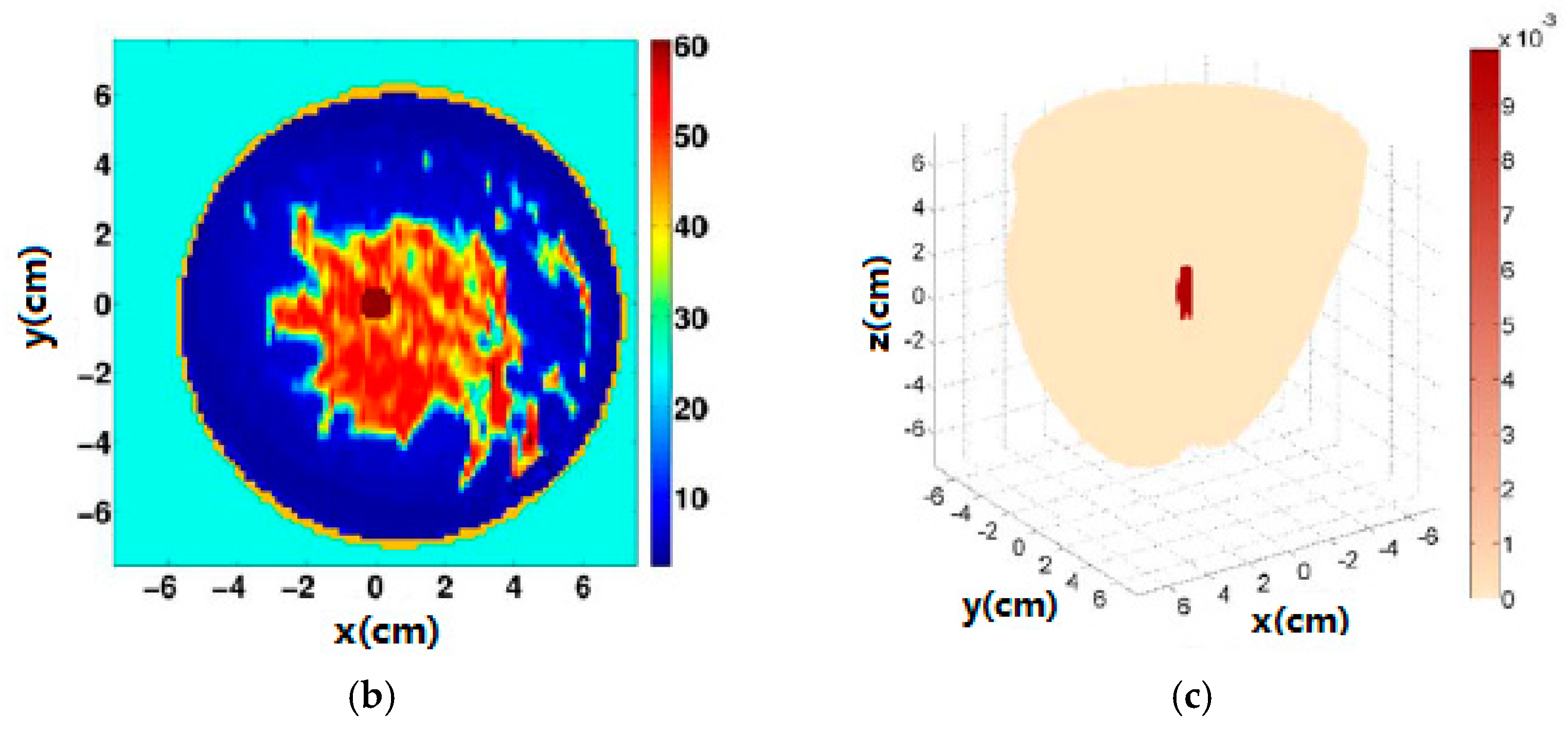
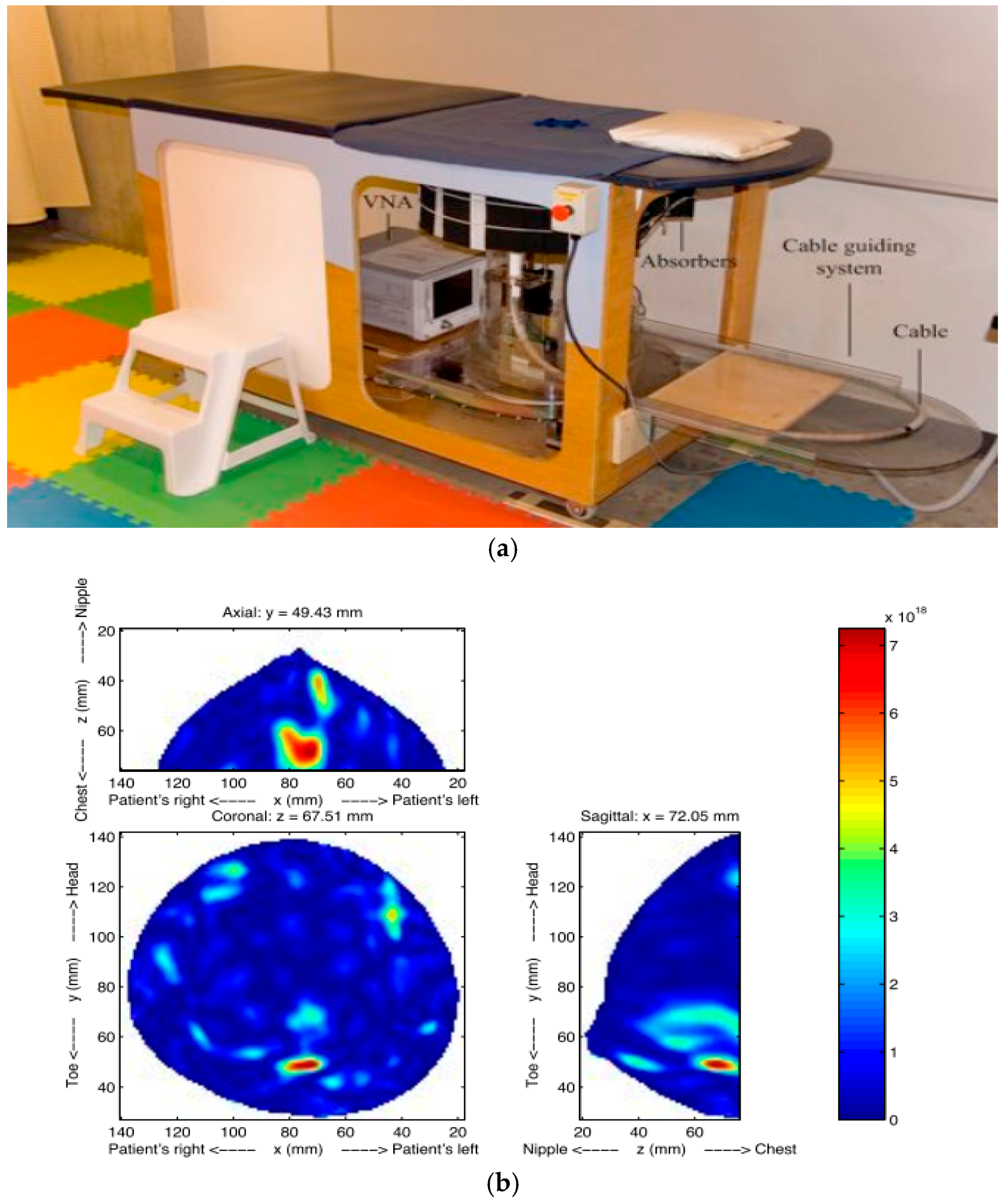
| Type | Use | Sensitivity * | Specificity * | limitations | Time |
|---|---|---|---|---|---|
| Mammography | Mass screening. Image bone, soft tissue and blood vessels all at the same time. Shadowing due to dense tissues | 67.8% | 75.0% | Ionizing radiation, low sensitivity and specificity, sensitivity drops with tissue density increases | few seconds |
| Ultrasound | Evaluate lumps found in mammography; Not suitable for bony structures | 83.0% | 34.0% | Low sensitivity; experienced operator is required during examination; low resolution image; | 10–20 min |
| MRI | Young women with high risk; Images small details of soft tissues | 94.4% | 26.4% | Some types of cancers cannot be detected such as ductal and lobular carcinoma; expensive; | 40–60 min |
| CT | To determine and image distant metastasis in a single exam | 91% | 93% | Low sensitivity; radiation risks; expensive scanner; | 5 min |
| PET | Functional imaging of biological processes. To image metastasis or response to therapy | 61.0% | 80.0% | Ionizing radiation, radioactive tracer injection | 90–240 min |
| Method | Dartmouth College [35,45,46,47] | University of Calgary [48] | University of Bristol [49,50] | McGill University [51,52,53] | Auckland University of Technology [54] |
|---|---|---|---|---|---|
| Sensor | 16 monopoles | 24 open-ended waveguides | 16 stacked-patch antennas | 16 wideband sensors | 16 open-ended waveguides |
| Sensor array | circular | cylindrical | spherical | hemispherical | spiral |
| Imaging | Microwave tomography | TSAR | UWB microwave radar imaging | UWB microwave radar imaging | HMI |
| Frequency | 0.5~3 GHz | 1.0~2.3 GHz | 4~10 GHz | 2~4 GHz | 12 GHz |
| Test object | Phantoms, patients | Phantoms, patients | Phantoms, patients | Phantoms, real patients | phantoms |
| Immersion medium | 0.9% saline water | canola oil | air | ultrasound gel | air |
| Image | 2D, 3D | 2D, 3D | 2D, 3D | 2D | 2D, 3D |
| Clinical trial | yes | yes | yes | yes | no |
| Biomarker | Technology Used for Discovery | Type |
|---|---|---|
| RS/DJ-1 | Serum profiling | Serum protein |
| CA15-3 | ||
| CA27-29 | ||
| HER-2 | ||
| p53 | Humoral response | autoantibody |
| HSP60 | ||
| HSP90 | ||
| MUC1 | ||
| -2-HS-Glycoprotein | Nipple aspirate fluid profiling | Ductal protein |
| Lipophilin B | ||
| β-Globin | ||
| Hemopexin | ||
| Vitamin D-binding protein |
| Target miRNA | Mechanism | Nanomaterial | Electrochemical Method | Linear Range | Detection Limit |
|---|---|---|---|---|---|
| let-7a | Polymerase extension/streptavidin/AP | AuNP | AuE/DPV | 100 fm~1 nm | 99.2 fm |
| let-7b | Nanoparticles catalyze oxidation of hydrazine OsO2 | NP | ITO/Amp | 0.30 pm~20 pm | 80 fm |
| let-7c | Peptide nucleic acid probe/polyaniline/H2O2 | RuO2NP | AuE/SWV | 5.0 fm~2 pm | 2.0 fm |
| miR-21 | Capture probe/aptamer/hemin | AuNP | AuE/EIS | 5 pm~5000 pm | 3.96 pm |
| Star trigon structure/endonuclease/MB | AuNP | GCE/SWV | 100 am~1 nm | 30 am | |
| LNA molecular beacon/streptavidin-HRP/HQ | GO/AuNP | GCE/Amp | 0.1 pm~7 pm | 0.06 pm | |
| TMB/HRP/Streptavidin-Poly-HRP80 | DNATN | AuE/Amp | 10 fm~10 nm | 10 fm | |
| 3D DNA stem-loop probe/ferrocene | AuNP/3D DNA | AuE/DPV | 100 pm~1 μm | 10 pm | |
| miR-24 | Oxidation signal of guanine | MWCNTs | GCE/DPV | 1 pm~1 nm | 1 pm |
| miR-141 | ELISA-like amplification/antibody/HRP/BQ | MWCNTs/GO | SPGE/SWV | 0 fm~1 nm | 10 fm |
| RNA-DNA antibodies/conducting polymer | GO | GCE/SWV | 1 fm~1 nm | 5 fm | |
| miR-122 | DNA Four-Way Junction/streptavidin | AuNP | SPCE/SWV | 10 am~1 fm | 2 am |
| miR-155 | Hairpin probe/hybridization chain reaction/MB | GO/AuNP | GCE/DPV | 10 fm~1 nm | 3.3 fm |
| Magnetic bead/ligase chain reaction/T4 ligase | PbS, CdS quantum dots | GCE/SWV | 50 fm~30 pm | 12 fm | |
| Nafion/thionine/H2O2 | PdNP | GCE/CV | 5.6 pm~56 mm | 1.87 pm | |
| capture Probe/OB | GO/GNR | GCE/DPV | 2 fm~8 pm | 0.6 fm |
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L. Early Diagnosis of Breast Cancer. Sensors 2017, 17, 1572. https://doi.org/10.3390/s17071572
Wang L. Early Diagnosis of Breast Cancer. Sensors. 2017; 17(7):1572. https://doi.org/10.3390/s17071572
Chicago/Turabian StyleWang, Lulu. 2017. "Early Diagnosis of Breast Cancer" Sensors 17, no. 7: 1572. https://doi.org/10.3390/s17071572
APA StyleWang, L. (2017). Early Diagnosis of Breast Cancer. Sensors, 17(7), 1572. https://doi.org/10.3390/s17071572





