Nucleic Acid Aptamers: An Emerging Tool for Biotechnology and Biomedical Sensing
Abstract
:1. Introduction

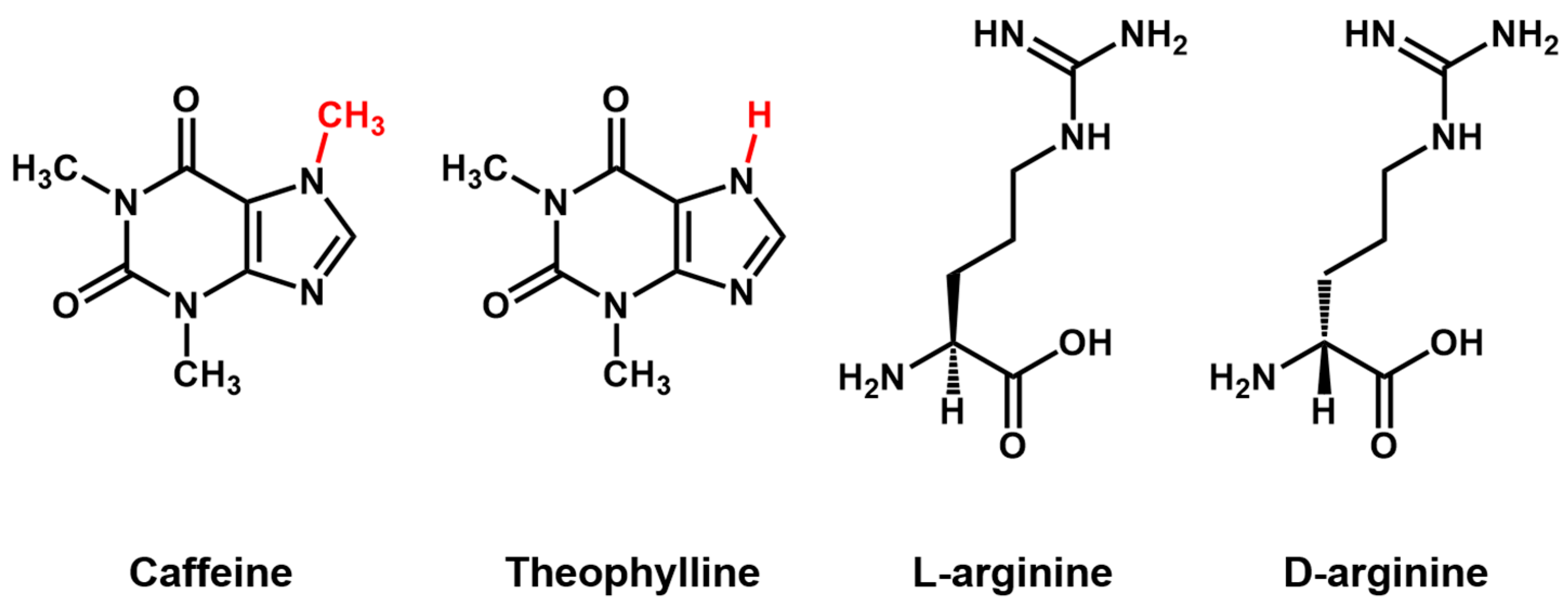
Aptamer Properties
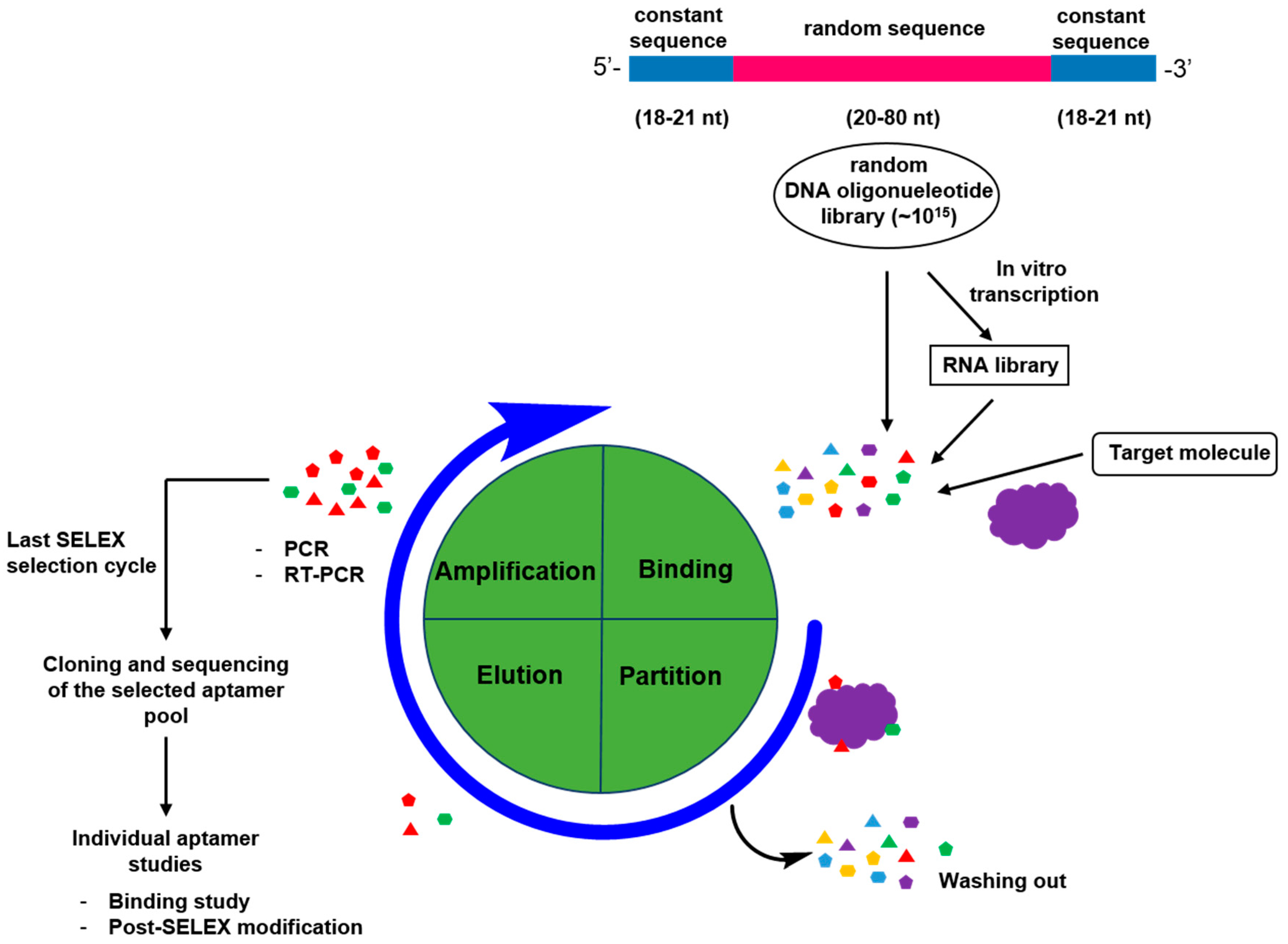
2. Generation of Aptamers
2.1. General Process for Aptamer Screening
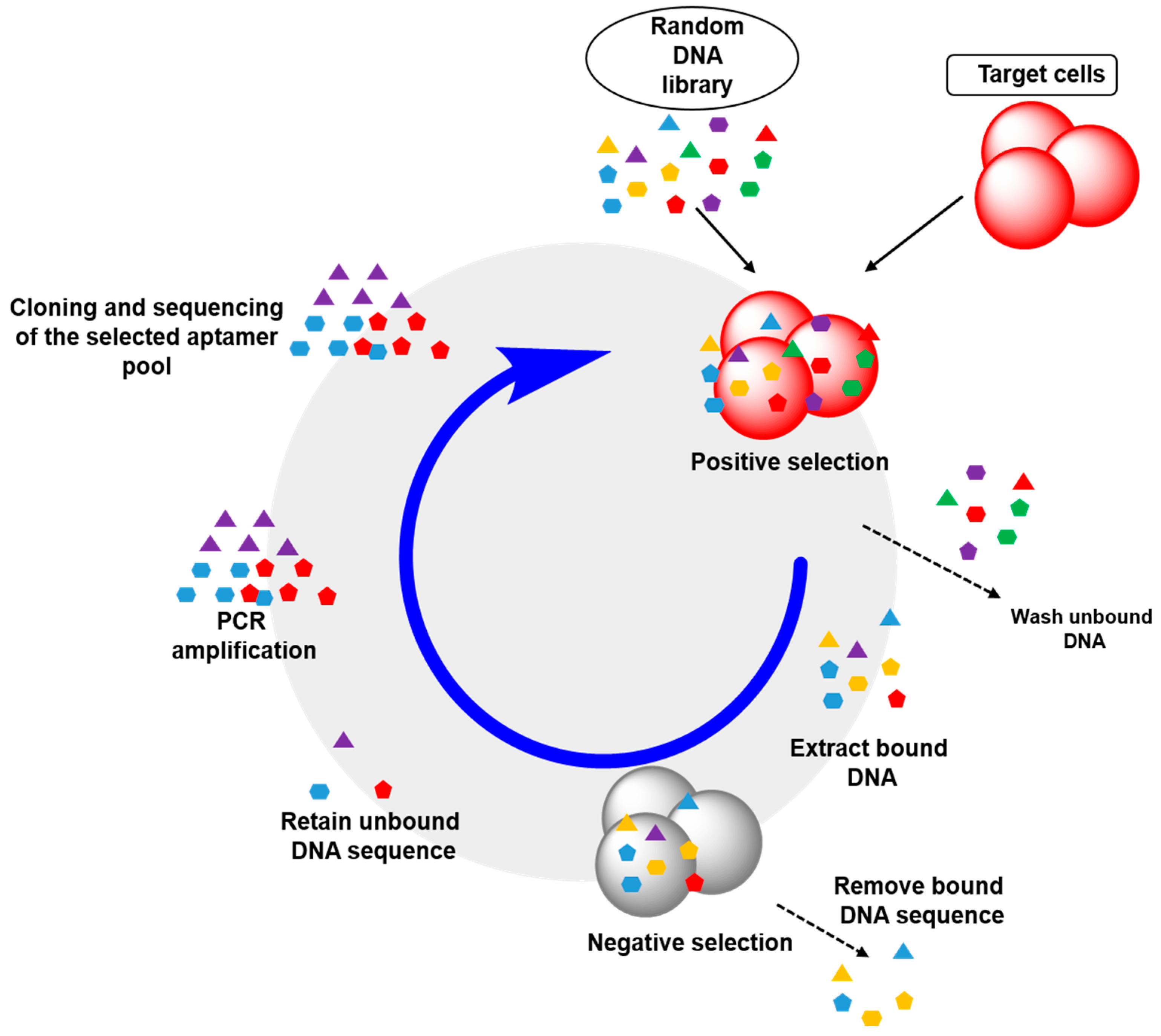
2.2. Cell-SELEX: Aptamers for Cell Membrane Analysis
2.3. Aptamer Truncation
3. Aptamers as Sensors for Biomarker Discovery
4. Aptamers as Molecular Imaging Probes
4.1. Aptamer-Based Molecular Beacons
4.2. Optical Molecular Imaging with Aptamer-Based Probes
4.3. Aptamer-Based Nanoimaging Agents for CT and MRI

5. Aptamers as Vehicles for Drug Delivery
5.1. Aptamer-Polymer Hybrid Delivery System
5.2. Aptamer-Liposome Hybrid Delivery System
5.3. Aptamer-Dendrimer Hybrid Delivery System
6. Aptamers as Potential Drugs
6.1. Therapeutic Aptamers in Cancer Therapy
6.2. Thrombin Binding Aptamers
7. Aptamer-Based Programmable Hydrogels
8. Aptamers in Precision Cancer Medicine
9. Future Perspective and Conclusions
Conflicts of Interest
References
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage t4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Feigon, J.; Dieckmann, T.; Smith, F.W. Aptamer structures from a to ζ. Chem. Biol. 1996, 3, 611–617. [Google Scholar] [CrossRef]
- Patel, D.J.; Suri, A.K.; Jiang, F.; Jiang, L.; Fan, P.; Kumar, R.A.; Nonin, S. Structure, recognition and adaptive binding in RNA aptamer complexes1. J. Mol. Biol. 1997, 272, 645–664. [Google Scholar] [CrossRef] [PubMed]
- Hermann, T.; Patel, D.J. Adaptive recognition by nucleic acid aptamers. Science 2000, 287, 820–825. [Google Scholar] [CrossRef] [PubMed]
- Piganeau, N.; Schroeder, R. Aptamer structures: A preview into regulatory pathways? Chem. Biol. 2003, 10, 103–104. [Google Scholar] [CrossRef]
- Hendeles, L.; Weinberger, M. Theophylline a “state of the art” review. Pharmacother. J. Hum. Pharmacol. Drug Ther. 1983, 3, 2–44. [Google Scholar]
- Jenison, R.; Gill, S.; Pardi, A.; Polisky, B. High-resolution molecular discrimination by RNA. Science 1994, 263, 1425–1429. [Google Scholar] [CrossRef] [PubMed]
- Geiger, A.; Burgstaller, P.; von der Eltz, H.; Roeder, A.; Famulok, M. RNA aptamers that bind L-arginine with sub-micromolar dissociation constants and high enantioselectivity. Nucleic Acids Res. 1996, 24, 1029–1036. [Google Scholar] [CrossRef] [PubMed]
- Argos, P.; Rossmann, M.G.; Grau, U.M.; Zuber, H.; Frank, G.; Tratschin, J.D. Thermal stability and protein structure. Biochemistry 1979, 18, 5698–5703. [Google Scholar] [CrossRef] [PubMed]
- Pontius, B.W.; Berg, P. Rapid renaturation of complementary DNA strands mediated by cationic detergents: A role for high-probability binding domains in enhancing the kinetics of molecular assembly processes. Proc. Natl. Acad. Sci. 1991, 88, 8237–8241. [Google Scholar] [CrossRef] [PubMed]
- SantaLucia, J.; Hicks, D. The thermodynamics of DNA structural motifs. Annu. Rev. Biophys. Biomol. Struct. 2004, 33, 415–440. [Google Scholar] [CrossRef] [PubMed]
- Chien, A.; Edgar, D.B.; Trela, J.M. Deoxyribonucleic acid polymerase from the extreme thermophile thermus aquaticus. J. Bacteriol. 1976, 127, 1550–1557. [Google Scholar] [PubMed]
- Brown, D.M.; Todd, A.R. 13. Nucleotides. Part x. Some observations on the structure and chemical behaviour of the nucleic acids. J. Chem. Soc. (Resumed) 1952, 52–58. [Google Scholar] [CrossRef]
- Beaucage, S.L.; Caruthers, M.H. Deoxynucleoside phosphoramidites—A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981, 22, 1859–1862. [Google Scholar] [CrossRef]
- Reese, C.B. Oligo- and poly-nucleotides: 50 years of chemical synthesis. Org. Biomol. Chem. 2005, 3, 3851–3868. [Google Scholar] [CrossRef] [PubMed]
- Kool, E.T. Preorganization of DNA: Design principles for improving nucleic acid recognition by synthetic oligonucleotides. Chem. Rev. 1997, 97, 1473–1488. [Google Scholar] [CrossRef] [PubMed]
- Ramzaeva, N.; Rosemeyer, H.; Leonard, P.; Mühlegger, K.; Bergmann, F.; von der Eltz, H.; Seela, F. Oligonucleotides functionalized by fluorescein and rhodamine dyes: Michael addition of methyl acrylate to 2′-deoxypseudouridine. Helv. Chim. Acta 2000, 83, 1108–1126. [Google Scholar] [CrossRef]
- Tung, C.-H.; Stein, S. Preparation and applications of peptide-oligonucleotide conjugates. Bioconjugate Chem. 2000, 11, 605–618. [Google Scholar] [CrossRef]
- Williams, B.A.R.; Chaput, J.C. Synthesis of peptide-oligonucleotide conjugates using a heterobifunctional crosslinker. In Current Protocols in Nucleic Acid Chemistry; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2001. [Google Scholar]
- Niemeyer, C.M. The developments of semisynthetic DNA-protein conjugates. Trends Biotechnol. 2002, 20, 395–401. [Google Scholar] [CrossRef]
- Ito, T.; Ueno, Y.; Komatsu, Y.; Matsuda, A. Synthesis, thermal stability and resistance to enzymatic hydrolysis of the oligonucleotides containing 5-(n-aminohexyl)carbamoyl-2′-o-methyluridines. Nucleic Acids Res. 2003, 31, 2514–2523. [Google Scholar] [CrossRef] [PubMed]
- Schoetzau, T.; Langner, J.; Moyroud, E.; Roehl, I.; Vonhoff, S.; Klussmann, S. Aminomodified nucleobases: Functionalized nucleoside triphosphates applicable for selex. Bioconjugate Chem. 2003, 14, 919–926. [Google Scholar] [CrossRef] [PubMed]
- Tennilä, T.; Antopolsky, M.; Azhayev, A.; Azhayeva, E. Peptide-oligonucleotide conjugates form stable and selective complexes with antibody and DNA. Bioconjugate Chem. 2008, 19, 1361–1367. [Google Scholar] [CrossRef] [PubMed]
- Kricka, L.J.; Fortina, P. Analytical ancestry: “Firsts” in fluorescent labeling of nucleosides, nucleotides, and nucleic acids. Clin. Chem. 2009, 55, 670–683. [Google Scholar] [CrossRef] [PubMed]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Pirrung, M.C. How to make a DNA chip. Angew. Chem. Int. Ed. 2002, 41, 1276–1289. [Google Scholar] [CrossRef]
- Sassolas, A.; Leca-Bouvier, B.D.; Blum, L.J. DNA biosensors and microarrays. Chem. Rev. 2008, 108, 109–139. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hao, C.; Ma, W.; Yong, Q.; Yan, W.; Kuang, H.; Wang, L.; Xu, C. Magnetic bead-based multiplex DNA sequence detection of genetically modified organisms using quantum dot-encoded silicon dioxide nanoparticles. J. Phys. Chem. C 2011, 115, 20134–20140. [Google Scholar] [CrossRef]
- Famulok, M.; Blind, M.; Mayer, G. Intramers as promising new tools in functional proteomics. Chem. Biol. 2001, 8, 931–939. [Google Scholar] [CrossRef]
- Burke, D.H.; Nickens, D.G. Expressing RNA aptamers inside cells to reveal proteome and ribonome function. Brief. Funct. Genomics Proteomics 2002, 1, 169–188. [Google Scholar] [CrossRef]
- Famulok, M.; Verma, S. In vivo-applied functional RNAs as tools in proteomics and genomics research. Trends Biotechnol. 2002, 20, 462–466. [Google Scholar] [CrossRef]
- Choi, K.H.; Park, M.W.; Lee, S.Y.; Jeon, M.-Y.; Kim, M.Y.; Lee, H.K.; Yu, J.; Kim, H.-J.; Han, K.; Lee, H.; et al. Intracellular expression of the t-cell factor-1 rna aptamer as an intramer. Mol. Cancer Ther. 2006, 5, 2428–2434. [Google Scholar] [CrossRef] [PubMed]
- Kwak, H.; Hwang, I.; Kim, J.H.; Kim, M.Y.; Yang, J.S.; Jeong, S. Modulation of transcription by the peroxisome proliferator-activated receptor δ-binding RNA aptamer in colon cancer cells. Mol. Cancer Ther. 2009, 8, 2664–2673. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C. Hybridoma technology for the generation of monoclonal antibodies. In Antibody Methods and Protocols; Proetzel, G., Ebersbach, H., Eds.; Humana Press: New York, NY, USA, 2012; Volume 901, pp. 117–135. [Google Scholar]
- Qin, C.-F.; Li, G.-C. Mammalian cell display technology coupling with aid induced shm in vitro: An ideal approach to the production of therapeutic antibodies. Int. Immunopharmacol. 2014, 23, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Sassanfar, M.; Szostak, J.W. An rna motif that binds ATP. Nature 1993, 364, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Mannironi, C.; di Nardo, A.; Fruscoloni, P.; Tocchini-Valentini, G.P. In vitro selection of dopamine RNA ligands. Biochemistry 1997, 36, 9726–9734. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.N.; Jensen, K.B.; Julin, C.M.; Weil, M.; Gold, L. High affinity ligands from in vitro selection: Complex targets. Proc. Natl. Acad. Sci. USA 1998, 95, 2902–2907. [Google Scholar] [CrossRef] [PubMed]
- Blank, M.; Weinschenk, T.; Priemer, M.; Schluesener, H. Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels: Selective targeting of endothelial regulatory protein pigpen. J. Biol. Chem. 2001, 276, 16464–16468. [Google Scholar] [CrossRef] [PubMed]
- Daniels, D.A.; Chen, H.; Hicke, B.J.; Swiderek, K.M.; Gold, L. A tenascin-c aptamer identified by tumor cell selex: Systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. USA 2003, 100, 15416–15421. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, M.; Yang, G.; Zhang, D.; Ding, H.; Wang, H.; Fan, M.; Shen, B.; Shao, N. Single-stranded DNA aptamers that bind differentiated but not parental cells: Subtractive systematic evolution of ligands by exponential enrichment. J. Biotechnol. 2003, 102, 15–22. [Google Scholar] [CrossRef]
- Mallikaratchy, P.; Stahelin, R.V.; Cao, Z.; Cho, W.; Tan, W. Selection of DNA ligands for protein kinase c-[small delta]. Chem. Commun. 2006, 3229–3231. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Shangguan, D.; Wang, K.; Shi, H.; Sefah, K.; Mallikratchy, P.; Chen, H.W.; Li, Y.; Tan, W. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007, 79, 4900–4907. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.W.; Medley, C.D.; Sefah, K.; Shangguan, D.; Tang, Z.; Meng, L.; Smith, J.E.; Tan, W. Molecular recognition of small-cell lung cancer cells using aptamers. ChemMedChem 2008, 3, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Sefah, K.; Tang, Z.W.; Shangguan, D.H.; Chen, H.; Lopez-Colon, D.; Li, Y.; Parekh, P.; Martin, J.; Meng, L.; Phillips, J.A.; et al. Molecular recognition of acute myeloid leukemia using aptamers. Leukemia 2009, 23, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Parekh, P.; Turner, P.; Moyer, R.W.; Tan, W. Generating aptamers for recognition of virus-infected cells. Clin. Chem. 2009, 55, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Swiderski, P.; Li, H.; Zhang, J.; Neff, C.P.; Akkina, R.; Rossi, J.J. Selection, characterization and application of new RNA HIV GP 120 aptamers for facile delivery of dicer substrate sirnas into hiv infected cells. Nucleic Acids Res. 2009, 37, 3094–3109. [Google Scholar] [CrossRef] [PubMed]
- Mi, J.; Liu, Y.; Rabbani, Z.N.; Yang, Z.; Urban, J.H.; Sullenger, B.A.; Clary, B.M. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 2010, 6, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Parekh, P.; Tang, Z.; Turner, P.C.; Moyer, R.W.; Tan, W. Aptamers recognizing glycosylated hemagglutinin expressed on the surface of vaccinia virus-infected cells. Anal. Chem. 2010, 82, 8642–8649. [Google Scholar] [CrossRef] [PubMed]
- Bayrac, A.T.; Sefah, K.; Parekh, P.; Bayrac, C.; Gulbakan, B.; Oktem, H.A.; Tan, W. In vitro selection of DNA aptamers to glioblastoma multiforme. ACS Chem. Neurosci. 2011, 2, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.-L.; Lu, C.-H.; Song, X.-Y.; Yang, H.-H.; Wang, X.-R. Bioresponsive controlled release using mesoporous silica nanoparticles capped with aptamer-based molecular gate. J. Am. Chem. Soc. 2011, 133, 1278–1281. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhao, Y.; He, D.; Wang, K.; Xu, F.; Tang, J. ATP-responsive controlled release system using aptamer-functionalized mesoporous silica nanoparticles. Langmuir 2012, 28, 12909–12915. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sefah, K.; Altman, M.B.; Chen, T.; You, M.; Zhao, Z.; Huang, C.Z.; Tan, W. Aptamer-conjugated nanorods for targeted photothermal therapy of prostate cancer stem cells. Chem.—Asian J. 2013, 8, 2417–2422. [Google Scholar] [CrossRef] [PubMed]
- Peng Ho, S.; Britton, D.H.O.; Stone, B.A.; Behrens, D.L.; Leffet, L.M.; Hobbs, F.W.; Miller, J.A.; Trainor, G.L. Potent antisense oligonucleotides to the human multidrug resistance-1 mRNA are rationally selected by mapping rna-accessible sites with oligonucleotide libraries. Nucleic Acids Res. 1996, 24, 1901–1907. [Google Scholar]
- Li, M.; Lin, N.; Huang, Z.; Du, L.; Altier, C.; Fang, H.; Wang, B. Selecting aptamers for a glycoprotein through the incorporation of the boronic acid moiety. J. Am. Chem. Soc. 2008, 130, 12636–12638. [Google Scholar] [CrossRef] [PubMed]
- Mehedi Masud, M.; Kuwahara, M.; Ozaki, H.; Sawai, H. Sialyllactose-binding modified DNA aptamer bearing additional functionality by selex. Bioorg. Med. Chem. 2004, 12, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Kusser, W. Chemically modified nucleic acid aptamers for in vitro selections: Evolving evolution. Rev. Mol. Biotechnol. 2000, 74, 27–38. [Google Scholar] [CrossRef]
- Keefe, A.D.; Cload, S.T. Selex with modified nucleotides. Curr. Opin. Chem. Biol. 2008, 12, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Pagratis, N.C.; Bell, C.; Chang, Y.-F.; Jennings, S.; Fitzwater, T.; Jellinek, D.; Dang, C. Potent 2[prime]-amino-, and 2[prime]-fluoro-2[prime]- deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat. Biotechnol. 1997, 15, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, M. Progress in chemically modified nucleic acid aptamers. In Chemical Biology of Nucleic Acids; Erdmann, V.A., Markiewicz, W.T., Barciszewski, J., Eds.; Springer Berlin Heidelberg: Berlin, Germany, 2014; pp. 243–270. [Google Scholar]
- Kluszmann, S.; Nolte, A.; Bald, R.; Erdmann, V.A.; Furste, J.P. Mirror-image RNA that binds d-adenosine. Nat. Biotechnol. 1996, 14, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Eulberg, D.; Klussmann, S. Spiegelmers: Biostable aptamers. ChemBioChem 2003, 4, 979–983. [Google Scholar] [CrossRef] [PubMed]
- Vater, A.; Klussmann, S. Turning mirror-image oligonucleotides into drugs: The evolution of spiegelmer® therapeutics. Drug Discov. Today 2015, 20, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Vallée-Bélisle, A.; Plaxco, K.W. Structure-switching biosensors: Inspired by nature. Curr. Opin. Struct. Biol. 2010, 20, 518–526. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Stormo, G.D. Combining selex with quantitative assays to rapidly obtain accurate models of protein–DNA interactions. Nucleic Acids Res. 2005, 33. [Google Scholar] [CrossRef] [PubMed]
- Tombelli, S.; Minunni, M.; Luzi, E.; Mascini, M. Aptamer-based biosensors for the detection of HIV-1 Tat protein. Bioelectrochemistry 2005, 67, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Eulberg, D.; Buchner, K.; Maasch, C.; Klussmann, S. Development of an automated in vitro selection protocol to obtain rna-based aptamers: Identification of a biostable substance p antagonist. Nucleic Acids Res. 2005, 33, e45. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S. Methods developed for selex. Anal. Bioanal. Chem. 2007, 387, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, H.; Ding, H.; Huang, Y.; Cao, X.; Yang, G.; Li, J.; Xie, Z.; Meng, Y.; Li, X.; et al. Identification of an aptamer targeting hnRNP a1 by tissue slide-based selex. J. Pathol. 2009, 218, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Gold, L.; Polisky, B.; Uhlenbeck, O.; Yarus, M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 1995, 64, 763–797. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [PubMed]
- Zhou, J.; Soontornworajit, B.; Snipes, M.P.; Wang, Y. Structural prediction and binding analysis of hybridized aptamers. J. Mol. Recognit. 2011, 24, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Tang, Z.; Mallikaratchy, P.; Xiao, Z.; Tan, W. Optimization and modifications of aptamers selected from live cancer cell lines. ChemBioChem 2007, 8, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Battig, M.; Wang, Y. Aptamer-based molecular recognition for biosensor development. Anal. Bioanal. Chem. 2010, 398, 2471–2480. [Google Scholar] [CrossRef] [PubMed]
- Rockey, W.M.; Hernandez, F.J.; Huang, S.-Y.; Cao, S.; Howell, C.A.; Thomas, G.S.; Liu, X.Y.; Lapteva, N.; Spencer, D.M.; McNamara, J.O.; et al. Rational truncation of an rna aptamer to prostate-specific membrane antigen using computational structural modeling. Nucleic Acid Ther. 2011, 21, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Hirota, M.; Waugh, S.M.; Murakami, I.; Suzuki, T.; Muraguchi, M.; Shibamori, M.; Ishikawa, Y.; Jarvis, T.C.; Carter, J.D.; et al. Chemically modified DNA aptamers bind interleukin-6 with high affinity and inhibit signaling by blocking its interaction with interleukin-6 receptor. J. Biol. Chem. 2014, 289, 8706–8719. [Google Scholar] [CrossRef] [PubMed]
- Drory Retwitzer, M.; Polishchuk, M.; Churkin, E.; Kifer, I.; Yakhini, Z.; Barash, D. RNAPattMatch: A web server for RNA sequence/structure motif detection based on pattern matching with flexible gaps. Nucleic Acids Res. 2015, 43. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Gan, L.; Jiang, L.; Zhang, X.; Yang, X.; Chen, M.; Lan, X. Neutralization of staphylococcal enterotoxin b by an aptamer antagonist. Antimicrob. Agents Chemother. 2015, 59, 2072–2077. [Google Scholar] [CrossRef] [PubMed]
- Green, L.S.; Jellinek, D.; Jenison, R.; Östman, A.; Heldin, C.-H.; Janjic, N. Inhibitory DNA ligands to platelet-derived growth factor b-chain. Biochemistry 1996, 35, 14413–14424. [Google Scholar] [CrossRef] [PubMed]
- Sayer, N.M.; Cubin, M.; Rhie, A.; Bullock, M.; Tahiri-Alaoui, A.; James, W. Structural determinants of conformationally selective, prion-binding aptamers. J. Biol. Chem. 2004, 279, 13102–13109. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Hatcher, K.L.; Bartz, J.C.; Chen, S.G.; Skinner, P.; Richt, J.; Liu, H.; Sreevatsan, S. Selection and characterization of DNA aptamers against prpsc. Exp. Biol. Med. 2011, 236, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Katilius, E.; Flores, C.; Woodbury, N.W. Exploring the sequence space of a DNA aptamer using microarrays. Nucleic Acids Res. 2007, 35, 7626–7635. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.; Soo Oh, S.; Nie, J.; Stewart, R.; Eisenstein, M.; Chambers, J.; Marth, J.D.; Walker, F.; Thomson, J.A.; Soh, H.T. Quantitative selection and parallel characterization of aptamers. Proc. Natl. Acad. Sci. USA 2013, 110, 18460–18465. [Google Scholar] [CrossRef] [PubMed]
- Shigdar, S.; Qiao, L.; Zhou, S.-F.; Xiang, D.; Wang, T.; Li, Y.; Lim, L.Y.; Kong, L.; Li, L.; Duan, W. RNA aptamers targeting cancer stem cell marker cd133. Cancer Lett. 2013, 330, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Ciardiello, F.; Tortora, G. EGFR antagonists in cancer treatment. New Engl. J. Med. 2008, 358, 1160–1174. [Google Scholar] [CrossRef] [PubMed]
- Mahlknecht, G.; Maron, R.; Mancini, M.; Schechter, B.; Sela, M.; Yarden, Y. Aptamer to ErbB-2/HER2 enhances degradation of the target and inhibits tumorigenic growth. Proc. Natl. Acad. Sci. USA 2013, 110, 8170–8175. [Google Scholar] [CrossRef] [PubMed]
- Chadd, H.E.; Chamow, S.M. Therapeutic antibody expression technology. Curr. Opin. Biotechnol. 2001, 12, 188–194. [Google Scholar] [CrossRef]
- Hudis, C.A. Trastuzumab—Mechanism of action and use in clinical practice. New Engl. J. Med. 2007, 357, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Mallikaratchy, P.; Tang, Z.; Kwame, S.; Meng, L.; Shangguan, D.; Tan, W. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in burkitt’s lymphoma cells. Mol. Cell. Proteomics 2007, 6, 2230–2238. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Cao, Z.; Meng, L.; Mallikaratchy, P.; Sefah, K.; Wang, H.; Li, Y.; Tan, W. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J. Proteome Res. 2008, 7, 2133–2139. [Google Scholar] [CrossRef] [PubMed]
- Van Simaeys, D.; Turek, D.; Champanhac, C.; Vaizer, J.; Sefah, K.; Zhen, J.; Sutphen, R.; Tan, W. Identification of cell membrane protein stress-induced phosphoprotein 1 as a potential ovarian cancer biomarker using aptamers selected by cell systematic evolution of ligands by exponential enrichment. Anal. Chem. 2014, 86, 4521–4527. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Kramer, F.R. Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol. 1996, 14, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Fang, X.; Schuster, S.M.; Tan, W. Molecular beacons: A novel approach to detect protein-DNA interactions. Angew. Chem. Int. Ed. 2000, 39, 1049–1052. [Google Scholar] [CrossRef]
- Hamaguchi, N.; Ellington, A.; Stanton, M. Aptamer beacons for the direct detection of proteins. Anal. Biochem. 2001, 294, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Fang, X.; Tan, W. Molecular aptamer beacons for real-time protein recognition. Biochem. Biophys. Res. Commun. 2002, 292, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Nutiu, R.; Li, Y. Structure-switching signaling aptamers. J. Am. Chem. Soc. 2003, 125, 4771–4778. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-M.; Wu, Z.; Liu, S.-J.; Chu, X. Structure-switching aptamer triggering hybridization chain reaction on the cell surface for activatable theranostics. Anal. Chem. 2015. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Cater, S.F.; Ellington, A.D. Quantum-dot aptamer beacons for the detection of proteins. ChemBioChem 2005, 6, 2163–2166. [Google Scholar] [CrossRef] [PubMed]
- Bagalkot, V.; Zhang, L.; Levy-Nissenbaum, E.; Jon, S.; Kantoff, P.W.; Langer, R.; Farokhzad, O.C. Quantum dot-aptamer conjugates for synchronous cancer imaging, therapy, and sensing of drug delivery based on bi-fluorescence resonance energy transfer. Nano Lett. 2007, 7, 3065–3070. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D. Quantum dot-nucleic acid/aptamer bioconjugate-based fluorimetric biosensors. Biochem. Soc. Trans. 2012, 40, 635–639. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, C.; Qian, M.; Zhao, X.S. Aptamer biosensor for protein detection using gold nanoparticles. Anal. Biochem. 2008, 373, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Song, K.-M.; Cho, M.; Jo, H.; Min, K.; Jeon, S.H.; Kim, T.; Han, M.S.; Ku, J.K.; Ban, C. Gold nanoparticle-based colorimetric detection of kanamycin using a DNA aptamer. Anal. Biochem. 2011, 415, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; He, Y.; Xing, X.-J.; Tan, D.-D.; Lin, Y.; Pang, D.-W.; Tang, H.-W. A gold nanoparticle-based label free colorimetric aptasensor for adenosine deaminase detection and inhibition assay. Analyst 2015, 140, 1572–1577. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-Q.; Wang, Y.-S.; Xue, J.-H.; He, Y.; Yang, H.-X.; Liang, J.; Shi, L.-F.; Xiao, X.-L. A gold nanoparticles-modified aptamer beacon for urinary adenosine detection based on structure-switching/fluorescence-“turning on” mechanism. J. Pharm. Biomed. Anal. 2012, 70, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Bai, W.; Niu, S.; Zhu, C.; Yang, S.; Chen, A. Highly sensitive colorimetric detection of 17[bgr]-estradiol using split DNA aptamers immobilized on unmodified gold nanoparticles. Sci. Rep. 2014, 4. [Google Scholar]
- Coons, A.H.; Creech, H.J.; Jones, R.N. Immunological properties of an antibody containing a fluorescent group. Exp. Biol. Med. 1941, 47, 200–202. [Google Scholar] [CrossRef]
- Engvall, E.; Perlmann, P. Enzyme-linked immunosorbent assay (elisa) quantitative assay of immunoglobulin g. Immunochemistry 1971, 8, 871–874. [Google Scholar] [CrossRef]
- Van Weemen, B.K.; Schuurs, A.H.W.M. Immunoassay using antigen-enzyme conjugates. FEBS Lett. 1971, 15, 232–236. [Google Scholar] [CrossRef]
- Ramos-Vara, J.A.; Miller, M.A. When tissue antigens and antibodies get along: Revisiting the technical aspects of immunohistochemistry—The red, brown, and blue technique. Vet. Pathol. Online 2014, 51, 42–87. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.K.; Griffiths, C.; Lea, S.M.; James, W. Structural characterization of an anti-gp120 rna aptamer that neutralizes r5 strains of HIV-1. RNA 2005, 11, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Z.; Bagalkot, V.; Vasilliou, C.C.; Gu, F.; Alexis, F.; Zhang, L.; Shaikh, M.; Yuet, K.; Cima, M.J.; Langer, R.; et al. Superparamagnetic iron oxide nanoparticle-aptamer bioconjugates for combined prostate cancer imaging and therapy. ChemMedChem 2008, 3, 1311–1315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jia, X.; Lv, X.-J.; Deng, Y.-L.; Xie, H.-Y. Fluorescent quantum dot-labeled aptamer bioprobes specifically targeting mouse liver cancer cells. Talanta 2010, 81, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Bernard, E.; Beking, M.; Rajamanickam, K.; Tsai, E.; DeRosa, M. Target binding improves relaxivity in aptamer–gadolinium conjugates. J. Biol. Inorg. Chem. 2012, 17, 1159–1175. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Dong, J.; Hu, X.; Gong, W.; Li, J.; Shen, J.; Tian, H.; Wang, J. A covalent approach for site-specific RNA labeling in mammalian cells. Angew. Chem. Int. Ed. 2015, 54, 4597–4602. [Google Scholar] [CrossRef] [PubMed]
- Charlton, J.; Sennello, J.; Smith, D. In vivo imaging of inflammation using an aptamer inhibitor of human neutrophil elastase. Chem. Biol. 1997, 4, 809–816. [Google Scholar] [CrossRef]
- Mi, J.; Zhang, X.; Giangrande, P.H.; McNamara Ii, J.O.; Nimjee, S.M.; Sarraf-Yazdi, S.; Sullenger, B.A.; Clary, B.M. Targeted inhibition of αvβ3 integrin with an rna aptamer impairs endothelial cell growth and survival. Biochem. Biophys. Res. Commun. 2005, 338, 956–963. [Google Scholar] [CrossRef] [PubMed]
- Farokhzad, O.C.; Cheng, J.; Teply, B.A.; Sherifi, I.; Jon, S.; Kantoff, P.W.; Richie, J.P.; Langer, R. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 6315–6320. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Yang, X.; Dobrucki, L.W.; Chaudhury, I.; Yin, Q.; Yao, C.; Lezmi, S.; Helferich, W.G.; Fan, T.M.; Cheng, J. Aptamer-functionalized, ultra-small, monodisperse silica nanoconjugates for targeted dual-modal imaging of lymph nodes with metastatic tumors. Angew. Chem. Int. Ed. 2012, 51, 12721–12726. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.-K.; Kim, B.; Choi, Y.; Ro, Y.; Cho, E.-J.; Lee, J.H.; Ryu, S.-H.; Suh, J.-S.; Haam, S.; Huh, Y.-M. Aptamer-conjugated magnetic nanoparticles enable efficient targeted detection of integrin αvβ3 via magnetic resonance imaging. J. Biomed. Mater. Res. Part A 2014, 102, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; He, X.; Wang, K.; Wu, X.; Ye, X.; Guo, Q.; Tan, W.; Qing, Z.; Yang, X.; Zhou, B. Activatable aptamer probe for contrast-enhanced in vivo cancer imaging based on cell membrane protein-triggered conformation alteration. Proc. Natl. Acad. Sci. USA 2011, 108, 3900–3905. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ji, X.; Zhang, Y.; Zhou, G.; Ke, X.; Wang, H.; Tinnefeld, P.; He, Z. One-pot synthesized aptamer-functionalized CdTe:Zn2+ quantum dots for tumor-targeted fluorescence imaging in vitro and in vivo. Anal. Chem. 2013, 85, 5843–5849. [Google Scholar] [CrossRef] [PubMed]
- Melancon, M.P.; Zhou, M.; Zhang, R.; Xiong, C.; Allen, P.; Wen, X.; Huang, Q.; Wallace, M.; Myers, J.N.; Stafford, R.J.; et al. Selective uptake and imaging of aptamer- and antibody-conjugated hollow nanospheres targeted to epidermal growth factor receptors overexpressed in head and neck cancer. ACS Nano 2014, 8, 4530–4538. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.A.; Shi, H.; He, X.; Wang, K.; Tang, J.; Chen, M.; Ye, X.; Xu, F.; Lei, Y. A versatile activatable fluorescence probing platform for cancer cells in vitro and in vivo based on self-assembled aptamer/carbon nanotube ensembles. Anal. Chem. 2014, 86, 9271–9277. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Yang, J.; Hwang, M.; Choi, J.; Kim, H.-O.; Jang, E.; Lee, J.; Ryu, S.-H.; Suh, J.-S.; Huh, Y.-M.; et al. Aptamer-modified magnetic nanoprobe for molecular mr imaging of VEGFR2 on angiogenic vasculature. Nanoscale Res. Lett. 2013, 8, 399. [Google Scholar] [CrossRef] [PubMed]
- Farokhzad, O.C.; Jon, S.; Khademhosseini, A.; Tran, T.-N.T.; LaVan, D.A.; Langer, R. Nanoparticle-aptamer bioconjugates: A new approach for targeting prostate cancer cells. Cancer Res. 2004, 64, 7668–7672. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Gao, X.; Su, L.; Xia, H.; Gu, G.; Pang, Z.; Jiang, X.; Yao, L.; Chen, J.; Chen, H. Aptamer-functionalized peg-plga nanoparticles for enhanced anti-glioma drug delivery. Biomaterials 2011, 32, 8010–8020. [Google Scholar] [CrossRef] [PubMed]
- Lü, J.-M.; Wang, X.; Marin-Muller, C.; Wang, H.; Lin, P.H.; Yao, Q.; Chen, C. Current advances in research and clinical applications of plga-based nanotechnology. Expert Rev. Mol. Diagn. 2009, 9, 325–341. [Google Scholar] [CrossRef] [PubMed]
- Knop, K.; Hoogenboom, R.; Fischer, D.; Schubert, U.S. Poly(ethylene glycol) in drug delivery: Pros and cons as well as potential alternatives. Angew. Chem. Int. Ed. 2010, 49, 6288–6308. [Google Scholar] [CrossRef] [PubMed]
- Danhier, F.; Ansorena, E.; Silva, J.M.; Coco, R.; Le Breton, A.; Préat, V. Plga-based nanoparticles: An overview of biomedical applications. J. Control. Release 2012, 161, 505–522. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, Y. Virosomes: Evolution of the liposome as a targeted drug delivery system. Adv. Drug Deliv. Rev. 2000, 43, 197–205. [Google Scholar] [CrossRef]
- Torchilin, V.P. Recent advances with liposomes as pharmaceutical carriers. Nat. Rev. Drug Discov. 2005, 4, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Platzgummer, M.; Kreismayr, G.; Quendler, H.; Stiegler, G.; Ferko, B.; Vecera, G.; Vorauer-Uhl, K.; Katinger, H. Gmp production of liposomes—A new industrial approach. J. Liposome Res. 2006, 16, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Tang, L.; Yang, X.; Hwang, K.; Wang, W.; Yin, Q.; Wong, N.Y.; Dobrucki, L.W.; Yasui, N.; Katzenellenbogen, J.A.; et al. Selective delivery of an anticancer drug with aptamer-functionalized liposomes to breast cancer cells in vitro and in vivo. J. Mater. Chem. B 2013, 1, 5288–5297. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Tong, R.; Mishra, A.; Xu, W.; Wong, G.C.L.; Cheng, J.; Lu, Y. Reversible cell-specific drug delivery with aptamer-functionalized liposomes. Angew. Chem. Int. Ed. 2009, 48, 6494–6498. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Song, C.; Jiang, C.; Shen, X.; Qiao, Q.; Hu, Y. Nucleolin targeting as1411 modified protein nanoparticle for antitumor drugs delivery. Mol. Pharm. 2013, 10, 3555–3563. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Guo, B.; Wu, H.; Shao, N.; Li, D.; Liu, J.; Dang, L.; Wang, C.; Li, H.; Li, S.; et al. Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference-based bone anabolic strategy. Nat. Med. 2015, 21, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Tomalia, D.A.; Baker, H.; Dewald, J.; Hall, M.; Kallos, G.; Martin, S.; Roeck, J.; Ryder, J.; Smith, P. A new class of polymers: Starburst-dendritic macromolecules. Polym. J. 1985, 17, 117–132. [Google Scholar] [CrossRef]
- Tomalia, D.A. Birth of a new macromolecular architecture: Dendrimers as quantized building blocks for nanoscale synthetic polymer chemistry. Prog. Polym. Sci. 2005, 30, 294–324. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Q.; Chang, H.; Cheng, Y. Surface-engineered dendrimers in gene delivery. Chem. Rev. 2015, 115, 5274–5300. [Google Scholar] [CrossRef] [PubMed]
- Svenson, S.; Tomalia, D.A. Dendrimers in biomedical applications—Reflections on the field. Adv. Drug Deliv. Rev. 2005, 57, 2106–2129. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Soontornworajit, B.; Martin, J.; Sullenger, B.A.; Gilboa, E.; Wang, Y. A hybrid DNA aptamer–dendrimer nanomaterial for targeted cell labeling. Macromol. Biosci. 2009, 9, 831–835. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Soontornworajit, B.; Wang, Y. A temperature-responsive antibody-like nanostructure. Biomacromolecules 2010, 11, 2087–2093. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.-H.; An, S.; Yu, M.K.; Kwon, H.-K.; Im, S.-H.; Jon, S. Targeted chemoimmunotherapy using drug-loaded aptamer–dendrimer bioconjugates. J. Control. Release 2011, 155, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Weihrauch, M.R.; Ansén, S.; Jurkiewicz, E.; Geisen, C.; Xia, Z.; Anderson, K.S.; Gracien, E.; Schmidt, M.; Wittig, B.; Diehl, V.; et al. Phase i/ii combined chemoimmunotherapy with carcinoembryonic antigen-derived HLA-a2-restricted cap-1 peptide and irinotecan, 5-fluorouracil, and leucovorin in patients with primary metastatic colorectal cancer. Clin. Cancer Res. 2005, 11, 5993–6001. [Google Scholar] [CrossRef] [PubMed]
- Najar, H.M.; Dutz, J.P. Topical cpg enhances the response of murine malignant melanoma to dacarbazine. J. Investig. Dermatol. 2008, 128, 2204–2210. [Google Scholar] [CrossRef] [PubMed]
- Millward, M.; Underhill, C.; Lobb, S.; McBurnie, J.; Meech, S.J.; Gomez-Navarro, J.; Marshall, M.A.; Huang, B.; Mather, C.B. Phase i study of tremelimumab (cp-675[thinsp]206) plus pf-3512676 (cpg 7909) in patients with melanoma or advanced solid tumours. Br. J. Cancer 2013, 108, 1998–2004. [Google Scholar] [CrossRef] [PubMed]
- Dam, D.H.M.; Lee, R.C.; Odom, T.W. Improved in vitro efficacy of gold nanoconstructs by increased loading of g-quadruplex aptamer. Nano Lett. 2014, 14, 2843–2848. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Salmasi, Z.; Hashemi, M.; Mosaffa, F.; Abnous, K.; Ramezani, M. Single-walled carbon nanotubes functionalized with aptamer and piperazine-polyethylenimine derivative for targeted sirna delivery into breast cancer cells. Int. J. Pharm. 2015, 485, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Taghdisi, S.M.; Lavaee, P.; Ramezani, M.; Abnous, K. Reversible targeting and controlled release delivery of daunorubicin to cancer cells by aptamer-wrapped carbon nanotubes. Eur. J. Pharm. Biopharm. 2011, 77, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Tseng, Y.-T.; Suo, G.; Chen, L.; Yu, J.; Chiu, W.-J.; Huang, C.-C.; Lin, C.-H. Photothermal therapeutic response of cancer cells to aptamer-gold nanoparticle-hybridized graphene oxide under nir illumination. ACS Appl. Mater. Interfaces 2015, 7, 5097–5106. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.K.; Kim, D.; Lee, I.-H.; So, J.-S.; Jeong, Y.Y.; Jon, S. Image-guided prostate cancer therapy using aptamer-functionalized thermally cross-linked superparamagnetic iron oxide nanoparticles. Small 2011, 7, 2241–2249. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-H.; Xu, X.-D.; Jia, H.-Z.; Lei, Q.; Luo, G.-F.; Cheng, S.-X.; Zhuo, R.-X.; Zhang, X.-Z. Therapeutic nanomedicine based on dual-intelligent functionalized gold nanoparticles for cancer imaging and therapy in vivo. Biomaterials 2013, 34, 8798–8807. [Google Scholar] [CrossRef] [PubMed]
- Gragoudas, E.S.; Adamis, A.P.; Cunningham, E.T.; Feinsod, M.; Guyer, D.R. Pegaptanib for neovascular age-related macular degeneration. New Engl. J. Med. 2004, 351, 2805–2816. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.L.; Colquitt, J.; Clegg, A.J.; Jones, J. Pegaptanib and ranibizumab for neovascular age-related macular degeneration: A systematic review. Br. J. Ophthalmol. 2007, 91, 1177–1182. [Google Scholar] [CrossRef] [PubMed]
- Novack, G.D. Pharmacotherapy for the treatment of choroidal neovascularization due to age-related macular degeneration. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J. Predicting the uncertain future of aptamer-based diagnostics and therapeutics. Molecules 2015, 20, 6866–6887. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, P.; Kurniawan, H.; Byrne, M.E.; Wower, J. Therapeutic RNA aptamers in clinical trials. Eur. J. Pharm. Sci. 2013, 48, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhu, X.; Lu, P.Y.; Rosato, R.R.; Tan, W.; Zu, Y. Oligonucleotide aptamers: New tools for targeted cancer therapy. Mol. Ther. Nucleic Acids 2014, 3, e182. [Google Scholar] [CrossRef] [PubMed]
- Butte, M.J.; Keir, M.E.; Phamduy, T.B.; Sharpe, A.H.; Freeman, G.J. Programmed death-1 ligand 1 interacts specifically with the b7–1 costimulatory molecule to inhibit t cell responses. Immunity 2007, 27, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. Pd-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef] [PubMed]
- Walunas, T.L.; Lenschow, D.J.; Bakker, C.Y.; Linsley, P.S.; Freeman, G.J.; Green, J.M.; Thompson, C.B.; Bluestone, J.A. Ctla-4 can function as a negative regulator of t cell activation. Immunity 1994, 1, 405–413. [Google Scholar] [CrossRef]
- Krummel, M.F.; Allison, J.P. Cd28 and ctla-4 have opposing effects on the response of t cells to stimulation. J. Exp. Med. 1995, 182, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Alegre, M.-L.; Frauwirth, K.A.; Thompson, C.B. T-cell regulation by cd28 and ctla-4. Nat. Rev. Immunol. 2001, 1, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, R.A.; Feldman, A.L.; Wada, D.A.; Yang, Z.-Z.; Comfere, N.I.; Dong, H.; Kwon, E.D.; Novak, A.J.; Markovic, S.N.; Pittelkow, M.R.; et al. B7-h1 (pd-l1, cd274) Suppresses Host Immunity in T-cell Lymphoproliferative Disorders. Blood 2009, 114, 2149–2158. [Google Scholar] [CrossRef] [PubMed]
- Steidl, C.; Shah, S.P.; Woolcock, B.W.; Rui, L.; Kawahara, M.; Farinha, P.; Johnson, N.A.; Zhao, Y.; Telenius, A.; Neriah, S.B.; et al. Mhc class ii transactivator ciita is a recurrent gene fusion partner in lymphoid cancers. Nature 2011, 471, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Rodig, S.; Juszczynski, P.; Ouyang, J.; Sinha, P.; O’Donnell, E.; Neuberg, D.; Shipp, M.A. Constitutive ap-1 activity and ebv infection induce pd-l1 in hodgkin lymphomas and posttransplant lymphoproliferative disorders: Implications for targeted therapy. Clin. Cancer Res. 2012, 18, 1611–1618. [Google Scholar] [CrossRef] [PubMed]
- O’Day, S.J.; Hamid, O.; Urba, W.J. Targeting cytotoxic t-lymphocyte antigen-4 (ctla-4). Cancer 2007, 110, 2614–2627. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Ghiringhelli, F. What is the role of cytotoxic t lymphocyte–associated antigen 4 blockade in patients with metastatic melanoma? Oncologist 2009, 14, 848–861. [Google Scholar] [CrossRef] [PubMed]
- Hamid, O.; Robert, C.; Daud, A.; Hodi, F.S.; Hwu, W.-J.; Kefford, R.; Wolchok, J.D.; Hersey, P.; Joseph, R.W.; Weber, J.S.; et al. Safety and tumor responses with lambrolizumab (anti–pd-1) in melanoma. New Engl. J. Med. 2013, 369, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Peng, C.; Sosman, J.A. Nivolumab in melanoma: Latest evidence and clinical potential. Ther. Adv. Med. Oncol. 2015, 7, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Bracarda, S.; Altavilla, A.; Hamzaj, A.; Sisani, M.; Marrocolo, F.; del Buono, S.; Danielli, R. Immunologic checkpoints blockade in renal cell, prostate, and urothelial malignancies. Semin. Oncol. 2015, 42, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Sundar, R.; Cho, B.-C.; Brahmer, J.R.; Soo, R.A. Nivolumab in nsclc: Latest evidence and clinical potential. Ther. Adv. Med. Oncol. 2015, 7, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Chames, P.; Van Regenmortel, M.; Weiss, E.; Baty, D. Therapeutic antibodies: Successes, limitations and hopes for the future. Br. J. Pharmacol. 2009, 157, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Harding, F.A.; Stickler, M.M.; Razo, J.; DuBridge, R.B. The immunogenicity of humanized and fully human antibodies: Residual immunogenicity resides in the CDR regions. mAbs 2010, 2, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.L.; Dhimolea, E.; Reichert, J.M. Development trends for human monoclonal antibody therapeutics. Nat. Rev. Drug Discov. 2010, 9, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Sherbenou, D.W.; Behrens, C.R.; Su, Y.; Wolf, J.L.; Martin Iii, T.G.; Liu, B. The development of potential antibody-based therapies for myeloma. Blood Rev. 2015, 29, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, T.C.I.; Gardener, M.J.; Williams, W.A. Discovery of functional antibodies targeting ion channels. J. Biomol. Screen. 2015, 20, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Santulli-Marotto, S.; Nair, S.K.; Rusconi, C.; Sullenger, B.; Gilboa, E. Multivalent rna aptamers that inhibit ctla-4 and enhance tumor immunity. Cancer Res. 2003, 63, 7483–7489. [Google Scholar] [PubMed]
- Prodeus, A.; Abdul-Wahid, A.; Fischer, N.W.; Huang, E.H.B.; Cydzik, M.; Gariepy, J. Targeting the pd-1/pd-l1 immune evasion axis with DNA aptamers as a novel therapeutic strategy for the treatment of disseminated cancers. Mol. Ther. Nucleic Acids 2015, 4, e237. [Google Scholar] [CrossRef] [PubMed]
- Vinay, D.S.; Kwon, B.S. Role of 4-1bb in immune responses. Semin. Immunol. 1998, 10, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Sica, G.; Chen, L. Modulation of the immune response through 4-1bb. In Cancer Gene Therapy; Habib, N., Ed.; Springer US: New York, NY, USA, 2002; Volume 465, pp. 355–362. [Google Scholar]
- Croft, M. Co-stimulatory members of the tnfr family: Keys to effective t-cell immunity? Nat. Rev. Immunol. 2003, 3, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Watts, T.H. TNF/TNFR family members in costimulation of t cell responses. Annu. Rev. Immunol. 2005, 23, 23–68. [Google Scholar] [CrossRef] [PubMed]
- McNamara, J.O.; Kolonias, D.; Pastor, F.; Mittler, R.S.; Chen, L.; Giangrande, P.H.; Sullenger, B.; Gilboa, E. Multivalent 4–1bb binding aptamers costimulate cd8(+) t cells and inhibit tumor growth in mice. J. Clin. Investig. 2008, 118, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Di Cera, E. Thrombin as procoagulant and anticoagulant. J. Thromb. Haemost. 2007, 5, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Mann, K.G. Biochemistry and physiology of blood coagulation. Thromb. Haemost. 1999, 82, 165–174. [Google Scholar] [PubMed]
- Di Cera, E. Thrombin. Mol. Aspects Med. 2008, 29, 203–254. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, B.I.; Quinlan, D.J.; Eikelboom, J.W. Novel oral factor Xa and thrombin inhibitors in the management of thromboembolism. Annu. Rev. Med. 2011, 62, 41–57. [Google Scholar] [CrossRef] [PubMed]
- Depta, J.P.; Bhatt, D.L. New approaches to inhibiting platelets and coagulation. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 373–397. [Google Scholar] [CrossRef] [PubMed]
- Stubbs, M.T.; Bode, W. The clot thickens: Clues provided by thrombin structure. Trends Biochem. Sci. 1995, 20, 23–28. [Google Scholar] [CrossRef]
- Davie, E.W.; Kulman, J.D. An overview of the structure and function of thrombin. Semin. Thromb. Hemost. 2006, 32, 003–015. [Google Scholar] [CrossRef] [PubMed]
- Macaya, R.F.; Schultze, P.; Smith, F.W.; Roe, J.A.; Feigon, J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. 1993, 90, 3745–3749. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.Y.; Krawczyk, S.H.; Bischofberger, N.; Swaminathan, S.; Bolton, P.H. The tertiary structure of a DNA aptamer which binds to and inhibits thrombin determines activity. Biochemistry 1993, 32, 11285–11292. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.Y.; McCurdy, S.; Shea, R.G.; Swaminathan, S.; Bolton, P.H. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry 1993, 32, 1899–1904. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Merlino, A.; Randazzo, A.; Novellino, E.; Mazzarella, L.; Sica, F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012, 40, 8119–8128. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, B.; Yan, J.; Yuan, Y.; An, L.; Guan, Y. Structure variations of tba g-quadruplex induced by 2′-o-methyl nucleotide in K+ and Ca2+ environments. Acta Biochim. Biophys. Sin. 2014, 46, 837–850. [Google Scholar] [CrossRef] [PubMed]
- Macaya, R.F.; Waldron, J.A.; Beutel, B.A.; Gao, H.; Joesten, M.E.; Yang, M.; Patel, R.; Bertelsen, A.H.; Cook, A.F. Structural and functional characterization of potent antithrombotic oligonucleotides possessing both quadruplex and duplex motifs. Biochemistry 1995, 34, 4478–4492. [Google Scholar] [CrossRef] [PubMed]
- Nallagatla, S.R.; Heuberger, B.; Haque, A.; Switzer, C. Combinatorial synthesis of thrombin-binding aptamers containing iso-guanine. J. Comb. Chem. 2009, 11, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Virgilio, A.; Petraccone, L.; Scuotto, M.; Vellecco, V.; Bucci, M.; Mayol, L.; Varra, M.; Esposito, V.; Galeone, A. 5-hydroxymethyl-2′-deoxyuridine residues in the thrombin binding aptamer: Investigating anticoagulant activity by making a tiny chemical modification. ChemBioChem 2014, 15, 2427–2434. [Google Scholar] [CrossRef] [PubMed]
- De Tito, S.; Morvan, F.; Meyer, A.; Vasseur, J.-J.; Cummaro, A.; Petraccone, L.; Pagano, B.; Novellino, E.; Randazzo, A.; Giancola, C.; et al. Fluorescence enhancement upon g-quadruplex folding: Synthesis, structure, and biophysical characterization of a dansyl/cyclodextrin-tagged thrombin binding aptamer. Bioconjugate Chem. 2013, 24, 1917–1927. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, A.; Hernandez, F.J.; Rasmussen, L.M.; Vester, B.; Wengel, J. Improved thrombin binding aptamer by incorporation of a single unlocked nucleic acid monomer. Nucleic Acids Res. 2011, 39, 1155–1164. [Google Scholar] [CrossRef] [PubMed]
- Griffin, L.; Tidmarsh, G.; Bock, L.; Toole, J.; Leung, L. In vivo Anticoagulant Properties of a Novel Nucleotide-Based Thrombin Inhibitor and Demonstration of Regional Anticoagulation in Extracorporeal Circuits. Blood 1993, 81, 3271–3276. [Google Scholar] [PubMed]
- Li, W.; Kaplan, A.; Grant, G.; Toole, J.; Leung, L. A Novel Nucleotide-Based Thrombin Inhibitor Inhibits Clot-Bound Thrombin and Reduces Arterial Platelet Thrombus Formation. Blood 1994, 83, 677–682. [Google Scholar] [PubMed]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2004, 56, 555–583. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Castanares, M.; Mukherjee, A.; Lupold, S.E. Nucleic acid aptamers: Clinical applications and promising new horizons. Curr. Med. Chem. 2011, 18, 4206–4214. [Google Scholar] [CrossRef] [PubMed]
- Vlieghe, P.; Lisowski, V.; Martinez, J.; Khrestchatisky, M. Synthetic therapeutic peptides: Science and market. Drug Discov. Today 2010, 15, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Deitchman, A.N.; Derendorf, H. Measuring drug distribution in the critically ill patient. Adv. Drug Deliv. Rev. 2014, 77, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.-N.; Zhang, C.-Q.; Wang, W.; Wang, P.C.; Zhou, J.-P.; Liang, X.-J. Ph-responsive mesoporous silica nanoparticles employed in controlled drug delivery systems for cancer treatment. Cancer Biol. Med. 2014, 11, 34–43. [Google Scholar] [PubMed]
- Belbekhouche, S.; Reinicke, S.; Espeel, P.; Du Prez, F.E.; Eloy, P.; Dupont-Gillain, C.; Jonas, A.M.; Demoustier-Champagne, S.; Glinel, K. Polythiolactone-based redox-responsive layers for the reversible release of functional molecules. ACS Appl. Mater. Interfaces 2014, 6, 22457–22466. [Google Scholar] [CrossRef] [PubMed]
- Agostini, A.; Mondragón, L.; Pascual, L.; Aznar, E.; Coll, C.; Martínez-Máñez, R.; Sancenón, F.; Soto, J.; Marcos, M.D.; Amorós, P.; et al. Design of enzyme-mediated controlled release systems based on silica mesoporous supports capped with ester-glycol groups. Langmuir 2012, 28, 14766–14776. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wang, T.; Perche, F.; Taigind, A.; Torchilin, V.P. Enhanced anticancer activity of nanopreparation containing an mmp2-sensitive peg-drug conjugate and cell-penetrating moiety. Proc. Natl. Acad. Sci. USA 2013, 110, 17047–17052. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Sumerlin, B.S. Glucose-sensitivity of boronic acid block copolymers at physiological pH. ACS Macro Lett. 2012, 1, 529–532. [Google Scholar] [CrossRef]
- Sinha, A.; Chakraborty, A.; Jana, N.R. Dextran-gated, multifunctional mesoporous nanoparticle for glucose-responsive and targeted drug delivery. ACS Appl. Mater. Interfaces 2014, 6, 22183–22191. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Cambre, J.N.; Sumerlin, B.S. Triply-responsive boronic acidblock copolymers: Solution self-assembly induced by changes in temperature, pH, or sugar concentration. Chem. Commun. 2009, 2106–2108. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhao, Y.; Zhao, J.; Han, M.; Zhang, A.; Wang, X. Codendrimer from polyamidoamine (pamam) and oligoethylene dendron as a thermosensitive drug carrier. Bioconjugate Chem. 2014, 25, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Du, J.-Z.; Long, H.-Y.; Yuan, Y.-Y.; Song, M.-M.; Chen, L.; Bi, H.; Wang, J. Micelle-to-vesicle morphological transition via light-induced rapid hydrophilic arm detachment from a star polymer. Chem. Commun. 2012, 48, 1257–1259. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Teh, C.; Sreejith, S.; Zhu, L.; Kwok, A.; Fang, W.; Ma, X.; Nguyen, K.T.; Korzh, V.; Zhao, Y. Functional mesoporous silica nanoparticles for photothermal-controlled drug delivery in vivo. Angew. Chem. Int. Ed. 2012, 51, 8373–8377. [Google Scholar] [CrossRef] [PubMed]
- Mura, S.; Nicolas, J.; Couvreur, P. Stimuli-responsive nanocarriers for drug delivery. Nat. Mater. 2013, 12, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, M.; Akimoto, J.; Okano, T. Polymeric micelles with stimuli-triggering systems for advanced cancer drug targeting. J. Drug Target. 2014, 22, 584–599. [Google Scholar] [CrossRef] [PubMed]
- Crucho, C.I.C. Stimuli-responsive polymeric nanoparticles for nanomedicine. ChemMedChem 2015, 10, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Fu, X.; Fu, W.; Li, Z. Biodegradable stimuli-responsive polypeptide materials prepared by ring opening polymerization. Chem. Soc. Rev. 2015, 44, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Altunbas, A.; Pochan, D. Peptide-based and polypeptide-based hydrogels for drug delivery and tissue engineering. In Peptide-Based Materials; Deming, T., Ed.; Springer Berlin Heidelberg: Berlin, Germany, 2012; Volume 310, pp. 135–167. [Google Scholar]
- Cui, H.; Zhuang, X.; He, C.; Wei, Y.; Chen, X. High performance and reversible ionic polypeptide hydrogel based on charge-driven assembly for biomedical applications. Acta Biomater. 2015, 11, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Jha, A.K.; Harrington, D.A.; Farach-Carson, M.C.; Jia, X. Hyaluronic acid-based hydrogels: From a natural polysaccharide to complex networks. Soft Matter 2012, 8, 3280–3294. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Bakaic, E.; Hoare, T.; Cranston, E.D. Injectable polysaccharide hydrogels reinforced with cellulose nanocrystals: Morphology, rheology, degradation, and cytotoxicity. Biomacromolecules 2013, 14, 4447–4455. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, T.; Liu, D.; Wang, Z. Fabricating three-dimensional hydrogel oligonucleotide microarrays to detect single nucleotide polymorphisms. Anal. Methods 2013, 5, 285–290. [Google Scholar] [CrossRef]
- Jung, Y.K.; Kim, J.; Mathies, R.A. Microfluidic linear hydrogel array for multiplexed single nucleotide polymorphism (SNP) detection. Anal. Chem. 2015, 87, 3165–3170. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-C.; Anseth, K. Peg hydrogels for the controlled release of biomolecules in regenerative medicine. Pharm. Res. 2009, 26, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Hoare, T.R.; Kohane, D.S. Hydrogels in drug delivery: Progress and challenges. Polymer 2008, 49, 1993–2007. [Google Scholar] [CrossRef]
- Soontornworajit, B.; Zhou, J.; Shaw, M.T.; Fan, T.-H.; Wang, Y. Hydrogel functionalization with DNA aptamers for sustained pdgf-bb release. Chem. Commun. 2010, 46, 1857–1859. [Google Scholar] [CrossRef] [PubMed]
- Soontornworajit, B.; Zhou, J.; Wang, Y. A hybrid particle-hydrogel composite for oligonucleotide-mediated pulsatile protein release. Soft Matter 2010, 6, 4255–4261. [Google Scholar] [CrossRef]
- Soontornworajit, B.; Zhou, J.; Snipes, M.P.; Battig, M.R.; Wang, Y. Affinity hydrogels for controlled protein release using nucleic acid aptamers and complementary oligonucleotides. Biomaterials 2011, 32, 6839–6849. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, N.; Li, S.; Battig, M.R.; Wang, Y. Programmable hydrogels for controlled cell catch and release using hybridized aptamers and complementary sequences. J. Am. Chem. Soc. 2012, 134, 15716–15719. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Zhang, Z.; Soontornworajit, B.; Zhou, J.; Wang, Y. Cell adhesion on an artificial extracellular matrix using aptamer-functionalized peg hydrogels. Biomaterials 2012, 33, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Kurzrock, R.; Giles, F.J. Precision oncology for patients with advanced cancer: The challenges of malignant snowflakes. Cell Cycle 2015, in press. [Google Scholar] [CrossRef] [PubMed]
- Desmond-Hellmann, S. Toward Precision Medicine: A New Social Contract? Sci. Transl. Med. 2012, 4. [Google Scholar] [CrossRef] [PubMed]
- Ashley, E.A. The precision medicine initiative: A new national effort. JAMA 2015, 313, 2119–2120. [Google Scholar] [CrossRef] [PubMed]
- Iacobuzio-Donahue, C.A.; Maitra, A.; Shen-Ong, G.L.; van Heek, T.; Ashfaq, R.; Meyer, R.; Walter, K.; Berg, K.; Hollingsworth, M.A.; Cameron, J.L.; et al. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am. J. Pathol. 2002, 160, 1239–1249. [Google Scholar] [CrossRef]
- Yin, P.; Xu, G. Metabolomics for tumor marker discovery and identification based on chromatography–mass spectrometry. Expert Rev. Mol. Diagn. 2013, 13, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-Y.; Cheng, C.-C.; Wang, J.-Y.; Wu, D.-C.; Hsieh, J.-S.; Lee, S.-C.; Wang, W.-M. Discovery of tumor markers for gastric cancer by proteomics. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-L.; Wu, Y.-C.; Su, L.-J.; Huang, Y.-J.; Charoenkwan, P.; Chen, W.-L.; Lee, H.-C.; Chu, W.C.-C.; Ho, S.-Y. Discovery of prognostic biomarkers for predicting lung cancer metastasis using microarray and survival data. BMC Bioinform. 2015, 16, 54. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J.; Sturgeon, C.M.; Sölétormos, G.; Barak, V.; Molina, R.; Hayes, D.F.; Diamandis, E.P.; Bossuyt, P.M.M. Validation of new cancer biomarkers: A position statement from the european group on tumor markers. Clin. Chem. 2015, 61, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Mitri, Z.; Constantine, T.; O’Regan, R. The HER2 receptor in breast cancer: Pathophysiology, clinical use, and new advances in therapy. Chemother. Res. Pract. 2012, 2012, 743193. [Google Scholar] [CrossRef] [PubMed]
- Gomella, L.G.; Liu, X.S.; Trabulsi, E.J.; Kelly, W.K.; Myers, R.; Showalter, T.; Dicker, A.; Wender, R. Screening for prostate cancer: The current evidence and guidelines controversy. Can. J. Urol. 2011, 18, 5875–5883. [Google Scholar] [PubMed]
- Bast, R.C.; Badgwell, D.; Lu, Z.; Marquez, R.; Rosen, D.; Liu, J.; Baggerly, K.A.; Atkinson, E.N.; Skates, S.; Zhang, Z.; et al. New tumor markers: CA125 and beyond. Int. J. Gynecol. Cancer 2005, 15, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.J.; Stinchcombe, T.E.; Der, C.J.; Socinski, M.A. Personalized medicine in non-small-cell lung cancer: Is K-Ras a useful marker in selecting patients for epidermal growth factor receptor-targeted therapy? J. Clin. Oncol. 2010, 28, 4769–4777. [Google Scholar] [CrossRef] [PubMed]
- Zlobec, I.; Bihl, M.P.; Schwarb, H.; Terracciano, L.; Lugli, A. Clinicopathological and protein characterization of braf- and k-ras-mutated colorectal cancer and implications for prognosis. Int. J. Cancer 2010, 127, 367–380. [Google Scholar] [PubMed]
- Krueger, K.E.; Srivastava, S. Posttranslational protein modifications: Current implications for cancer detection, prevention, and therapeutics. Mol. Cell. Proteomics 2006, 5, 1799–1810. [Google Scholar] [CrossRef] [PubMed]
- Karve, T.M.; Cheema, A.K. Small changes huge impact: The role of protein posttranslational modifications in cellular homeostasis and disease. J. Amino Acids 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Drolet, D.W.; Moon-McDermott, L.; Romig, T.S. An enzyme-linked oligonucleotide assay. Nat. Biotechnol. 1996, 14, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.-Y.; Lu, Y.-P.; Mihai Grumezescu, A.; Han Ho, F.; Kao, Y.-H.; Yang, Y.-S.; Yang, C.-H. Tumor marker detection by aptamer-functionalized graphene oxide. Curr. Org. Chem. 2013, 17, 132–136. [Google Scholar] [CrossRef]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Ostroff, R.M.; Bigbee, W.L.; Franklin, W.; Gold, L.; Mehan, M.; Miller, Y.E.; Pass, H.I.; Rom, W.N.; Siegfried, J.M.; Stewart, A.; et al. Unlocking biomarker discovery: Large scale application of aptamer proteomic technology for early detection of lung cancer. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Lv, Y.; Chen, T.; Zhang, X.; Wang, K.; Tan, W. Nucleic acid aptamers for living cell analysis. Annu. Rev. Anal. Chem. 2014, 7, 405–426. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Huang, P.-J.; Ding, J.; Liu, J. Aptamer-based biosensors for biomedical diagnostics. Analyst 2014, 139, 2627–2640. [Google Scholar] [CrossRef] [PubMed]
- Toh, S.Y.; Citartan, M.; Gopinath, S.C.B.; Tang, T.-H. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens. Bioelectron. 2015, 64, 392–403. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ku, T.-H.; Zhang, T.; Luo, H.; Yen, T.M.; Chen, P.-W.; Han, Y.; Lo, Y.-H. Nucleic Acid Aptamers: An Emerging Tool for Biotechnology and Biomedical Sensing. Sensors 2015, 15, 16281-16313. https://doi.org/10.3390/s150716281
Ku T-H, Zhang T, Luo H, Yen TM, Chen P-W, Han Y, Lo Y-H. Nucleic Acid Aptamers: An Emerging Tool for Biotechnology and Biomedical Sensing. Sensors. 2015; 15(7):16281-16313. https://doi.org/10.3390/s150716281
Chicago/Turabian StyleKu, Ti-Hsuan, Tiantian Zhang, Hua Luo, Tony M. Yen, Ping-Wei Chen, Yuanyuan Han, and Yu-Hwa Lo. 2015. "Nucleic Acid Aptamers: An Emerging Tool for Biotechnology and Biomedical Sensing" Sensors 15, no. 7: 16281-16313. https://doi.org/10.3390/s150716281
APA StyleKu, T.-H., Zhang, T., Luo, H., Yen, T. M., Chen, P.-W., Han, Y., & Lo, Y.-H. (2015). Nucleic Acid Aptamers: An Emerging Tool for Biotechnology and Biomedical Sensing. Sensors, 15(7), 16281-16313. https://doi.org/10.3390/s150716281





