Population Genetic Structure: Where, What, and Why?
Abstract
1. Introduction
2. Where?
2.1. Species Concepts
2.2. Population
2.2.1. Population’s Role in Evolution and the Major Ecological Relationships Between Populations
2.2.2. Population Size
2.2.3. Population Types
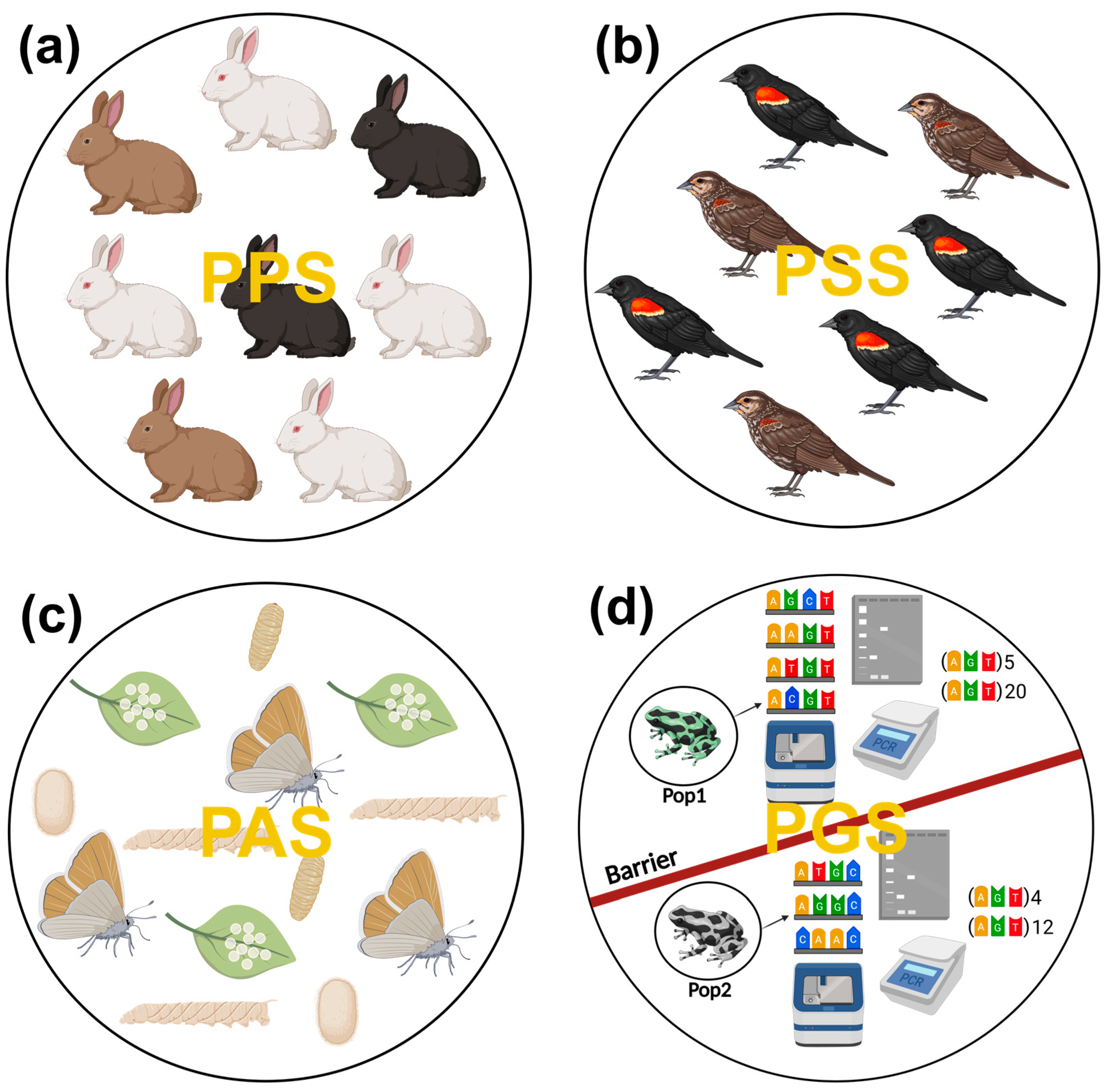
2.3. Population Structures
2.3.1. Problematics of Population Morphological Structure
2.3.2. Population Ethological Structure
2.3.3. Additional Population Structures?
2.3.4. The Unique Role of Population Genetic Structure
3. What?
3.1. DNA, Its Variations, and Genetic Diversity
3.1.1. DNA Diversity and Terminology
3.1.2. Variations Within DNA Sequences
3.1.3. Modern Approaches Used to Reveal and Research Variations Within DNA Sequences
3.1.4. Problematics of the Interpretation of the Meaning of DNA Variations
3.1.5. From DNA Sequence Variations to Genomes
3.1.6. From Genomes to Pangenomes
3.1.7. From the DNA Level to the Chromosome Level
3.1.8. Crucial Open Questions Regarding Genetic Diversity
3.2. Intraspecific Genetic Diversity Within the Context of Microevolution
3.2.1. Genetic Diversity, Adaptation, Survival, and Evolutionary Potential
3.2.2. Intraspecific Genetic Diversity and Evolutionary Forces
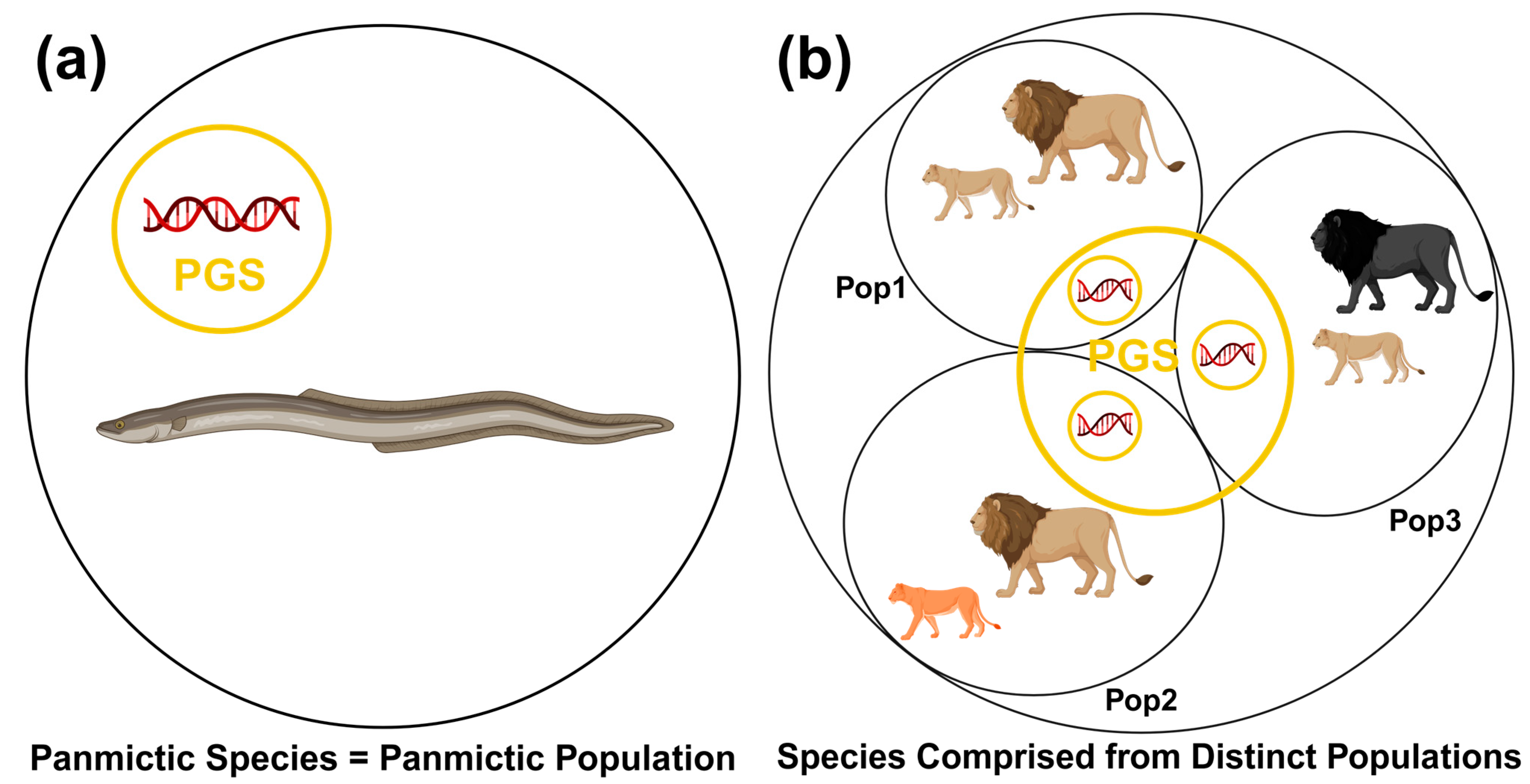
3.3. From Intraspecific Genetic Diversity to Population Genetic Structure
3.3.1. Toward a Greater Understanding of the Population Genetic Structure of a Species
3.3.2. Problems with Non-Standardized Definitions and a Possible Solution
3.3.3. Population Genetic Structure as a Dimensional Genetic Mosaic
3.3.4. Population Genetic Structure and Ethology
4. Why?
4.1. Why Is Population Genetic Structure Meaningful in the Context of Microevolution?
4.2. Why Is Population Genetic Structure Meaningful in the Context of Speciation?
4.3. Why Is Population Genetic Structure Meaningful in the Context of Conservation Genetics?
4.4. Why Is Population Genetic Structure Meaningful in the Context of Sustainable Use of Resources?
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adams, C.I.; Knapp, M.; Gemmell, N.J.; Jeunen, G.J.; Bunce, M.; Lamare, M.D.; Taylor, H.R. Beyond biodiversity: Can environmental DNA (eDNA) cut it as a population genetics tool? Genes 2019, 10, 192. [Google Scholar] [CrossRef]
- Torres-Romero, E.J.; Fisher, J.T.; Nijman, V.; He, F.; Eppley, T.M. Accelerated human-induced extinction crisis in the world’s freshwater mammals. Glob. Environ. Change Adv. 2024, 2, 100006. [Google Scholar] [CrossRef]
- Sadler, D.E.; Watts, P.C.; Uusi-Heikkilä, S. The riddle of how fisheries influence genetic diversity. Fishes 2023, 8, 510. [Google Scholar] [CrossRef]
- Gippoliti, S.; Robovský, J.; Angelici, F.M. Taxonomy and translocations of African mammals: A plea for a cautionary approach. Conservation 2021, 1, 121–136. [Google Scholar] [CrossRef]
- Díaz, S.; Malhi, Y. Biodiversity: Concepts, patterns, trends, and perspectives. Annu. Rev. Environ. Resourc. 2022, 47, 31–63. [Google Scholar] [CrossRef]
- Minamoto, T. Environmental DNA analysis for macro-organisms: Species distribution and more. DNA Res. 2022, 29, dsac018. [Google Scholar] [CrossRef]
- Laikre, L.; Palm, S.; Ryman, N. Genetic population structure of fishes: Implications for coastal zone management. AMBIO A J. Hum. Environ. 2005, 34, 111–119. [Google Scholar] [CrossRef]
- Weinbaum, K.Z.; Brashares, J.S.; Golden, C.D.; Getz, W.M. Searching for sustainability: Are assessments of wildlife harvests behind the times? Ecol. Lett. 2013, 16, 99–111. [Google Scholar] [CrossRef]
- Goymann, W.; Küblbeck, M. The second warning to humanity—Why ethology matters? Ethology 2020, 126, 1–9. [Google Scholar] [CrossRef]
- Gissi, E.; Schiebinger, L.; Santoleri, R.; Micheli, F. Sex analysis in marine biological systems: Insights and opportunities. Front. Ecol. Environ. 2023, 21, 324–332. [Google Scholar] [CrossRef]
- Li, L.; Shao, H.; Mikheev, P.B.; Zhang, Z.; Jin, H.; Lu, W. Age, Growth, Sex Composition, and Diet of the Burbot, Lota lota, the Only Freshwater Species of the Family Lotidae in the Amur (Heilongjiang) River, Northeast China. Fishes 2024, 9, 428. [Google Scholar] [CrossRef]
- Ngwava, J.M.; Xiao, F.; Malonza, P.K.; Bwong, B.A.; Shi, H.T. Reproductive ecology of the critically endangered pancake tortoise (Malacochersus tornieri) in the wild. Wildl. Biol. 2024, 2024, e01181. [Google Scholar] [CrossRef]
- Shaw, R.E.; Farquharson, K.A.; Bruford, M.W.; Coates, D.J.; Elliott, C.P.; Mergeay, J.; Ottewell, K.M.; Segelbacher, G.; Hoban, S.; Hvilsom, C.; et al. Global meta-analysis shows action is needed to halt genetic diversity loss. Nature 2025, 638, 704–710. [Google Scholar] [CrossRef]
- Coates, D.J.; Byrne, M.; Moritz, C. Genetic Diversity and Conservation Units: Dealing With the Species-Population Continuum in the Age of Genomics. Front. Ecol. Evol. 2018, 6, 165. [Google Scholar] [CrossRef]
- Krofel, M.; Hatlauf, J.; Bogdanowicz, W.; Campbell, L.A.D.; Godinho, R.; Jhala, Y.V.; Kitchener, A.C.; Koepfli, K.P.; Moehlman, P.; Senn, H.; et al. Towards resolving taxonomic uncertainties in wolf, dog and jackal lineages of Africa, Eurasia and Australasia. J. Zool. 2022, 316, 155–168. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Dudash, M.R.; Eldridge, M.D.B.; Fenster, C.B.; Lacy, R.C.; Ryder, O.A. Implications of different species concepts for conserving biodiversity. Biol. Conserv. 2012, 153, 25–31. [Google Scholar] [CrossRef]
- Aldhebiani, A.Y. Species concept and speciation. Saudi J. Biol. Sci. 2018, 25, 437–440. [Google Scholar] [CrossRef]
- Dupré, J. Speciation and species: A process perspective. Evol. J. Linn. Soc. 2024, 3, kzae020. [Google Scholar] [CrossRef]
- Gao, L.; Rieseberg, L.H. While neither universally applicable nor practical operationally, the biological species concept continues to offer a compelling framework for studying species and speciation. Natl. Sci. Rev. 2020, 7, 1398–1400. [Google Scholar] [CrossRef]
- Feng, Y.; Ryan, U.M.; Xiao, L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018, 34, 997–1011. [Google Scholar] [CrossRef]
- Hoban, S.; Paz-Vinas, I.; Shaw, R.E.; Castillo-Reina, L.; da Silva, J.M.; DeWoody, J.A.; Ekblom, R.; Fedorca, A.; Forester, B.R.; Funk, W.C.; et al. DNA-based studies and genetic diversity indicator assessments are complementary approaches to conserving evolutionary potential. Conserv. Genet. 2024, 25, 1147–1153. [Google Scholar] [CrossRef]
- Hirschfeld, M.; Dudgeon, C.; Sheaves, M.; Barnett, A. Barriers in a sea of elasmobranchs: From fishing for populations to testing hypotheses in population genetics. Glob. Ecol. Biogeogr. 2021, 30, 2147–2163. [Google Scholar] [CrossRef]
- Schleimer, A.; Frantz, A.C. Landscape influence on pollinator population genetic connectivity. Insect. Conserv. Divers. 2025, 18, 285–302. [Google Scholar] [CrossRef]
- Selkoe, K.A.; Watson, J.R.; White, C.; Horin, T.B.; Iacchei, M.; Mitarai, S.; Siegel, D.A.; Steven, D.; Toonen, R.J. Taking the chaos out of genetic patchiness: Seascape genetics reveals ecological and oceanographic drivers of genetic patterns in three temperate reef species. Mol. Ecol. 2010, 19, 3708–3726. [Google Scholar] [CrossRef] [PubMed]
- Groeneveld, M.J.; Klein, J.D.; Bennett, R.H.; Abdulla, A.S.; Bond, M.E.; Ebert, D.A.; Fernando, S.M.; Gledhill, K.S.; Jaquemet, S.; Kiszka, J.J.; et al. Population genetic structure of bottlenose and whitespotted wedgefishes from the Southwest Indian Ocean using a dual marker approach. Endanger. Species Res. 2024, 53, 409–427. [Google Scholar] [CrossRef]
- Sul, J.H.; Martin, L.S.; Eskin, E. Population structure in genetic studies: Confounding factors and mixed models. PLoS Genet. 2018, 14, e1007309. [Google Scholar] [CrossRef]
- Hohenlohe, P.A.; Funk, W.C.; Rajora, O.P. Population genomics for wildlife conservation and management. Mol. Ecol. 2021, 30, 62–82. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, M.; Gao, Z.; Ma, H.; Chong, Y.; Hong, J.; Wu, J.; Wu, D.; Xi, D.; Deng, W. Advances in Whole Genome Sequencing: Methods, Tools, and Applications in Population Genomics. Int. J. Mol. Sci. 2025, 26, 372. [Google Scholar] [CrossRef]
- Frank, S.C.; Ordiz, A.; Gosselin, J.; Hertel, A.; Kindberg, J.; Leclerc, M.; Pelletier, F.; Steyaert, S.M.J.G.; Støen, O.G.; Van de Walle, J.; et al. Indirect effects of bear hunting: A review from Scandinavia. Ursus 2017, 28, 150–164. [Google Scholar] [CrossRef]
- Jones, O.R.; Wang, J. A comparison of four methods for detecting weak genetic structure from marker data. Ecol. Evol. 2012, 2, 1048–1055. [Google Scholar] [CrossRef]
- Theodorakis, C.W.; Meyer, M.A.; Okay, O.; Yakan, S.D.; Schramm, K.W. Contamination acts as a genotype-dependent barrier to gene flow, causing genetic erosion and fine-grained population subdivision in Mussels from the Strait of Istanbul. Ecotoxicology 2024, 33, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Clark, Z.S.; Butcher, P.A.; Weeks, A.R.; Huveneers, C.; Toomey, M.; Holland, O.J.; Fish, J.J.; Sherman, C.D.H.; Blower, D.C.; Miller, A.D. Genomic assessment of Australian White Sharks (Carcharodon carcharias) challenges previous evidence of population subdivision. Divers. Distrib. 2025, 31, e13946. [Google Scholar] [CrossRef]
- Lončar, V.; Kralj, J.; Stronen, A.V.; Grgurević, M.; Pavlinec, Ž.; Jurinović, L.; Svetličić, I.; Buzan, E.; Piro, S.; Herrmann, C.; et al. High genetic diversity yet weak population genetic structure in European common terns. Sci. Rep. 2024, 14, 29173. [Google Scholar] [CrossRef]
- Viana, J.; Evanno, G.; Audet, C.; Teletchea, F. Fine-Scale Genetic Structure of Small Fish Populations in Islands: The Case of Brook Charr Salvelinus fontinalis (Mitchill, 1814) in Saint-Pierre and Miquelon (France). Evol. Appl. 2025, 18, e70041. [Google Scholar] [CrossRef]
- Silva, P.; López-Bao, J.V.; Llaneza, L.; Álvares, F.; Lopes, S.; Cortés, Y.; García, E.; Palavios, V.; Rio-Maior, H.; Ferrand, N.; et al. Cryptic population structure reveals low dispersal in Iberian wolves. Sci. Rep. 2018, 8, 14108. [Google Scholar] [CrossRef] [PubMed]
- García-Castro, K.L.; Márquez, E.J. Temporal-scale assessment of population genetics of the freshwater fish Prochilodus magdalenae in an area impacted by construction of a dam. Hydrobiologia 2024, 851, 1513–1531. [Google Scholar] [CrossRef]
- Noguerales, V.; Cordero, P.J.; Ortego, J. Hierarchical genetic structure shaped by topography in a narrow-endemic montane grasshopper. BMC Evol. Biol. 2016, 16, 96. [Google Scholar] [CrossRef]
- Ragauskas, A.; Butkauskas, D.; Bianchini, M.L. Distinct matrilines in the panmictic population of the European eel Anguilla anguilla? Aquat. Living Resour. 2017, 30, 21. [Google Scholar] [CrossRef]
- Sanz, N.; Araguas, R.M.; Giampiccolo, M.; Duchi, A. Native population structure beyond hatchery introgression in the endemic Sicilian trout. Diversity 2023, 15, 274. [Google Scholar] [CrossRef]
- Nester, T.L.; López-Solano, A.; Perea, S.; Doadrio, I. Genomic population structure and diversity of the Endangered Aphanius iberus: Strategies for killifish conservation. Conserv. Genet. 2024, 26, 263–277. [Google Scholar] [CrossRef]
- Verkuil, Y.I.; Piersma, T.; Jukema, J.; Hooijmeijer, J.C.; Zwarts, L.; Baker, A.J. The interplay between habitat availability and population differentiation: A case study on genetic and morphological structure in an inland wader (Charadriiformes). Biol. J. Linn. 2012, 106, 641–656. [Google Scholar] [CrossRef]
- Mueller, S.A.; Merondun, J.; Lečić, S.; Wolf, J.B. Epigenetic variation in light of population genetic practice. Nat. Commun. 2025, 16, 1028. [Google Scholar] [CrossRef] [PubMed]
- Miguel-Tomé, S.; Llinás, R.R. Broadening the definition of a nervous system to better understand the evolution of plants and animals. Plant Signal. Behav. 2021, 16, 1927562. [Google Scholar] [CrossRef] [PubMed]
- Mallatt, J.; Blatt, M.R.; Draguhn, A.; Robinson, D.G.; Taiz, L. Debunking a myth: Plant consciousness. Protoplasma 2021, 258, 459–476. [Google Scholar] [CrossRef]
- Ralls, K.; Ballou, J.D.; Dudash, M.R.; Eldridge, M.D.; Fenster, C.B.; Lacy, R.C.; Sunnucks, P.; Frankham, R. Call for a paradigm shift in the genetic management of fragmented populations. Conserv. Lett. 2018, 11, e12412. [Google Scholar] [CrossRef]
- Hauser, L.; Carvalho, G.R. Paradigm shifts in marine fisheries genetics: Ugly hypotheses slain by beautiful facts. Fish Fish. 2008, 9, 333–362. [Google Scholar] [CrossRef]
- Mayr, E. Systematics and the Origin of Species; The Columbia Classics in Evolution Series; Columbia University Press: New York, NY, USA, 1942. [Google Scholar]
- Mayr, E. Animal Species and Evolution; Harvard University Press: Cambridge, MA, USA, 1963. [Google Scholar]
- Templeton, A.R. The meaning of species and speciation: A genetic perspective. In Speciation and Its Consequences; Otte, D., Endler, J.A., Eds.; Sinauer Associates: Sunderland, MA, USA, 1989; pp. 3–27. [Google Scholar]
- Paterson, H.E.H. The recognition concept of species. In Species and Speciation; Vrba, E.S., Ed.; Cambridge University Press: Cambridge, UK, 1985; pp. 21–29. [Google Scholar]
- Van Valen, L. Ecological species, multispecies, and oaks. Taxon 1976, 25, 233–239. [Google Scholar] [CrossRef]
- Baverstock, P.R.; Moritz, C. Molecular genetics and evolution. In Evolutionary Biology; Hecht, M.K., Ross, C.W., Eds.; Springer: Boston, MA, USA, 1996; pp. 89–122. [Google Scholar]
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Simpson, G.G. Principles of Animal Taxonomy; Columbia University Press: New York, NY, USA, 1961. [Google Scholar]
- Sokal, R.R.; Sneath, P.H.A. Principles of Numerical Taxonomy; W.H. Freeman: San Francisco, CA, USA, 1963. [Google Scholar]
- Mallet, J. A species definition for the modern synthesis. Trends Ecol. Evol. 1995, 10, 294–299. [Google Scholar] [CrossRef]
- Sneath, P.H.A. Polythetic classification: Conceptions and methods. In Classification and Communication; Warren, J.W., Ed.; Academic Press: London, UK, 1976; pp. 129–150. [Google Scholar]
- Wheeler, Q.D.; Meier, R. Species Concepts and Phylogenetic Theory; Columbia University Press: New York, NY, USA, 2000. [Google Scholar]
- Blaxter, M.L.; Mann, J.; Chapman, T.; Thomas, F.; Whitton, C.; Floyd, R.; Abebe, E. Defining operational taxonomic units using DNA barcode data. Philos. Trans. R. Soc. B 2005, 360, 1935–1943. [Google Scholar] [CrossRef]
- Huse, S.M.; Dethlefsen, L.; Huber, J.A.; Mark Welch, D.; Relman, D.A.; Sogin, M.L. Exploring microbial diversity and taxonomy using high-throughput DNA sequencing. Curr. Opin. Microbiol. 2010, 13, 591–597. [Google Scholar]
- Nelson, G.; Platnick, N.I. Systematics and Biogeography: Cladistics and Vicariance; Columbia University Press: New York, NY, USA, 1981. [Google Scholar]
- Wiley, E.O. The evolutionary species concept reconsidered. Syst. Zool. 1978, 27, 17–26. [Google Scholar] [CrossRef]
- Cracraft, J. Species concepts and speciation analysis. In Current Ornithology; Johnston, R.F., Ed.; Springer: New York, NY, USA, 1983; pp. 159–187. [Google Scholar] [CrossRef]
- De Queiroz, K. Species concepts and species delimitation. Syst. Biol. 2007, 56, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Hague, E.L.; McWhinnie, L.H. Narwhal, beluga and bowhead whale responses to marine vessel traffic: A systematic map. Ocean. Coast. Manag. 2024, 255, 107251. [Google Scholar] [CrossRef]
- Berryman, A.A. Population: A central concept for ecology? Oikos 2002, 97, 439–442. [Google Scholar] [CrossRef]
- Waples, R.S.; Gaggiotti, O. What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol. Ecol. 2006, 15, 1419–1439. [Google Scholar] [CrossRef]
- Wells, J.V.; Richmond, M.E. Populations, metapopulations, and species populations: What are they and who should care? Wildl. Soc. Bull. 1995, 23, 458–462. Available online: https://www.jstor.org/stable/3782955 (accessed on 4 August 2025).
- Pfeifer, M.A.; Henle, K.; Settele, J. Populations with explicit borders in space and time: Concept, terminology, and estimation of characteristic parameters. Acta Biotheor. 2007, 55, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Carbone, I.; Kohn, L. Inferring process from pattern in fungal population genetics. Appl. Mycol. Biotechnol. 2004, 4, 30. [Google Scholar]
- Bergek, S.; Sundblad, G.; Björklund, M. Population differentiation in perch Perca fluviatilis: Environmental effects on gene flow? J. Fish Biol. 2010, 76, 1159–1172. [Google Scholar] [CrossRef]
- Ellegren, H.; Galtier, N. Determinants of genetic diversity. Nat. Rev. Genet. 2016, 17, 422–433. [Google Scholar] [CrossRef]
- Moya, A.; Holmes, E.C.; González-Candelas, F. The population genetics and evolutionary epidemiology of RNA viruses. Nat. Rev. Microbiol. 2004, 2, 279–288. [Google Scholar] [CrossRef]
- Leone, A.; Arnaud-Haond, S.; Babbucci, M.; Bargelloni, L.; Coscia, I.; Damalas, D.; Delord, C.; Franch, R.; Garibaldi, F.; Macias, D.; et al. Population genomics of the blue shark, Prionace glauca, reveals different populations in the Mediterranean Sea and the Northeast Atlantic. Evol. Appl. 2024, 17, e70005. [Google Scholar] [CrossRef]
- Balkenhol, N.; Waits, L.P.; Dezzani, R.J. Statistical approaches in landscape genetics: An evaluation of methods for linking landscape and genetic data. Ecography 2009, 32, 818–830. [Google Scholar] [CrossRef]
- Fraser, D.J.; Lippé, C.; Bernatchez, L. Consequences of unequal population size, asymmetric gene flow and sex-biased dispersal on population structure in brook charr (Salvelinus fontinalis). Mol. Ecol. 2004, 13, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Crookes, S.; Shaw, P.W. Isolation by distance and non-identical patterns of gene flow within two river populations of the freshwater fish Rutilus rutilus (L. 1758). Conserv. Genet. 2016, 17, 861–874. [Google Scholar] [CrossRef]
- Parsons, M.A.; Newsome, T.M.; Young, J.K. The consequences of predators without prey. Front. Ecol. Environ. 2022, 20, 31–39. [Google Scholar] [CrossRef]
- Faulks, L.; Svanbäck, R.; Eklöv, P.; Östman, Ö. Genetic and morphological divergence along the littoral–pelagic axis in two common and sympatric fishes: Perch, Perca fluviatilis (Percidae) and roach, Rutilus rutilus (Cyprinidae). Biol. J. Linn. Soc. 2015, 114, 929–940. [Google Scholar] [CrossRef]
- Frainer, A.; McKie, B.G.; Amundsen, P.-A.; Knudsen, R.; Lafferty, K.D. Parasitism and the Biodiversity–Functioning Relationship. Trends Ecol. Evol. 2018, 33, 260–268. [Google Scholar] [CrossRef]
- Pérez-Sorribes, L.; Villar-Yanez, P.; Smeds, L.; Mergeay, J. Comparing genetic Ne reconstructions over time with long-time wolf monitoring data in two populations. Evol. Appl. 2024, 17, e70022. [Google Scholar] [CrossRef]
- Besson, M.; Feeney, W.E.; Gache, C.; O’Brien, D.A.; Berthe, C.; Cowan, Z.L.; Brooker, R.M.; Laudet, V.; Lecchini, D. Anemone bleaching impacts the larval recruitment success of an anemone-associated fish. Coral Reefs 2023, 42, 195–203. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Katz, M.E.; Knoll, A.H.; Quigg, A.; Raven, J.A.; Schofield, O.; Taylor, F.J.R. The evolution of modern eukaryotic phytoplankton. Science 2004, 305, 354–360. [Google Scholar] [CrossRef]
- Connell, J.H.; Slatyer, R.O. Mechanisms of succession in natural communities and their role in community stability and organization. Am. Nat. 1977, 111, 1119–1144. [Google Scholar] [CrossRef]
- Fedorca, A.; Mergeay, J.; Akinyele, A.O.; Albayrak, T.; Biebach, I.; Brambilla, A.; Burger, P.A.; Buzan, E.; Curik, I.; Gargiulo, R.; et al. Dealing with the complexity of effective population size in conservation practice. Evol. Appl. 2024, 17, e70031. [Google Scholar] [CrossRef]
- Çiftci, Y.; Okumuş, İ. Fish population genetics and applications of molecular markers to fisheries and aquaculture: I-Basic principles of fish population genetics. Turk. J. Fish. Aquat. Sci. 2002, 2, 145–155. [Google Scholar]
- Godoy, E.M.D.; Camargo, T.R.; Toniato, M.; Proença, D.C.; Obando, J.M.C.; Roubach, R.; Gallardo, P.; Bueno, G.W. Framework for Estimating Environmental Carrying Capacity in Diverse Climatic Conditions and Fish Farming Production in Neotropical Reservoirs. Sustainability 2025, 17, 5282. [Google Scholar] [CrossRef]
- Strier, K.B.; Anthony, R.I. Abrupt Demographic Change Affects Projected Population Size: Implications for an Endangered Species in a Protected Area. Ecology 2025, 106, e4487. [Google Scholar] [CrossRef] [PubMed]
- Sibly, R.M.; Barker, D.; Denham, M.C.; Hone, J.; Pagel, M. On the regulation of populations of mammals, birds, fish, and insects. Science 2005, 309, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.I.; Smart, J.J.; Rigby, C.L.; White, W.T.; Chin, A.; Baje, L.; Simpfendorfer, C.A. Intraspecific demography of the silky shark (Carcharhinus falciformis): Implications for fisheries management. ICES J. Mar. Sci. 2020, 77, 241–255. [Google Scholar] [CrossRef]
- Delord, C.; Arnaud-Haond, S.; Leone, A.; Rolland, J.; Nikolic, N. Unraveling the Complexity of the Ne/Nc Ratio for Conservation of Large and Widespread Pelagic Fish Species: Current Status and Challenges. Evol. Appl. 2024, 17, e70020. [Google Scholar] [CrossRef]
- Casillas, S.; Barbadilla, A. Molecular population genetics. Genetics 2017, 205, 1003–1035. [Google Scholar] [CrossRef] [PubMed]
- Gargiulo, R.; Budde, K.B.; Heuertz, M. Mind the lag: Understanding genetic extinction debt for conservation. Trends Ecol. Evol. 2024, 40, 228–237. [Google Scholar] [CrossRef]
- Forester, B.R.; Beever, E.A.; Darst, C.; Szymanski, J.; Funk, W.C. Linking evolutionary potential to extinction risk: Applications and future directions. Front. Ecol. Environ. 2022, 20, 507–515. [Google Scholar] [CrossRef]
- David, A.A. Reconsidering panmixia: The erosion of phylogeographic barriers due to anthropogenic transport and the incorporation of biophysical models as a solution. Front. Mar. Sci. 2018, 5, 280. [Google Scholar] [CrossRef]
- Avise, J.C. Catadromous eels continue to be slippery research subjects. Mol. Ecol. 2011, 20, 1317–1319. [Google Scholar] [CrossRef]
- Tambovtseva, V.G.; Samusenok, V.P.; Yur’ev, A.L.; Korostelev, N.B.; Khlystov, V.S.; Matveev, A.N.; Alekseyev, S.S. Contrasting levels of sympatric divergence within lacustrine Arctic charr Salvelinus alpinus forms flock: High differentiation between size forms, low differentiation between seasonal races. Hydrobiologia 2024, 852, 3523–3540. [Google Scholar] [CrossRef]
- Laikre, L.; Olsson, F.; Jansson, E.; Hössjer, O.; Ryman, N. Metapopulation effective size and conservation genetic goals for the Fennoscandian wolf (Canis lupus) population. Heredity 2016, 117, 279–289. [Google Scholar] [CrossRef]
- Chen, Y.S.; Su, Y.C.; Pan, W. Effect of spatial constraints on Hardy-Weinberg equilibrium. Sci. Rep. 2016, 6, 19297. [Google Scholar] [CrossRef]
- Andrello, M.; Bevacqua, D.; Maes, G.E.; De Leo, G.A. An integrated genetic-demographic model to unravel the origin of genetic structure in European eel (Anguilla anguilla L.). Evol. Appl. 2011, 4, 517–533. [Google Scholar] [CrossRef]
- Baltazar-Soares, M.; Eizaguirre, C. Does asymmetric gene flow among matrilines maintain the evolutionary potential of the European eel? Ecol. Evol. 2016, 6, 5305–5320. [Google Scholar] [CrossRef] [PubMed]
- Foote, M. The evolution of morphological diversity. Annu. Rev. Ecol. Syst. 1997, 28, 129–152. [Google Scholar] [CrossRef]
- Tellería, J.L.; De La Hera, I.; Perez-Tris, J. Morphological variation as a tool for monitoring bird populations: A review. Ardeola 2013, 60, 191–224. [Google Scholar] [CrossRef]
- Peichel, C.L.; Marques, D.A. The genetic and molecular architecture of phenotypic diversity in sticklebacks. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20150486. [Google Scholar] [CrossRef]
- Svanbäck, R.; Eklöv, P. Genetic variation and phenotypic plasticity: Causes of morphological variation in Eurasian perch. Evol. Ecol. Res. 2006, 8, 37–49. [Google Scholar]
- Starodubaitė, M.; Sruoga, A.; Butkauskas, D.; Potapov, M.; Evsikov, V. Phenetic studies of the species Talpa europaea L. in Lithuania. Vet. Med. Zoot. 2010, 53, 61–70. [Google Scholar]
- Zakharov, V.M.; Shadrina, E.G.; Trofimov, I.E. Fluctuating asymmetry, developmental noise and developmental stability: Future prospects for the population developmental biology approach. Symmetry 2020, 12, 1376. [Google Scholar] [CrossRef]
- Gül, S.; Dursun, C.; Tabak, C.; Büyüksofuoğlu, S.; Özdemir, N. Age Structure, Body Size, and Sexual Dimorphism in a High-Altitude Population of Pelophylax ridibundus (Pallas, 1771). Animals 2024, 14, 3230. [Google Scholar] [CrossRef]
- Gümüş, B.; Gümüş, E.; Balaban, M.O. Image analysis to determine length-weight and area-weight relationships, and color differences in scaled carp and mirror carp grown in fiberglass and concrete tanks. Turk. J. Fish. Aquat. Sci. 2022, 23, TRJFAS21260. [Google Scholar] [CrossRef]
- Bartels, T. Variations in the morphology, distribution, and arrangement of feathers in domesticated birds. J. Exp. Zool. B Mol. Dev. Evol. 2003, 298, 91–108. [Google Scholar] [CrossRef]
- Sandberg, O.M.; Schultz, A.; Guðmundsdóttir, R.; Skúlason, S. ‘Species’ Is Not the (Only) Unit of Biodiversity: A Process-Philosophical Perspective on Conservation Concepts. Mar. Ecol. 2024, 46, e12857. [Google Scholar] [CrossRef]
- Rawat, S.; Benakappa, S.; Kumar, J.; Naik, K.; Pandey, G.; Pema, C.W. Identification of fish stocks based on truss morphometric: A review. J. Fish. Life Sci. 2017, 2, 9–14. [Google Scholar]
- Reimchen, T.E.; Cox, K.D. Differential Temperature Preferences of Vertebral Phenotypes in Threespine Stickleback (Gasterosteus aculeatus). Can. J. Zool. 2016, 94, 1–5. [Google Scholar] [CrossRef]
- Ewnetu, S.; Girma, Z. Population estimate and social organization of the Southern Gelada (Theropithecus gelada obscurus, Heuglin 1863) in Abune Yosef Zigit community conservation area, North Wollo, Ethiopia. Isr. J. Ecol. Evol. 2024, 1, 1–9. [Google Scholar] [CrossRef]
- Pouey-Santalou, V.; Moreno-Godoy, P.; Ducret, M.; Dramet, M.; Toulot, A.; Eriksson, I.; De Weerdt, J. The French connection: Multiple records of Strait of Gibraltar killer whales (Orcinus orca) (2003–2023) in the Bay of Biscay, France. Mar. Mammal Sci. 2025, 41, e13207. [Google Scholar] [CrossRef]
- Zamudio, K.R.; Bell, R.C.; Mason, N.A. Phenotypes in phylogeography: Species’ traits, environmental variation, and vertebrate diversification. Proc. Natl. Acad. Sci. USA 2016, 113, 8041–8048. [Google Scholar] [CrossRef]
- Johnston, J.D. Physiological responses to food intake throughout the day. Nutr. Res. Rev. 2014, 27, 107–118. [Google Scholar] [CrossRef]
- Vetere, A.; Capasso, M.; Di Ianni, F. Sex determination in reptiles: A review. Animals 2025, 15, 168. [Google Scholar] [CrossRef] [PubMed]
- Kalous, L.; Kuříková, P.; Kohout, J.; Rylková, K.; Petrtýl, M.; Čech, M. Differences in spatial communities of European perch (Perca fluviatilis Linnaeus, 1758) fry in a canyon-shaped reservoir are not attributable to genetics. J. Appl. Ichthyol. 2017, 33, 306–313. [Google Scholar] [CrossRef]
- Biswas, C.; Chakraborty, S.; Munilkumar, S.; Gireesh-Babu, P.; Sawant, P.B.; Chadha, N.K.; Krishna, G.; Dasgupta, S. Effect of high temperature during larval and juvenile stages on masculinization of common carp (Cyprinus carpio, L.). Aquaculture 2021, 530, 735803. [Google Scholar] [CrossRef]
- Bull, J.J. Sex determination in reptiles. Q. Rev. Biol. 1980, 55, 3–21. [Google Scholar] [CrossRef]
- Hildebrandt, J.P. Ecology Meets Physiology: Phenotypic Plasticity and the Ability of Animals to Adjust to Changing Environmental Conditions. Physiologia 2023, 3, 366–380. [Google Scholar] [CrossRef]
- Edeline, E. Adaptive phenotypic plasticity of eel diadromy. Mar. Ecol. Prog. Ser. 2007, 341, 229–232. [Google Scholar] [CrossRef]
- Andrews, R.M. Patterns of growth in reptiles. In Biology of the Reptilia; Gans, C., Pough, F.H., Eds.; Academic Press: New York, NY, USA, 1982; Volume 13, pp. 273–320. [Google Scholar]
- Choi, W.I.; Lee, D.H.; Jung, J.B.; Park, Y.S. Oak decline syndrome in Korean forests: History, biology, and prospects for Korean oak wilt. Forests 2022, 13, 964. [Google Scholar] [CrossRef]
- Athayde, A.; Cantor, M.; Cardoso, J.; Francisco, A.; Santos, F.P.D.; Crespo, H.; de Morais, M.V.; da Cruz Albaladejo, M.; Neto, H.G.; Siciliano, S. Movements and social behavior of killer whales (Orcinus orca) off the Brazilian coast. Front. Mar. Sci. 2023, 10, 1206796. [Google Scholar] [CrossRef]
- Cope, O.L.; Burkle, L.A.; Croy, J.R.; Mooney, K.A.; Yang, L.H.; Wetzel, W.C. The role of timing in intraspecific trait ecology. Trends Ecol. Evol. 2022, 37, 997–1005. [Google Scholar] [CrossRef]
- Nakazawa, T.; Matsumoto, T.K.; Katsuhara, K.R. When is lethal deceptive pollination maintained? A population dynamics approach. Ann. Bot. 2024, 134, 665–682. [Google Scholar] [CrossRef]
- Anderson, J.R. Comparative thanatology. Curr. Biol. 2016, 26, R553–R556. [Google Scholar] [CrossRef] [PubMed]
- Gromov, V.S. Relationship between the social structure and potential reproductive success in muroid rodents (rodentia, myomorpha). Biol. Bull. 2021, 48, 1740–1746. [Google Scholar] [CrossRef]
- Alcázar-Treviño, J.; Johnson, M.; Arranz, P.; Warren, V.E.; Pérez-González, C.J.; Marques, T.; Madsen, P.T.; Aguilar de Soto, N. Deep-diving beaked whales dive together but forage apart. Proc. R. Soc. B 2021, 288, 20201905. [Google Scholar] [CrossRef] [PubMed]
- Azizeh, T.R.; Sprogis, K.R.; Soley, R.; Nielsen, M.L.; Uhart, M.M.; Sironi, M.; Marón, C.F.; Behder, L.; Madsen, P.T.; Christiansen, F. Acute and chronic behavioral effects of kelp gull micropredation on southern right whale mother-calf pairs off Península Valdés, Argentina. Mar. Ecol. Prog. Ser. 2021, 668, 133–148. [Google Scholar] [CrossRef]
- Mrusczok, M.T.; Zwamborn, E.; von Schmalensee, M.; Ramallo, S.R.; Stefansson, R.A. First Account of Apparent Alloparental Care of a Long-Finned Pilot Whale Calf (Globicephala melas) by a Female Killer Whale (Orcinus orca). Can. J. Zool. 2023, 101, 288–293. [Google Scholar] [CrossRef]
- Le Bihan, D. From Black Holes Entropy to Consciousness: The Dimensions of the Brain Connectome. Entropy 2023, 25, 1645. [Google Scholar] [CrossRef]
- Williams, H. Mechanisms of cultural evolution in the songs of wild bird populations. Front. Psychol. 2021, 12, 643343. [Google Scholar] [CrossRef]
- Zhao, Y.; Seluanov, A.; Gorbunova, V. Revelations about aging and disease from unconventional vertebrate model organisms. Annu. Rev. Genet. 2021, 55, 135–159. [Google Scholar] [CrossRef]
- Giles, D.A.; Teman, S.J.; Ellis, S.; Ford, J.K.; Shields, M.W.; Hanson, M.B.; Emmons, C.K.; Cottrell, P.E.; Baird, R.W.; Osborne, R.W.; et al. Harassment and killing of porpoises (“phocoenacide”) by fish-eating southern resident killer whales (Orcinus orca). Mar. Mamm. Sci. 2024, 40, e13073. [Google Scholar] [CrossRef]
- Reznikova, Z. Ants’ Personality and Its Dependence on Foraging Styles: Research Perspectives. Front. Ecol. Evol. 2021, 9, 661066. [Google Scholar] [CrossRef]
- Whiten, A. Cultural Evolution in Animals. Annu. Rev. Ecol. Evol. Syst. 2019, 50, 27–48. [Google Scholar] [CrossRef]
- Crespo-Picazo, J.L.; Rubio-Guerri, C.; Jiménez, M.A.; Aznar, F.J.; Marco-Cabedo, V.; Melero, M.; Sánchez-Vizcaíno, J.M.; Gozalbes, P.; García-Párraga, D. Bottlenose dolphins (Tursiops truncatus) aggressive behavior towards other cetacean species in the western Mediterranean. Sci. Rep. 2021, 11, 21582. [Google Scholar] [CrossRef] [PubMed]
- Wright, T.F.; Derryberry, E.P. Defining the multidimensional phenotype: New opportunities to integrate the behavioral ecology and behavioral neuroscience of vocal learning. Neurosci. Biobehav. Rev. 2021, 125, 328–338. [Google Scholar] [CrossRef]
- Gonçalves, A.; Biro, D. Comparative thanatology, an integrative approach: Exploring sensory/cognitive aspects of death recognition in vertebrates and invertebrates. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170263. [Google Scholar] [CrossRef]
- Kashimada, K.; Koopman, P. Sry: The master switch in mammalian sex determination. Development 2010, 137, 3921–3930. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Jin, J.; Li, X.; Gedman, G.; Pelan, S.; Rhie, A.; Jiang, C.; Fedrigo, O.; Howe, K.; Phillippy, A.M.; et al. Chromosome-level echidna genome illuminates evolution of multiple sex chromosome system in monotremes. GigaScience 2025, 14, giae112. [Google Scholar] [CrossRef]
- Wagner, G.P.; Zhang, J. The pleiotropic structure of the genotype–phenotype map: The evolvability of complex organisms. Nat. Rev. Genet. 2011, 12, 204–213. [Google Scholar] [CrossRef]
- Bog, M.; Appenroth, K.J.; Sree, K.S. Duckweed (Lemnaceae): Its molecular taxonomy. Front. Sustain. Food Syst. 2019, 3, 117. [Google Scholar] [CrossRef]
- Moore, L.J.; Petrovan, S.O.; Bates, A.J.; Hicks, H.L.; Baker, P.J.; Perkins, S.E.; Yarnell, R.W. Demographic Effects of Road Mortality on Mammalian Populations: A Systematic Review. Biol. Rev. 2023, 98, 1033–1050. [Google Scholar] [CrossRef]
- Crews, D.; Bergeron, J.M.; Bull, J.J.; Flores, D.; Tousignant, A.; Skipper, J.K.; Wibbels, T. Temperature-dependent sex determination in reptiles: Proximate mechanisms, ultimate outcomes, and practical applications. Dev. Genet. 1994, 15, 297–312. [Google Scholar] [CrossRef]
- Janzen, F.J.; Paukstis, G.L. Environmental sex determination in reptiles: Ecology, evolution, and experimental design. Q. Rev. Biol. 1991, 66, 149–179. [Google Scholar] [CrossRef] [PubMed]
- Laikre, L.; Schwartz, M.K.; Waples, R.S.; Ryman, N. Compromising genetic diversity in the wild: Unmonitored large-scale release of plants and animals. Trends Ecol. Evol. 2010, 25, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.; Hoban, S.; Hunter, M.; Paz-Vinas, I.; Garroway, C.J. Genetic diversity and IUCN Red List status. Conserv. Biol. 2023, 37, e14064. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.J.; Cordes, J.F. DNA marker technologies and their applications in aquaculture genetics. Aquaculture 2004, 238, 1–37. [Google Scholar] [CrossRef]
- Brower, A.V.; DeSalle, R. DNA barcodes in taxonomic descriptions. In DNA Barcoding: Methods and Protocols; Kress, W.J., Erickson, D.L., Eds.; Springer: New York, NY, USA, 2024; pp. 105–115. [Google Scholar]
- Wu, J. Similarity-weighted entropy for quantifying genetic diversity in viral quasispecies. Virus Evol. 2025, 11, veaf029. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V. A virocentric perspective on the evolution of life. Curr. Opin. Virol. 2013, 3, 546–557. [Google Scholar] [CrossRef] [PubMed]
- Shorter, J.; Lindquist, S. Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 2005, 6, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Brockhurst, M.A.; Harrison, E.; Hall, J.P.; Richards, T.; McNally, A.; MacLean, C. The ecology and evolution of pangenomes. Curr. Biol. 2019, 29, R1094–R1103. [Google Scholar] [CrossRef] [PubMed]
- Tiddy, I.C.; Schneider, K.; Elmer, K.R. Environmental correlates of adaptive diversification in postglacial freshwater fishes. J. Fish Biol. 2024, 104, 517–535. [Google Scholar] [CrossRef]
- Seifertová, M.; Bryja, J.; Vyskočilová, M.; Martínková, N.; Šimková, A. Multiple Pleistocene refugia and post-glacial colonization in the European chub (Squalius cephalus) revealed by combined use of nuclear and mitochondrial markers. J. Biogeogr. 2012, 39, 1024–1040. [Google Scholar] [CrossRef]
- Hadly, E.A.; Ramakrishnan, U.; Chan, Y.L.; van Tuinen, M.; O’Keefe, K.; Spaeth, P.A.; Conroy, C.J. Genetic response to climatic change: Insights from ancient DNA and phylochronology. PLoS Biol. 2004, 2, e290. [Google Scholar] [CrossRef]
- Sawicki, J.; Krawczyk, K.; Paukszto, Ł.; Maździarz, M.; Kurzyński, M.; Szablińska-Piernik, J.; Szczecińska, M. Nanopore Sequencing Technology as an Emerging Tool for Diversity Studies of Plant Organellar Genomes. Diversity 2024, 16, 173. [Google Scholar] [CrossRef]
- Lichman, V.; Ozerov, M.; López, M.E.; Noreikiene, K.; Kahar, S.; Pukk, L.; Burimski, O.; Gross, R.; Vasemägi, A. Whole-genome analysis reveals phylogenetic and demographic history of Eurasian perch. J. Fish Biol. 2024, 105, 871–885. [Google Scholar] [CrossRef]
- Puertas, M.J.; González-Sánchez, M. Insertions of mitochondrial DNA into the nucleus—Effects and role in cell evolution. Genome 2020, 63, 365–374. [Google Scholar] [CrossRef]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Malathi, V.G.; Renuka Devi, P. ssDNA Viruses: Key Players in Global Virome. VirusDisease 2019, 30, 3–12. [Google Scholar] [CrossRef]
- Brown, T.A.; Clayton, D.A. Release of replication termination controls mitochondrial DNA copy number after depletion with 2′,3′-dideoxycytidine. Nucleic Acids Res. 2002, 30, 2004–2010. [Google Scholar] [CrossRef]
- Nicholls, T.J.; Minczuk, M. In D-loop: 40 years of mitochondrial 7S DNA. Exp. Gerontol. 2014, 56, 175–181. [Google Scholar] [CrossRef]
- Di Antonio, M.; Ponjavic, A.; Radzevičius, A.; Ranasinghe, R.T.; Catalano, M.; Zhang, X.; Shen, J.; Needham, L.M.; Lee, S.F.; Klenerman, D.; et al. Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat. Chem. 2020, 12, 832–837. [Google Scholar] [CrossRef]
- Wang, G.; Vasquez, K.M. Dynamic alternative DNA structures in biology and disease. Nat. Rev. Genet. 2023, 24, 211–234. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Lynch, M.; Crease, T.J. The analysis of population survey data on DNA sequence variation. Mol. Biol. Evol. 1990, 7, 377–394. [Google Scholar] [CrossRef]
- Lepais, O.; Chancerel, E.; Boury, C.; Salin, F.; Manicki, A.; Taillebois, L.; Dutech, C.; Aissi, A.; Bacles, C.F.E.; Daverat, F.; et al. Fast sequence-based microsatellite genotyping development workflow. PeerJ 2020, 8, e9085. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Wang, D.; Jia, Q. Evaluation of genetic diversity and construction of DNA fingerprinting in Polygonatum Mill. based on InDel molecular markers. Genet. Resour. Crop Evol. 2025, 72, 7453–7464. [Google Scholar] [CrossRef]
- Giuffra, E.; Tuggle, C.K.; Consortium, F. Functional annotation of animal genomes (FAANG): Current achievements and roadmap. Annu. Rev. Anim. Biosci. 2019, 7, 65–88. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef]
- Rakoczy-Trojanowska, M.; Bolibok, H. Characteristics and a comparison of three classes of microsatellite-based markers and their application in plants. Cell. Mol. Biol. Lett. 2004, 9, 221–238. [Google Scholar]
- Sharma, P.C.; Hüttel, B.; Winter, P.; Kahl, G.; Gardner, R.C.; Weising, K. The potential of microsatellites for hybridization-and polymerase chain reaction-based DNA fingerprinting of chickpea (Cicer arietinum L.) and related species. Electrophoresis 1995, 16, 1755–1761. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Li, Y.; Liu, X.; Ma, Y.; Jiang, L. A review of the pangenome: How it affects our understanding of genomic variation, selection and breeding in domestic animals? J. Anim. Sci. Biotechnol. 2023, 14, 73. [Google Scholar] [CrossRef]
- Zhao, Z.; Shi, T. Haplotype-Resolved Assembly in Polyploid Plants: Methods, Challenges, and Implications for Evolutionary and Breeding Research. Genes 2025, 16, 636. [Google Scholar] [CrossRef]
- Liao, I.J.Y.; Lu, T.M.; Chen, M.E.; Luo, Y.J. Spiralian genomics and the evolution of animal genome architecture. Brief. Funct. Genom. 2023, 22, 498–508. [Google Scholar] [CrossRef]
- Iannucci, A.; Makunin, A.I.; Lisachov, A.P.; Ciofi, C.; Stanyon, R.; Svartman, M.; Trifonov, V.A. Bridging the gap between vertebrate cytogenetics and genomics with single-chromosome sequencing (ChromSeq). Genes 2021, 12, 124. [Google Scholar] [CrossRef]
- Ma, X.-K.; Yu, Y.; Huang, T.; Zhang, D.; Tian, C.; Tang, W.; Luo, M.; Du, P.; Yu, G.; Yang, L. Bioinformatics Software Development: Principles and Future Directions. Innov. Life 2024, 2, 100083. [Google Scholar] [CrossRef]
- Durán-Fuentes, J.A.; Maronna, M.M.; Palacios-Gimenez, O.M.; Castillo, E.R.; Ryan, J.F.; Daly, M.; Stampar, S.N. Repeatome diversity in sea anemone genomics (Cnidaria: Actiniaria) based on the Actiniaria-REPlib library. BMC Genom. 2025, 26, 473. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H. Genome sequencing and population genomics in non-model organisms. Trends Ecol. Evol. 2014, 29, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Van Doornik, D.M.; Moran, P.; Rondeau, E.B.; Nichols, K.M.; Narum, S.R.; Campbell, M.R.; Clemento, A.J.; Hargrove, J.S.; Hess, J.E.; Horn, R.L.; et al. A new, standardized international Pacific Rim baseline for genetic stock identification (GSI) of Chinook Salmon. N. Am. J. Fish. Manag. 2024, 44, 857–869. [Google Scholar] [CrossRef]
- Jarvis, E.D.; Mirarab, S.; Aberer, A.J.; Li, B.; Houde, P.; Li, C.; Ho, S.Y.; Faircloth, B.C.; Nabholz, B.; Howard, J.T.; et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 2014, 346, 1320–1331. [Google Scholar] [CrossRef]
- Cardeñosa, D.; Babcock, E.A.; Shea, S.K.; Zhang, H.; Feldheim, K.A.; Gale, S.W.; Milis, D.; Chapman, D.D. Small sharks, big problems: DNA analysis of small fins reveals trade regulation gaps and burgeoning trade in juvenile sharks. Sci. Adv. 2024, 10, eadq6214. [Google Scholar] [CrossRef]
- Courtaillac, K.L.; Landschoff, J.; von der Heyden, S. Of Biogeography, Fishes and Kelp: Environmental DNA Metabarcoding the Great African Seaforest. Divers. Distrib. 2025, 31, e70045. [Google Scholar] [CrossRef]
- Rajora, O.; Rahman, M. Microsatellite DNA and RAPD fingerprinting, identification and genetic relationships of hybrid poplar (Populus x canadensis) cultivars. Theor. Appl. Genet. 2003, 106, 470–477. [Google Scholar] [CrossRef]
- Odah, M.A.A. Unlocking the genetic code: Exploring the potential of DNA barcoding for biodiversity assessment. AIMS Mol. Sci. 2023, 10, 263–294. [Google Scholar] [CrossRef]
- Wibowo, A.; Kurniawan, K.; Prakoso, V.A.; Ginanjar, R.; Rochman, F.; Zamroni, M.; Atminarso, D.; Sumarto, B.K.A.; Chadijah, A.; Irawan, D.; et al. Characterizing spatial patterns among freshwater fishes and shrimps of the Poso River (Sulawesi, Indonesia) using DNA barcoding. Aquat. Sci. 2025, 87, 2. [Google Scholar] [CrossRef]
- Carninci, P.; Hayashizaki, Y. Noncoding RNA transcription beyond annotated genes. Curr. Opin. Genet. Dev. 2007, 17, 139–144. [Google Scholar] [CrossRef]
- Galtier, N. Half a century of controversy: The neutralist/selectionist debate in molecular evolution. Genome Biol. Evol. 2024, 16, evae003. [Google Scholar] [CrossRef]
- Ewing, B.; Green, P. Analysis of expressed sequence tags indicates 35,000 human genes. Nat. Genet. 2000, 25, 232–234. [Google Scholar] [CrossRef]
- Hellsten, U.; Harland, R.M.; Gilchrist, M.J.; Hendrix, D.; Jurka, J.; Kapitonov, V.; Ovcharenko, I.; Putnam, N.H.; Shu, S.; Taher, L.; et al. The genome of the Western clawed frog Xenopus tropicalis. Science 2010, 328, 633–636. [Google Scholar] [CrossRef]
- Church, D.M.; Goodstadt, L.; Hillier, L.W.; Zody, M.C.; Goldstein, S.; She, X.; Bult, C.J.; Agarwala, R.; Cherry, J.L.; DiCuccio, M.; et al. The Mouse Genome Sequencing Consortium. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009, 7, e1000112. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S. The genetic signatures of noncoding RNAs. PLoS Genet. 2009, 5, e1000459. [Google Scholar] [CrossRef] [PubMed]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Gregory, T.R. The case for junk DNA. PLoS Genet. 2014, 10, e1004351. [Google Scholar] [CrossRef]
- Gayon, J. From Mendel to epigenetics: History of genetics. C. R. Biol. 2016, 339, 225–230. [Google Scholar] [CrossRef]
- Wang, J.; Al-Ouran, R.; Hu, Y.; Kim, S.Y.; Wan, Y.W.; Wangler, M.F.; Yamamoto, S.; Chao, H.T.; Comjean, A.; Mohr, S.E.; et al. MARRVEL: Integration of human and model organism genetic resources to facilitate functional annotation of the human genome. Am. J. Hum. Genet. 2017, 100, 843–853. [Google Scholar] [CrossRef]
- Yang, Z.; Su, B.; Cao, C.; Wen, J.R. Regulatory DNA sequence design with reinforcement learning. arXiv 2025, arXiv:2503.07981. [Google Scholar] [CrossRef]
- US DOE Joint Genome Institute. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Vasemägi, A.; Ozerov, M.; Noreikiene, K.; López, M.E.; Gårdmark, A. Unlocking the genome of perch—From genes to ecology and back again. Ecol. Freshw. Fish 2023, 32, 677–702. [Google Scholar] [CrossRef]
- Gregory, T.R. Synergy between sequence and size in large-scale genomics. Nat. Rev. Genet. 2005, 6, 699–708. [Google Scholar] [CrossRef]
- Combrink, L.L.; Golcher-Benavides, J.; Lewanski, A.L.; Rick, J.A.; Rosenthal, W.C.; Wagner, C.E. Population genomics of adaptive radiation. Mol. Ecol. 2025, 34, e17574. [Google Scholar] [CrossRef]
- Stam, M.; Tark-Dame, M.; Fransz, P. 3D genome organization: A role for phase separation and loop extrusion? Curr. Opin. Plant Biol. 2019, 48, 36–46. [Google Scholar] [CrossRef]
- Hose, J.; Zheng, Q.; Sharp, N.P.; Gasch, A.P. On the rate of aneuploidy reversion in a wild yeast model. Genetics 2025, 229, iyae196. [Google Scholar] [CrossRef]
- Caron, H.; Molino, J.F.; Sabatier, D.; Léger, P.; Chaumeil, P.; Scotti-Saintagne, C.; Frigério, J.M.; Scotti, I.; Franc, A.; Petit, R.J. Chloroplast DNA variation in a hyperdiverse tropical tree community. Ecol. Evol. 2019, 9, 4897–4905. [Google Scholar] [CrossRef]
- Liu, W.S. Mammalian sex chromosome structure, gene content, and function in male fertility. Annu. Rev. Anim. Biosci. 2019, 7, 103–124. [Google Scholar] [CrossRef]
- Cheng, L.; Song, D.; Yu, X.; Du, X.; Huo, T. Endangered Schizothoracin Fish in the Tarim River Basin Are Threatened by Introgressive Hybridization. Biology 2022, 11, 981. [Google Scholar] [CrossRef]
- Schubert, I.; Lysak, M.A. Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 2011, 27, 207–216. [Google Scholar] [CrossRef]
- Baltazar-Soares, M.; Balard, A.; Heckwolf, M.J. Epigenetic Diversity and the Evolutionary Potential of Wild Populations. Evol. Appl. 2024, 17, e70011. [Google Scholar] [CrossRef]
- Oyarieme, H.W.; Otiti, J.; Obros, O. A Review on Genetic Variations within and between Populations: A Population Genetic Perspective. Am. Res. J. Biosci. 2024, 9, 1–10. [Google Scholar] [CrossRef]
- Henry, L.P.; Bruijning, M.; Forsberg, S.K.; Ayroles, J.F. The microbiome extends host evolutionary potential. Nat. Commun. 2021, 12, 5141. [Google Scholar] [CrossRef]
- Crow, J.F. Population genetics history: A personal view. Annu. Rev. Genet. 1987, 21, 1–22. [Google Scholar] [CrossRef]
- Chong, R.A.; Mueller, R.L. Evolution along the mutation gradient in the dynamic mitochondrial genome of salamanders. Genome Biol. Evol. 2013, 5, 907–919. [Google Scholar] [CrossRef]
- Harris, K.; Nielsen, R. Error-prone polymerase activity causes multinucleotide mutations in humans. Genome Res. 2014, 24, 1445–1454. [Google Scholar] [CrossRef]
- Kondrashov, A.S. Deleterious mutations and the evolution of sexual reproduction. Nature 1988, 336, 435–440. [Google Scholar] [CrossRef]
- Angst, P.; Ameline, C.; Haag, C.R.; Ben-Ami, F.; Ebert, D.; Fields, P.D. Genetic drift shapes the evolution of a highly dynamic metapopulation. Mol. Biol. Evol. 2022, 39, msac264. [Google Scholar] [CrossRef]
- Gerstein, A.C.; Sharp, N.P. The population genetics of ploidy change in unicellular fungi. FEMS Microbiol. Rev. 2021, 45, fuab006. [Google Scholar] [CrossRef]
- Davis, C.D.; Epps, C.W.; Flitcroft, R.L.; Banks, M.A. Refining and defining riverscape genetics: How rivers influence population genetic structure. Wiley Interdiscip. Revs. Water 2018, 5, e1269. [Google Scholar] [CrossRef]
- Hendry, A.P.; Day, T. Population structure attributable to reproductive time: Isolation by time and adaptation by time. Mol. Ecol. 2005, 14, 901–916. [Google Scholar] [CrossRef]
- Woodruff, D.S. Populations, species, and conservation genetics. In Encyclopedia of Biodiversity; Elsevier: Amsterdam, The Netherlands, 2004; pp. 811–829. [Google Scholar] [CrossRef]
- Selkoe, K.A.; Gaines, S.D.; Caselle, J.E.; Warner, R.R. Current shifts and kin aggregation explain genetic patchiness in fish recruits. Ecology 2006, 87, 3082–3094. [Google Scholar] [CrossRef] [PubMed]
- Gregory, T.R. Understanding natural selection: Essential concepts and common misconceptions. Evol. Educ. Outreach. 2009, 2, 156–175. [Google Scholar] [CrossRef]
- Donoghue, H.D.; Taylor, G.M.; Stewart, G.R.; Lee, O.Y.C.; Wu, H.H.; Besra, G.S.; Minnikin, D.E. Positive diagnosis of ancient leprosy and tuberculosis using ancient DNA and lipid biomarkers. Diversity 2017, 9, 46. [Google Scholar] [CrossRef]
- Charlesworth, B.; Charlesworth, D. The genetic basis of inbreeding depression. Genet. Res. 1999, 74, 329–340. [Google Scholar] [CrossRef]
- Balkenhol, N.; Cushman, S.A.; Storfer, A.; Waits, L.P. Introduction to landscape genetics–concepts, methods, applications. In Landscape Genetics: Concepts, Methods, Application; John Wiley and Sons: Hoboken, NJ, USA, 2015; pp. 1–8. [Google Scholar] [CrossRef]
- Le Bihan, D. Scaling in the Brain. Brain Multiphysics 2024, 7, 100102. [Google Scholar] [CrossRef]
- Chanda, P.; Costa, E.; Hu, J.; Sukumar, S.; Van Hemert, J.; Walia, R. Information theory in computational biology: Where we stand today. Entropy 2020, 22, 627. [Google Scholar] [CrossRef]
- Ma, C.; Korniss, G.; Szymanski, B.K.; Gao, J. Universality of noise-induced resilience restoration in spatially-extended ecological systems. Commun. Phys. 2021, 4, 262. [Google Scholar] [CrossRef]
- Martínez, O.; Reyes-Valdés, M.H. Defining diversity, specialization, and gene specificity in transcriptomes through information theory. Proc. Natl. Acad. Sci. USA 2008, 105, 9709–9714. [Google Scholar] [CrossRef]
- Ishengoma, E. Vertebrate genomics and adaptation–Status and prospects in Africa. Mol. Ecol. 2023, 32, 3368–3381. [Google Scholar] [CrossRef]
- Ribolli, J.; Hoeinghaus, D.J.; Johnson, J.A.; Zaniboni-Filho, E.; de Freitas, P.D.; Galetti, P.M. Isolation-by-time population structure in potamodromous Dourado Salminus brasiliensis in southern Brazil. Conserv. Genet. 2017, 18, 67–76. [Google Scholar] [CrossRef]
- Merschel, A.G.; Beedlow, P.A.; Shaw, D.C.; Woodruff, D.R.; Lee, E.H.; Cline, S.P.; Reilly, M.J. An ecological perspective on living with fire in ponderosa pine forests of Oregon and Washington: Resistance, gone but not forgotten. Trees For. People 2021, 4, 100074. [Google Scholar] [CrossRef]
- Nicol, S.C. Energy homeostasis in monotremes. Front. Neurosci. 2017, 11, 195. [Google Scholar] [CrossRef]
- Van Rheede, T.; Bastiaans, T.; Boone, D.N.; Hedges, S.B.; de Jong, W.W.; Madsen, O. The platypus is in its place: Nuclear genes and indels confirm the sister group relation of monotremes and Therians. Mol. Biol. Evol. 2006, 23, 587–597. [Google Scholar] [CrossRef]
- Martin, H.C.; Batty, E.M.; Hussin, J.; Westall, P.; Daish, T.; Kolomyjec, S.; Piazza, P.; Bowden, R.; Hawkins, M.; Grant, T.; et al. Insights into platypus population structure and history from whole-genome sequencing. Mol. Biol. Evol. 2018, 35, 1238–1252. [Google Scholar] [CrossRef]
- Thorpe, J.; Osei-Owusu, I.A.; Avigdor, B.E.; Tupler, R.; Pevsner, J. Mosaicism in human health and disease. Annu. Rev. Genet. 2020, 54, 487–510. [Google Scholar] [CrossRef]
- Berthold, P. Genetic control of migratory behaviour in birds. Trends Ecol. Evol. 1991, 6, 254–257. [Google Scholar] [CrossRef]
- Swift, D.G.; O’Leary, S.J.; Grubbs, R.D.; Frazier, B.S.; Fields, A.T.; Gardiner, J.M.; Drymon, J.M.; Bethea, D.M.; Wiley, T.R.; Portnoy, D.S. Philopatry influences the genetic population structure of the blacktip shark (Carcharhinus limbatus) at multiple spatial scales. Mol. Ecol. 2023, 32, 4953–4970. [Google Scholar] [CrossRef]
- Cunningham, C.; Parra, J.E.; Coals, L.; Beltrán, M.; Zefania, S.; Székely, T. Social interactions predict genetic diversification: An experimental manipulation in shorebirds. Behav. Ecol. 2018, 29, 609–618. [Google Scholar] [CrossRef]
- Van Oers, K.; Drent, P.J.; De Goede, P.; Van Noordwijk, A.J. Realized heritability and repeatability of risk-taking behaviour in relation to avian personalities. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2004, 271, 65–73. [Google Scholar] [CrossRef]
- Blonder, B.; Morrow, C.B.; Maitner, B.; Harris, D.J.; Lamanna, C.; Violle, C.; Enquist, B.J.; Kerkhoff, A.J. New approaches for delineating n-dimensional hypervolumes. Methods Ecol. Evol. 2018, 9, 305–319. [Google Scholar] [CrossRef]
- Piñeros, V.J.; del, R. Pedraza-Marrón, C.; Betancourt-Resendes, I.; Calderón-Cortés, N.; Betancur-R, R.; Domínguez-Domínguez, O. Genome-wide species delimitation analyses of a silverside fish species complex in central Mexico indicate taxonomic over-splitting. BMC Ecol. Evol. 2022, 22, 108. [Google Scholar] [CrossRef]
- Morin, P.A.; Archer, F.I.; Foote, A.D.; Vilstrup, J.; Allen, E.E.; Wade, P.; Durban, J.; Parsons, K.; Pitman, R.; Li, L.; et al. Complete mitochondrial genome phylogeographic analysis of killer whales (Orcinus orca) indicates multiple species. Genome Res. 2010, 20, 908–916. [Google Scholar] [CrossRef]
- Sousa-Santos, C.; Collares-Pereira, M.J.; Almada, V.C. Evidence of extensive mitochondrial introgression with nearly complete substitution of the typical Squalius pyrenaicus-like mtDNA of the Squalius alburnoides complex (Cyprinidae) in an independent Iberian drainage. J. Fish Biol. 2006, 68, 292–301. [Google Scholar] [CrossRef]
- Laikre, L.; Larsson, L.C.; Palmé, A.; Charlier, J.; Josefsson, M.; Ryman, N. Potentials for monitoring gene level biodiversity: Using Sweden as an example. Biodivers. Conserv. 2008, 17, 893–910. [Google Scholar] [CrossRef]
- Mastretta-Yanes, A.; da Silva, J.M.; Grueber, C.E.; Castillo-Reina, L.; Köppä, V.; Forester, B.R.; Funk, W.C.; Heuertz, M.; Ishihama, F.; Jordan, R.; et al. Multinational Evaluation of Genetic Diversity Indicators for the Kunming-Montreal Global Biodiversity Framework. Ecol. Lett. 2024, 27, e14461. [Google Scholar] [CrossRef]
- Brown, A.H.D. Core collections: A practical approach to genetic resources management. Genome 1989, 31, 818–824. [Google Scholar] [CrossRef]
- Gu, R.; Fan, S.; Wei, S.; Li, J.; Zheng, S.; Liu, G. Developments on core collections of plant genetic resources: Do we know enough? Forests 2023, 14, 926. [Google Scholar] [CrossRef]
- Trense, D.; Schmidt, T.L.; Yang, Q.; Chung, J.; Hoffmann, A.A.; Fischer, K. Anthropogenic and natural barriers affect genetic connectivity in an Alpine butterfly. Mol. Ecol. 2021, 30, 114–130. [Google Scholar] [CrossRef]
- Pampoulie, C.; Berg, P.R.; Jentoft, S. Hidden but revealed: After years of genetic studies behavioural monitoring combined with genomics uncover new insight into the population dynamics of Atlantic cod in Icelandic waters. Evol. Appl. 2023, 16, 223–233. [Google Scholar] [CrossRef]

| Species Concept | Key Features | Temporal Focus | Asexual? | Ontology | Ref. |
|---|---|---|---|---|---|
| Biological (BSC) | Reproductive isolation | Horizontal | No | Realistic | [47,48] |
| Cohesion | Maintained by gene flow and demographic exchangeability | Horizontal | No | Realistic | [49] |
| Recognition | Defined by mating-recognition systems | Horizontal | No | Realistic | [50] |
| Ecological (ESC) | Species occupy unique ecological niches | Horizontal | Yes | Realistic | [51] |
| Genetic (GSC) | Measurable genetic differences (allele frequencies, nucleotide sequence divergence, etc.) | Horizontal | Yes | Realistic | [52,53] |
| Morphological (MSC) | Shared physical traits | Horizontal | Yes | Operational | [54] |
| Phenetic | Grouped by overall similarity of traits | Horizontal | Yes | Operational | [55] |
| Genotypic Cluster | Clusters of genetically similar individuals | Horizontal | Yes | Operational | [56] |
| Polythetic | Overlapping sets of traits with no single defining feature | Horizontal | Yes | Operational | [57] |
| Taxonomic | Based on expert judgment and multiple criteria (e.g., morphology, ecology, and genetics) | Horizontal | Yes | Pragmatic | [58] |
| OTU (Operational) | Defined by sequence similarity thresholds (e.g., 97%) | Horizontal | Yes | Pragmatic | [59,60] |
| Cladistic | Based on shared derived characters | Vertical | Yes | Realistic | [61] |
| Evolutionary (EvSC) | Lineages with their own evolutionary tendencies and fates | Vertical | Yes | Realistic | [62] |
| Phylogenetic (PSC) | Smallest diagnosable monophyletic group sharing a common ancestor | Vertical | Yes | Realistic | [63,64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ragauskas, A.; Maziliauskaitė, E.; Prakas, P.; Butkauskas, D. Population Genetic Structure: Where, What, and Why? Diversity 2025, 17, 584. https://doi.org/10.3390/d17080584
Ragauskas A, Maziliauskaitė E, Prakas P, Butkauskas D. Population Genetic Structure: Where, What, and Why? Diversity. 2025; 17(8):584. https://doi.org/10.3390/d17080584
Chicago/Turabian StyleRagauskas, Adomas, Evelina Maziliauskaitė, Petras Prakas, and Dalius Butkauskas. 2025. "Population Genetic Structure: Where, What, and Why?" Diversity 17, no. 8: 584. https://doi.org/10.3390/d17080584
APA StyleRagauskas, A., Maziliauskaitė, E., Prakas, P., & Butkauskas, D. (2025). Population Genetic Structure: Where, What, and Why? Diversity, 17(8), 584. https://doi.org/10.3390/d17080584









