Abstract
Protein degradation plays a fundamental role in maintaining protein homeostasis and ensures proper cellular function by regulating protein quality and quantity. Heat shock protein 100 (Hsp100), found in bacteria, plants, and fungi, is a unique chaperone family responsible for rescuing misfolded proteins from aggregated states in an ATP-dependent manner. To date, they are primarily known to mediate heat stress adaptation and enhance cellular survival under extreme conditions in higher plants and algae. Resting cyst formation in dinoflagellates is widely recognized as a response to adverse conditions, which offers an adaptive advantage to endure harsh environmental extremes that are unsuitable for vegetative cell growth and survival. In this study, based on a full-length cDNA sequence, we characterized an Hsp100 gene (SaHsp100) from the cosmopolitan bloom-forming dinoflagellate Scrippsiella acuminata, aiming to examine its life stage-specific expression patterns and preliminarily explore its potential functions. The qPCR results revealed that Hsp100 transcript levels were significantly elevated in newly formed resting cysts compared to vegetative cells and continued to increase during storage under simulated marine sediment conditions (darkness, low temperature, and anoxia). Parallel reaction monitoring (PRM)-based quantification further confirmed that Hsp100 protein levels were significantly higher in resting cysts than in vegetative cells and increased after three months of storage. These findings collectively highlighted the fundamental role of Hsp100 in the alteration of the life cycle and dormancy maintenance of S. acuminata, likely by enhancing stress adaptation and promoting cell survival through participation in proteostasis maintenance, particularly under natural sediment-like conditions that trigger severe abiotic stress. Our work deepens the current understanding of Hsp family members in dinoflagellates, paving the way for future investigations into their ecological relevance within this ecologically significant group.
1. Introduction
Dinoflagellates are a diverse and ubiquitous group of unicellular protists with major ecological impacts, as they are important worldwide primary producers, essential coral endosymbionts, and key contributors to harmful algal blooms (HABs) and biotoxins in the ocean [1]. So far, approximately 200 species of dinoflagellates are known to cause HABs, contributing to roughly 75% of global marine HAB events [2] and representing ~40% of all documented HAB-causing algal species [3]. Dinoflagellate HABs negatively impact various sectors including commercial and recreational fishing, aquaculture operations, and coastal tourism activities, while also posing risks to both human populations and marine ecosystems [4]. Mortality or physiological impairments, caused by toxin production, oxygen depletion, or physical clogging, are associated with the massive biomass accumulation during blooms [4,5]. The resting stage of dinoflagellates, also well-known as resting cysts, plays a critical role in dinoflagellate ecology, especially for HAB-causing species. Multiple evidence shows that resting cysts help dinoflagellates survive harsh conditions, promote genetic recombination, drive bloom dynamics, and enable population dispersal [6]. Moreover, the researchers routinely analyze the abundance and distribution characteristics of resting cysts in marine sediments to develop empirical models for predicting the potential outbreak intensity and affected range of algal blooms [7,8]. Although resting cyst formation plays a crucial ecological role in the life cycle of HAB-forming dinoflagellates, the genetic mechanisms governing this process remain poorly characterized. Key aspects such as the specific functional genes and molecular pathways involved are still not well understood [9]. Understanding these mechanisms is particularly challenging for resting cysts buried in natural sediments, where they are exposed to cold, dark, and hypoxic/anoxic conditions.
Proteostasis maintenance is critical for the physiology and survival of organisms [10]. Heat shock proteins (Hsps) are ubiquitous molecular chaperones that assist the folding of other proteins, either by facilitating proper folding or by stabilizing partially folded intermediates. Based on sequence homology and approximate molecular weight, Hsps are classified into the Hsp100, Hsp90, Hsp70, Hsp60, and small heat shock protein families [11]. While Hsp60/Hsp10 and Hsp70/Hsp40 help cells minimize protein aggregation by recognizing and refolding misfolded proteins into their biologically active conformations, protein disaggregation is primarily mediated by the Hsp100 family [12,13]. Hsp100/Clp (caseinolytic protease) proteins, belonging to the AAA+ (ATPases associated with diverse cellular activities) superfamily, are ATP-dependent chaperones found in bacteria, fungi, plants, and algae [11,13,14,15]. These proteins represent a unique chaperone family that rescues misfolded proteins from aggregated states in an ATP-dependent manner, often in cooperation with their cognate Hsp70 members [12,16]. Peptides released from these aggregates can either be refolded into their native state or directed to proteolytic degradation [13,17]. In contrast to the extensive characterization of other Hsps, Hsp100 proteins, originally identified in bacteria (ClpB) and yeast (Hsp104) in the late 1980s and early 1990s, generally received less attention. The heat shock inducibility of the Hsp100 gene and its protein product has been observed in nearly all plant species studied to date [13,17]. Moreover, accumulating evidence suggests that Hsp100 solubilizes and reactivates a broad range of aggregated proteins, thereby conferring thermotolerance and promoting cell survival under severe stress conditions [14,15,16].
While significant progress has been made in characterizing Hsp70 and Hsp90 families in dinoflagellates [18,19], the Hsp100 family has been largely overlooked, with current knowledge predominantly based on omics predictions. The Hsp100-annotated genes were identified in the transcriptomic studies of Symbiodinium sp. [20], Scrippsiella acuminata (formerly S. trochoidea) [21,22], and Alexandrium pacificum [23]. Differential expression of one Hsp100/Clp protein was observed in Prorocentrum donghaiense across different cell cycle phases, suggesting its role in maintaining proper cell cycle progression [24]. Significantly downregulated protein abundance of Hsp100/Clp was also reported in Alexandrium catenella under organic phosphorus resupply conditions, implying its involvement in enhancing cellular stress tolerance [25]. Additionally, our previous studies suggested a potential role of Hsp members in life stage transitions and the dormancy maintenance of dinoflagellates (see below). A comparative transcriptomic study of S. acuminata revealed a significant downregulation of three Hsp70-encoding genes in resting cysts [22]. A parallel expression pattern was found in Akashiwo sanguinea, where Hsp70 transcripts reached maximum levels in newly formed cysts but decreased as cysts matured [18]. The Hsp40 gene exhibited significantly elevated transcriptional activity in newly formed resting cysts of S. acuminata relative to both vegetative cells and mature cysts [26]. Conversely, Hsp90 gene expression displayed preferential upregulation during dormancy maintenance [19]. Additionally, Hsp60 and Hsp10 exhibited progressive upregulation from vegetative cells through cyst formation to long-term storage [27]. All these findings collectively demonstrate functional coordination among different Hsp members during dinoflagellate encystment and cyst dormancy.
The calcareous dinoflagellate S. acuminata is a cosmopolitan species that frequently forms blooms in global coastal waters. Due to its high propensity for resting cyst formation, this species has been established as a model organism for investigating dinoflagellates’ life history [9]. In this study, we characterized an Hsp100 gene (SaHsp100) from S. acuminata through molecular cloning, transcriptional profiling and protein abundance analysis to examine its life stage-specific expression patterns and potential functions. The obtained results advance our understanding of Hsps in dinoflagellates, providing a foundation for future investigations into their ecological significance in this ecologically important group.
2. Materials and Methods
2.1. Scrippsiella acuminata Culture Maintenance
The Scrippsiella acuminata strain IOCAS-St-1, first isolated from the Yellow Sea of China, was acquired from the Marine Biological Culture Collection Centre, which is affiliated with the Institute of Oceanology, at the Chinese Academy of Sciences [22]. For routine cultivation, the strain was kept in silicate-free f/2 medium [28]. The medium was made with natural seawater (salinity about 32) that had been pre-filtered with a 0.22 μm membrane filter (Millipore, Billerica, MA, USA) and autoclaved at 121 °C for 30 min. Culture batches were grown and maintained in an incubator with a temperature of 20 ± 1 °C and a 12 h light:12 h dark cycle. Cool white fluorescent lights supplied a photon flux of ~100 μmol photons m−2 s−1 for illumination. Immediately before inoculation, 3% final concentration of penicillin-streptomycin mixture (100×, Solarbio, Beijing, China) was added to the medium. Stock cultures were maintained in exponential growth by biweekly transfer to fresh f/2-Si medium.
2.2. Full-Length cDNA Cloning of SaHsp100
Approximately 106 exponentially growing vegetative cells were harvested by centrifugation for total RNA extraction. RNA was isolated using the Deng et al. [19] protocol, followed by DNase I treatment (QIAGEN, Hilden, Germany) to remove genomic DNA. RNA quality was verified by using both agarose gel electrophoresis and spectrophotometric analysis (NanoDropTM 1000, Thermo Fisher Scientific, Waltham, MA, USA). First-strand cDNA was synthesized from 1 μg total RNA using M-MLV reverse transcriptase (Takara, Tokyo, Japan) with random primers for subsequent PCR amplification. The cDNA fragment was amplified following the protocol described in Deng et al. [26] with specific primers Fr-F and Fr-R (Table 1), which were designed based on an Hsp100-like sequence identified in the transcriptomic data of S. acuminata (GenBank Accession No. SRP058465) [22].

Table 1.
Primer details.
Gene-specific primers were designed based on the obtained cDNA fragment for 3′ and 5′ RACE amplification (Table 1). The RACE template was synthesized from 1 μg total RNA using an anchor primer (Table 1). For 5′-RACE, reactions were performed using the dinoflagellate-specific forward primer DinoSL [29] and nested reverse primers (5R-outer and 5R-inner, Table 1). For 3′-RACE, forward primers 3R-outer and 3R-inner (Table 1) were used in combination with the reverse primer GeneRacer3 (Invitrogen, Karlsruhe, Germany). Amplified DNA fragments were electrophoresed onto 1% agarose gel, with target bands subsequently excised and purified using a commercial DNA extraction kit (TaKaRa, Tokyo, Japan). These fragments were then cloned into the pMD-19T vector system (TaKaRa, Tokyo, Japan) for sequencing analysis (TsingKe Biotech, Beijing, China).
2.3. Sequence Homology and Phylogenetic Analysis
The putative protein-coding region was identified using the Open Reading Frame (ORF) finder [30]. Sequence homology and functional predictions were obtained through NCBI BLAST (2.15.0) analysis (E-value cutoff: 1 × 10−6) [31]. Protein domain architecture was predicted using SMART [32] and validated using Pfam [33]. Theoretical molecular characteristics (molecular weight and isoelectric point) were determined using ProtParam (available via ExPASy) analysis [34].
The newly identified SaHsp100 sequence and thirty reference Hsp100 homologs (Supplementary Table S1) from the GenBank database were aligned using MAFFT v7.475 (G-INS-i algorithm) [35]. The alignment was manually refined in BioEdit v7.2.5 [36], followed by Bayesian inference analysis conducted with MrBayes 3.2.6. [37]. We ran four Markov chains (1 million generations, sampling every 100 generations), discarded the first 25% as burn-in, and built a 50% majority-rule consensus tree. Final tree visualization was performed using FigTree v1.4.4.
2.4. Resting Cysts Preparation and Sample Treatment
Resting cysts were produced following the method by Yue et al. [38] by transferring exponentially growing vegetative cells from 75 cm2 culture flasks (Sorfa, Suzhou, China) into N/P-limited f/2-Si medium (1000× diluted). Cultures were maintained under standard conditions and monitored every 48 h using a microscope (IX73, Olympus, Tokyo, Japan). After ≥30 days incubation, we harvested cysts when microscope examination confirmed sufficient yield. The cysts were then washed repeatedly with sterile filtered seawater until no motile cells (vegetative cells or planozygote) remained visable under a light microscope. The resting cysts used in this study had typical calcareous spines and contained red accumulation bodies. The harvested cysts were immediately used for sample treatments.
We cultured vegetative cells (~2 × 103 cells mL−1 initial density) in 500 mL Pyrex flasks containing 300 mL of medium under standard conditions, with Day 0 marking inoculation. On Day 5 (exponential phase, “V1”) and Day 10 (stationary phase, “V2”), we collected samples. Newly formed resting cysts were harvested separately as “C0” (Table 2). Resting cysts storage under conditions mimicking those in natural marine sediments (darkness, low temperature, and anoxia) for different periods (Table 2) was carried out referring to previous studies, as summarized below. Following the protocol of Li et al. [39], anoxic conditions were established by flushing the cyst-containing tubes with nitrogen gas, while dissolved oxygen levels were continuously monitored using an oxygen meter (Microx 4, PreSens, Regensburg, Germany). To visually confirm anoxia, the culture medium was supplemented with Na-resazurin (0.1% w/v) as a redox indicator, where color change indicates oxygen depletion [40]. We maintained the samples at a low temperature (4 ± 1 °C) using a refrigerator (without light). To ensure complete darkness, cyst-containing tubes were wrapped in aluminum foil before being stored for different periods (Table 2). We collected samples for qPCR analysis at designated time points (~5 × 104 cells/cysts per sample; n = 3 replicates). After brief contrifugation in 1.5 mL tubes, we flash-froze cysts in liquid nitrogen and then stored them at −80 °C until RNA extraction. Four samples, V1, V2, C0, and C3 were subjected to the PRM analysis. All cells in each sample (~106 cells; 3 biological replicates) were pelleted in centrifuge tubes and immediately stored at −80 °C until protein extraction. The RNA and protein were isolated from the same set of biological samples.

Table 2.
Experimental design in this study.
2.5. Transcriptional Profiles of SaHsp100 with qPCR Detection
A pair of gene-specific primers (q100-F and q100-R; Table 1) was designed using Primer Premier 5 to amplify a 121-bp fragment of SaHsp100. We extracted total RNA using an RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany), treated samples with an RNase-Free DNase Set (QIAGEN, Hilden, Germany) and measured RNA quality/quantity using a NanoDropTM 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). First-strand cDNA was reverse transcribed from approximately 100 ng of total RNA using random primers. The qPCR amplifications were performed using TB Green Premix Ex TaqTM II (TaKaRa, Tokyo, Japan) on the Eppendorf Mastercycler® ep realplex S (Eppendorf, Hamburg, Germany). Following reference gene validation studies [22], we selected the ubiquitin conjugating enzyme (UBC) (Table 1) as the internal control. Reverse transcription proceeded at 37 °C (15 min) with a final enzyme inactivation step at 85 °C (5 s). Each sample was analyzed in three biological replicates, each of which was accompanied by three technical replicates to ensure the reliability of the experimental results. Non-template reactions (NTC) were run as controls. The details of the primers and the qPCR results are displayed in the Supplementary Materials (Supplementary Table S2, Figures S2–S4). We calculated relative expression levels for six target genes using the 2−△△Ct method [41], normalizing to UBC internal control values and expressing results as unitless ratios. All data were expressed as mean ± SD. Statistical analyses were performed using one-way ANOVA followed by Tukey’s post hoc test (SPSS 24.0, Windows version), with statistical significance set at p < 0.05.
2.6. PRM-Based Quantification of Hsp100 Protein Levels in S. acuminata
2.6.1. Protein Extraction
The samples were frozen, pulverized in liquid nitrogen and homogenized in lysis buffer (7 M Urea, 2 M Thiourea, 4% CHAPS, 40 mM Tris-HCl, pH 8.5) containing protease inhibitors (1 mM PMSF and 2 mM EDTA). Following a 5 min incubation period, 10 mM DTT was added to reduce disulfide bonds, followed by sonication (200 W, 15 min) and centrifugation (30,000× g, 15 min, 4 °C) to clarify the lysate. Proteins were precipitated overnight at −20 °C with acetone containing 10% (v/v) TCA, washed three times with chilled acetone, and air-dried. The pellet was then dissolved in a modified lysis buffer (7 M urea, 2 M thiourea, 4% NP40, 20 mM Tris-HCl, pH 8.0–8.5). The resulting suspension underwent sonication (200 W, 15 min) followed by centrifugation (30,000× g, 15 min, 4 °C) to obtain the final protein extract. The supernatant was reduced with DTT (10 mM, 56 °C, 1 h) and alkylated with iodoacetamide (55 mM, dark, 1 h) before being reprecipitated with acetone. After the final centrifugation, the protein pellet was resuspended in 0.5 M TEAB, sonicated, and centrifuged to obtain a clear protein extract, which was quantified using the Bradford assay and stored at −80 °C for downstream applications.
2.6.2. Protein Digestion
A total of 100 μg of protein from each sample was subjected to tryptic digestion using Trypsin Gold (Promega, Madison, WI, USA) with a protein-to-protease ratio of 30:1 (w/w). The enzymatic digestion was performed at 37 °C for 16 h to ensure complete protein cleavage. After digestion, the generated peptides were concentrated by vacuum centrifugation and subsequently reconstituted in 0.5 M TEAB buffer for further downstream processing.
2.6.3. Liquid Chromatography (LC)-PRM-Mass Spectrometry (MS)
The PRM technique offers high sensitivity and accuracy for targeted peptide quantification through selective ion monitoring [42]. In this study, peptides of an Hsp100-annotated protein identified in Scrippsiella acuminata proteomics data (our unpublished data) were validated using a peptide uniqueness checker [43]. Then the peptide LVAEVQEAK was selected as the target peptide for the following experiments due to its specificity to the Hsp100-annotated protein. We also screened the full-length transcriptome of S. acuminata ([44]; NCBI accession number: PRJNA1127753) to confirm that no other Hsp100 isoforms express the same peptide. The target peptide was input into Skyline [45] and was used to determine the PRM parameters (m/z, charges, polarity, collision energy, retention time) for targeted peptide analysis. Following tryptic digestion, samples were spiked with 50 fmol β-galactosidase as an internal standard. PRM was performed on a QTRAP 6500 LC-MS/MS system (SCIEX, Framingham, MA, USA) interfaced with an LC-20AD nanoHPLC system (Shimadzu, Kyoto, Japan). The binary mobile phase consisted of (A) 0.1% aqueous formic acid and (B) 0.1% formic acid in 98% acetonitrile. Peptide separation was achieved on a C18 column (0.075 × 150 mm column, 3.6 μm particle size) at a flow rate of 300 nL/min, with elution performed via a gradient program of 5–30% solvent B for 38 min, 30–80% solvent B for 4 min, and maintenance at 80% for 8 min. The QTRAP 6500 mass spectrometer was operated with a spray voltage of 2400 V and a dwell time of 10 ms was used. To enhance specificity, multiple PRM transitions were monitored with unit resolution applied to both the Q1 and Q3 quadrupoles.
2.6.4. Data Analysis
Mass spectrometry data analysis was performed using Skyline software (22.2) [44] to process the raw files generated by QTRAP 6500 (SCIEX, Framingham, MA, USA). Peptide identification and chromatographic alignment were conducted using an iRT (indexed retention time) strategy [46] against a spectral library. Each peptide’s transitions were all utilized for quantification purposes, except in cases where matrix interference was detected. For label-free data normalization, β-galactosidase was spiked in as an internal reference [47]. Statistical analysis was performed using MSstats implementing a linear mixed-effects modeling approach [48], with an adjusted p-value maintaining the false discovery rate (FDR) below the 0.05 significant threshold. Proteins were deemed statistically significant if they had a p-value less than 0.05 and a fold change exceeding 1.5.
3. Results
3.1. The Full-Length cDNA Sequence of SaHsp100
The full-length cDNA sequence of SaHsp100 (3576 bp) was assembled by overlapping the 1720 bp 3′ RACE product, the 1087 bp 5′ RACE product, and a 1673 bp fragment amplified with primers Fr-F/Fr-R. The sequence comprises a 94 bp 5′ UTR, a 2835 bp ORF, and a 647 bp 3′ UTR containing a poly (A) tail. The ORF has a 58% G+C content and encodes a 944-amino acid protein with a predicted molecular weight of 102.73 kDa and an isoelectric point of 5.21 (Figure 1). The obtained sequence was deposited in GenBank with the accession number PV587988.

Figure 1.
The deduced amino acid sequences of SaHsp100 (GenBank Accession number: PV587988). Sequences are numbered on the left. The conserved ATP-binding sites are denoted with triangles. The amino acid residues constituting the hexamerization interface are highlighted with dots.
3.2. Homology and Phylogenetic Analysis of SaHsp100
BLAST analysis identified a conserved ATP-binding site in the deduced SaHsp100 protein sequence with eight functionally essential residues, including Val658, Thr695, Val697, Gly698, Lys699, Thr700, Glu701, and Asp765 (Figure 1). A total of nineteen amino acid residues (Arg674, Ala677, Gly678, Lys679, Asp752, Arg755, Arg756, Asp773, Leu777, Asp783, Arg802, Pro803, Glu804, Asn807, Arg808, Leu809, Asp810, Glu811, and Ile812) likely constitute the hexamerization interface (Figure 1). Sequence homology analysis revealed that SaHsp100 shared 60.9–68.8% amino acid sequence similarity with seven other dinoflagellate Hsp100 counterparts (Supplementary Figure S1). Phylogenetic reconstruction placed SaHsp100 into the dinoflagellate group, which formed a sister clade of green algal Hsp100 with high node support (BI 1.00) (Figure 2).
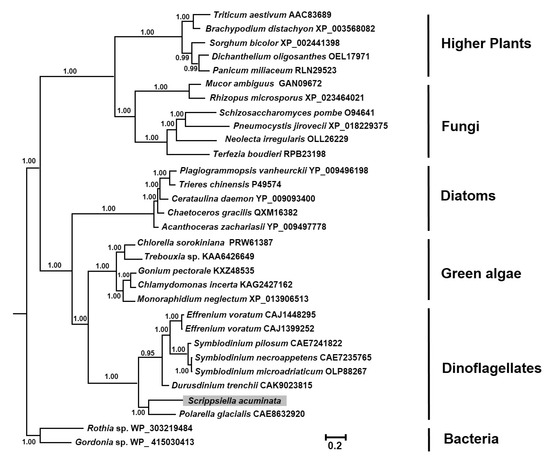
Figure 2.
Bayesian inference (BI) distance tree formulated from the alignment of the Hsp100 amino acid sequences. The numbers at each node indicate the BI posterior probabilities. For the thirty sequences retrieved from the GenBank database, each entry is labeled with its accession number following the corresponding species name (further details are available in Supplementary Table S1). The Hsp100 gene newly sequenced from Scrippsiella acuminata in this study is highlighted with a gray background.
3.3. Differential Transcription Profiles of SaHsp100 at Different Stages of Growth and Life Cycle
The transcription levels of SaHsp100 were compared across vegetative cells (exponential and stationary phases), newly formed resting cysts, and resting cysts stored under marine sediment-like conditions for varying durations. While no significant difference in SaHsp100 transcription was detected between vegetative cells at different growth stages (V1 and V2) (ANOVA, p > 0.05; Figure 3), significantly higher expression levels were observed in resting cysts compared to vegetative cells (ANOVA, p < 0.01; Figure 3). Newly formed resting cysts exhibited higher SaHsp100 transcription levels than vegetative cells, with levels showing a general increasing trend during storage under simulated marine sediment conditions (Figure 3).
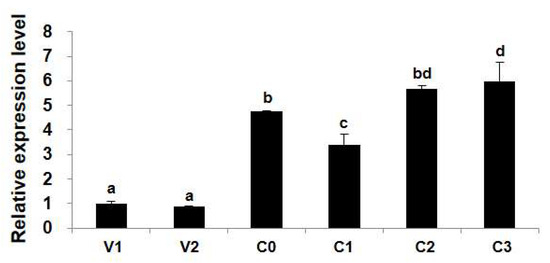
Figure 3.
The transcription levels of SaHsp100 in cells at different stages of growth and life cycle. The samples contain vegetative cells at the exponential and stationary growth stages, newly formed resting cysts, and resting cysts stored under natural marine sediment-like conditions for varying durations. Gene expression data are presented as normalized fold-change values relative to the vegetative cell control (V1, Day 5), with vertical bars indicating mean ± SD of triplicate measurements. Significant differential expression (p < 0.05, ANOVA with post hoc testing) is denoted by alphabetical superscripts.
3.4. Protein Levels of Hsp100 Determined Using the Method of PRM
The protein levels of Hsp100 were quantitatively analyzed by PRM across vegetative cells at the exponential and stationary growth stages, newly formed resting cysts, and resting cysts stored under natural marine sediment-like conditions for three months. All experiments were conducted using the target peptide LVAEVQEAK (Figure 4A). No significant difference was observed in protein abundance between vegetative cells at different growth stages (V1 and V2) (ANOVA, p > 0.05; Figure 4B). Hsp100 protein abundance was significantly higher in newly formed resting cysts than in vegetative cells (ANOVA, p < 0.05; Figure 4B) and showed a further significant increase after three months of storage under simulated marine sediment conditions (darkness, low temperature, and anoxia) (ANOVA, p < 0.05; Figure 4B).
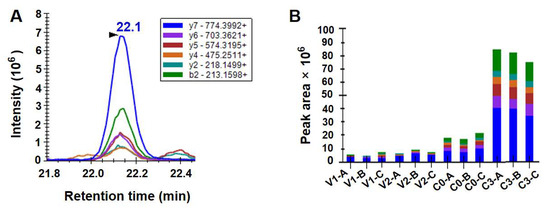
Figure 4.
PRM-based determination of Hsp100 protein levels in S. acuminata at different stages of growth and life cycle. All experiments were conducted using the target peptide LVAEVQEAK. (A) Overlays of PRM signals of all the individual product ions. The m/z ratios for the mass spectrometry spectra are shown in the legend. (B) Peak areas for the individual product ions corresponding to the target peptide in different samples. All samples were prepared in biological triplicates (labeled as -A, -B, and -C).
4. Discussion
The activation of Hsp synthesis is a universal adaptive mechanism in organisms ranging from bacteria to plants and animals, which enable them to survive and adapt to diverse environmental stresses. Extensive studies have documented that nearly all types of stressful cues can induce the expression of Hsp genes and/or the synthesis of Hsp proteins [11,49]. Meanwhile, many Hsps are constitutively expressed under normal conditions and participate in diverse physiological processes [49,50]. Unlike other Hsps that have been extensively characterized, Hsp100 proteins were not identified until the late 1980s and have generally received less research attention. To date, Hsp100 is primarily recognized for its decisive roles in heat stress adaptation and promoting cell survival under severe stress conditions in higher plants and algae [11,13,14,17]. In dinoflagellates, current knowledge of Hsp100 remains limited to omics predictions without structural or functional validation [20,22,23,24,25]. As of May 2025, the public GenBank database contains only about forty entries associated with Hsp100/Clp genes and/or proteins from a limited number of dinoflagellate species, predominantly those that have undergone whole-genome sequencing. No verified complete coding sequence of the Hsp100/Clp gene with its expression profiles under stress or developmental conditions has been characterized in dinoflagellates. Here, we characterized an Hsp100 gene from S. acuminata through full-length cDNA sequence isolation, aiming to preliminarily investigate its potential association with different life cycle stages of this HAB-forming dinoflagellate. The conserved sequence signatures and motifs of SaHsp100 suggest functional conservation similar to that observed in other well-characterized organisms, although these functions have not yet been explicitly demonstrated.
In this study, the relative transcript levels of SaHsp100 were quantified in vegetative cells at the exponential stage, stationary stage, and in resting cysts that were newly formed and maintained in dormancy for different periods. The experimental design, in which cysts were stored at 4 ± 1 °C under anoxic and dark conditions, was intended to simulate the natural environment that cysts typically experience in marine sediments. Because Hsps are rapidly upregulated in response to environmental insults [18,50], the absence of significant differences in SaHsp100 transcription among vegetative cells at different growth stages implied that vegetative cells were well adapted to variations in the physicochemical conditions of the culture medium during prolonged incubation. The transcript levels were significantly elevated in newly formed resting cysts relative to vegetative cells and continued to rise during storage in simulated marine sediment conditions. PRM-based quantification further confirmed that Hsp100 protein levels in resting cysts were significantly higher than in vegetative cells and increased during three months of storage. Together, these results suggest the potential involvement of Hsp100 chaperones in the life stage transitions and dormancy maintenance of S. acuminata. Cellular proteins are highly prone to denaturation under stressful conditions [10]. Because denatured proteins are toxic to cellular function, organisms have evolved diverse mechanisms to address this issue. These mechanisms operate constitutively and are further induced upon stress perception across species, ranging from bacteria to higher plants and metazoans [13]. Hsp100 chaperones have been documented to possess the unique ability to facilitate proper folding of nascent proteins and stabilize or refold misfolded ones, thus preventing aberrant aggregation [11,14,15]. Given the current lack of well-established information regarding the signalling pathways involved in encystment and dormancy, it remains unclear whether Hsp100 directly interacts with encystment-specific factors or merely acts as a bystander in general stress buffering. Resting cyst formation is widely recognized as a response to adverse conditions, which offers an adaptive advantage to endure harsh environmental extremes (e.g., darkness, low temperature, and anoxia) that are unsuitable for the growth and/or survival of vegetative cells [6]. Therefore, the upregulation of Hsp100 at both transcriptional and translational levels may play a crucial role in maintaining proteostasis by preventing protein aggregation and resolving misfolded proteins during the resting stage of S. acuminata, particularly under natural sediment-like conditions that trigger severe abiotic stress. Our findings collectively highlight the possible fundamental role of Hsp100 in the alteration of the life cycle and dormancy maintenance of S. acuminata, likely by enhancing stress adaptation and promoting cell survival through participation in proteostasis maintenance. To fully elucidate the functional roles of Hsp100 in dinoflagellates, systematic studies examining both gene expression and protein synthesis patterns across different species under varying environmental stresses and growth stages are needed. Such investigations will provide crucial insights into this distinctive chaperone system in these ecologically important marine microorganisms.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d17080519/s1, Table S1: Details for the Hsp100 sequences used in the phylogenetic analysis. Table S2: Details of the qPCR primer set used for the SaHsp100 gene. Figure S1: Alignment and comparison of Hsp100-deduced amino acid sequences from Scrippsiella acuminata with other registered counterparts. Figure S2: The qPCR amplification curves of hsp100 generated with serial 2-fold dilutions of templates (E = 100.7%). Figure S3: The standard curve for amplification efficiency determination. Figure S4: Melting curve analysis revealed a single peak with a melting temperature (Tm) of 89 °C.
Author Contributions
Conceptualization, Y.D. and Y.Z.T.; methodology, F.L.; software, L.S.; validation, H.Z. and C.S.; formal analysis, Z.H.; investigation, F.L.; resources, Y.D.; data curation, L.S.; writing—original draft preparation, F.L.; writing—review and editing, Y.D.; visualization, L.S.; supervision, Y.D.; project administration, Y.Z.T.; funding acquisition, F.L. and L.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant numbers 42406214 and 42206215.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data are contained within the article and Supplementary Materials.
Acknowledgments
We would like to express our gratitude to the anonymous reviewers for their constructive suggestions and comments. We acknowledge financial support from the National Science Foundation of China, grant numbers 42406214 and 42206215.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| HAB | Harmful algal bloom |
| Hsp100 | Heat shock protein 100 |
| PRM | Parallel reaction monitoring |
| RACE | Rapid amplification of cDNA ends |
| ORF | Open Reading Frame |
References
- Taylor, F.; Hoppenrath, M.; Saldarriaga, J.F. Dinoflagellate diversity and distribution. Biodivers. Conserv. 2008, 17, 407–418. [Google Scholar] [CrossRef]
- Smayda, T.J. Harmful algal blooms: Their ecophysiology and general relevance to phytoplankton blooms in the sea. Limnol. Oceanogr. 1997, 42, 1137–1153. [Google Scholar] [CrossRef]
- Jeong, H.J.; Kang, H.C.; Lim, A.S.; Jang, S.H.; Lee, K.; Lee, S.Y.; Ok, J.H.; You, J.H.; Kim, J.H.; Lee, K.H. Feeding diverse prey as an excellent strategy of mixotrophic dinoflagellates for global dominance. Sci. Adv. 2021, 7, eabe4214. [Google Scholar] [CrossRef]
- Geng, H.-X.; Kong, F.-Z.; Wang, J.-X.; Zhang, Q.-C.; Li, F.; Hong, X.; Song, M.-J.; Lian, Z.; Cai, Y.-L.; Yu, R.-C. An unusual winter bloom of dinoflagellates with notable damage to kelp cultivation around Shandong peninsula, China. Mar. Environ. Res. 2024, 201, 106687. [Google Scholar] [CrossRef]
- Hallegraeff, G.M.; Anderson, D.M.; Belin, C.; Bottein, M.-Y.D.; Bresnan, E.; Chinain, M.; Enevoldsen, H.; Iwataki, M.; Karlson, B.; McKenzie, C.H. Perceived global increase in algal blooms is attributable to intensified monitoring and emerging bloom impacts. Commun. Earth Environ. 2021, 2, 117. [Google Scholar] [CrossRef]
- Bravo, I.; Figueroa, R.I. Towards an ecological understanding of dinoflagellate cyst functions. Microorganisms 2014, 2, 11–32. [Google Scholar] [CrossRef]
- Anderson, D.M.; Keafer, B.A.; Kleindinst, J.L.; McGillicuddy, D.J., Jr.; Martin, J.L.; Norton, K.; Pilskaln, C.H.; Smith, J.L.; Sherwood, C.R.; Butman, B. Alexandrium fundyense cysts in the Gulf of Maine: Long-term time series of abundance and distribution, and linkages to past and future blooms. Deep Sea Res. Part II Top. Stud. Oceanogr. 2014, 103, 6–26. [Google Scholar] [CrossRef]
- Rodríguez-Villegas, C.; Figueroa, R.I.; Baldrich, Á.M.; Pérez-Santos, I.; Díaz, M.; Tomasetti, S.J.; Seguel, M.; Álvarez, G.; Salgado, P.; Díaz, P.A. Small and patchy is enough: An example about how toxic HAB events can spread through low resting cyst loads. Harmful Algae 2023, 129, 102495. [Google Scholar] [CrossRef]
- Deng, Y.; Yue, C.; Yang, H.; Li, F.; Hu, Z.; Shang, L.; Chai, Z.; Lin, S.; Tang, Y.Z. Broad active metabolic pathways, autophagy, and antagonistic hormones regulate dinoflagellate cyst dormancy in marine sediments. Sci. Adv. 2025, 11, eads7789. [Google Scholar] [CrossRef]
- Morimoto, R.I. The Heat Shock Response: Systems Biology of Proteotoxic Stress in Aging and Disease; Cold Spring Harbor Symposia on Quantitative Biology: New York, NY, USA, 2011; pp. 91–99. [Google Scholar]
- Al-Whaibi, M.H. Plant heat-shock proteins: A mini review. J. King. Saud. Univ. Sci. 2011, 23, 139–150. [Google Scholar] [CrossRef]
- Saibil, H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Bio. 2013, 14, 630–642. [Google Scholar] [CrossRef]
- Kumar, R.; Khungar, L.; Shimphrui, R.; Tiwari, L.D.; Tripathi, G.; Sarkar, N.K.; Agarwal, S.-K.; Agarwal, M.; Grover, A. AtHsp101 research sets course of action for the genetic improvement of crops against heat stress. J. Plant. Biochem. Biot. 2020, 29, 715–732. [Google Scholar] [CrossRef]
- Santos, T.F.; Pereira, H.; Schüler, L.; Maia, I.B.; Jacinto, R.; Bombo, G.; Pinheiro, F.; Barreira, L.; Varela, J. Enhancement of heat tolerance by salt stress in Tetraselmis striata CTP4: Impacts on HSP gene expression, pigments, and proximal composition. J. Appl. Phycol. 2025, 37, 287–301. [Google Scholar] [CrossRef]
- Agarwal, M.; Katiyar-Agarwal, S.; Grover, A. Plant Hsp100 proteins: Structure, function and regulation. Plant Sci. 2002, 163, 397–405. [Google Scholar] [CrossRef]
- Mogk, A.; Kummer, E.; Bukau, B. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front. Mol. Biosci. 2015, 2, 22. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.C.; Grover, A. ClpB/Hsp100 proteins and heat stress tolerance in plants. Crit. Rev. Biotechnol. 2016, 36, 862–874. [Google Scholar] [CrossRef]
- Deng, Y.; Hu, Z.; Zhan, Z.; Ma, Z.; Tang, Y. Differential expressions of an Hsp70 gene in the dinoflagellate Akashiwo sanguinea in response to temperature stress and transition of life cycle and its implications. Harmful Algae 2015, 50, 57–64. [Google Scholar] [CrossRef]
- Deng, Y.; Li, F.; Hu, Z.; Yue, C.; Tang, Y.Z. Expression patterns of the heat shock protein 90 (Hsp90) gene suggest its possible involvement in maintaining the dormancy of dinoflagellate resting cysts. Int. J. Mol. Sci. 2021, 22, 11054. [Google Scholar] [CrossRef]
- Parkinson, J.E.; Baumgarten, S.; Michell, C.T.; Baums, I.B.; LaJeunesse, T.C.; Voolstra, C.R. Gene expression variation resolves species and individual strains among coral-associated dinoflagellates within the genus Symbiodinium. Genome Biol. Evol. 2016, 8, 665–680. [Google Scholar] [CrossRef]
- Kretschmann, J.; Elbrächter, M.; Zinssmeister, C.; Soehner, S.; Kirsch, M.; Kusber, W.-H.; Gottschling, M. Taxonomic clarification of the dinophyte Peridinium acuminatum Ehrenb.,≡ Scrippsiella acuminata, comb. nov. (Thoracosphaeraceae, Peridiniales). Phytotaxa 2015, 220, 239–256. [Google Scholar] [CrossRef]
- Deng, Y.; Hu, Z.; Shang, L.; Peng, Q.; Tang, Y.Z. Transcriptomic analyses of Scrippsiella trochoidea reveals processes regulating encystment and dormancy in the life cycle of a dinoflagellate, with a particular attention to the role of abscisic acid. Front. Microbiol. 2017, 8, 2450. [Google Scholar] [CrossRef]
- Wang, H.; Kim, H.; Ki, J.S. Transcriptome survey, molecular identification, and expression analysis of stress-responsive genes in the toxic dinoflagellate Alexandrium pacificum under algicidal agents and metal stresses. J. Appl. Phycol. 2021, 33, 3139–3151. [Google Scholar] [CrossRef]
- Wang, D.Z.; Zhang, Y.J.; Zhang, S.F.; Lin, L.; Hong, H.S. Quantitative proteomic analysis of cell cycle of the dinoflagellate Prorocentrum donghaiense (Dinophyceae). PLoS ONE 2013, 8, 63659. [Google Scholar] [CrossRef]
- Zhang, S.F.; Chen, Y.; Xie, Z.X.; Zhang, H.; Lin, L.; Wang, D.-Z. Unraveling the molecular mechanism of the response to changing ambient phosphorus in the dinoflagellate Alexandrium catenella with quantitative proteomics. J. Proteom. 2019, 196, 141–149. [Google Scholar] [CrossRef]
- Deng, Y.; Hu, Z.; Shang, L.; Chai, Z.; Tang, Y.Z. Transcriptional responses of the heat shock protein 20 (Hsp20) and 40 (Hsp40) genes to temperature stress and alteration of life cycle stages in the harmful alga Scrippsiella trochoidea (Dinophyceae). Biology 2020, 9, 408. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Hu, Z.; Chai, Z.; Tang, Y.Z. Molecular cloning of heat shock protein 60 (Hsp60) and 10 (Hsp10) genes from the cosmopolitan and harmful dinoflagellate Scrippsiella trochoidea and their differential transcriptions responding to temperature stress and alteration of life cycle. Mar. Biol. 2019, 166, 7. [Google Scholar] [CrossRef]
- Guillard, R.R. Culture of Phytoplankton for Feeding Marine Invertebrates. In Culture of Marine Invertebrate Animals: Proceedings—1st Conference on Culture of Marine Invertebrate Animals Greenport; Plenum Press: New York, NY, USA, 1975; pp. 29–60. [Google Scholar]
- Zhang, H.; Hou, Y.; Miranda, L.; Campbell, D.A.; Sturm, N.R.; Gaasterland, T.; Lin, S. Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl. Acad. Sci. USA 2007, 104, 4618–4623. [Google Scholar] [CrossRef]
- Rombel, I.T.; Sykes, K.F.; Rayner, S.; Johnston, S.A. ORF-FINDER: A vector for high-throughput gene identification. Gene 2002, 282, 33–41. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.M. The Proteomics Protocols Handbook; Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Yue, C.; Chai, Z.; Hu, Z.; Shang, L.; Deng, Y.; Tang, Y.Z. Deficiency of nitrogen but not phosphorus triggers the life cycle transition of the dinoflagellate Scrippsiella acuminata from vegetative growth to resting cyst formation. Harmful Algae 2022, 118, 102312. [Google Scholar] [CrossRef]
- Li, F.; Yue, C.; Deng, Y.; Tang, Y.Z. Characterizing the status of energetic metabolism of dinoflagellate resting cysts under mock conditions of marine sediments via physiological and transcriptional measurements. Int. J. Mol. Sci. 2022, 23, 15033. [Google Scholar] [CrossRef]
- Xiao, J.; Zhang, Y.; Wang, J.; Yu, W.; Wang, W.; Ma, X. Monitoring of cell viability and proliferation in hydrogel-encapsulated system by resazurin assay. Appl. Biochem. Biotechnol. 2010, 162, 1996–2007. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Zakrajsek, B.A. Effect of experimental treatment on housekeeping gene expression: Validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods 2000, 46, 69–81. [Google Scholar] [CrossRef]
- Peterson, A.C.; Russell, J.D.; Bailey, D.J.; Westphall, M.S.; Coon, J.J. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteom. 2012, 11, 1475–1488. [Google Scholar] [CrossRef]
- Schaeffer, M.; Gateau, A.; Teixeira, D.; Michel, P.-A.; Zahn-Zabal, M.; Lane, L. The neXtProt peptide uniqueness checker: A tool for the proteomics community. Bioinformatics 2017, 33, 3471–3472. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Yue, C.; Deng, Y.; Tang, Y.Z. Full-length transcriptome analysis of a bloom-forming dinoflagellate Scrippsiella acuminata (Dinophyceae). Sci. Data 2025, 12, 352. [Google Scholar] [CrossRef]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef]
- Escher, C.; Reiter, L.; MacLean, B.; Ossola, R.; Herzog, F.; Chilton, J.; MacCoss, M.J.; Rinner, O. Using i RT, a normalized retention time for more targeted measurement of peptides. Proteomics 2012, 12, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Aguilar, J.; Molloy, M.P. Label-free selected reaction monitoring enables multiplexed quantitation of S100 protein isoforms in cancer cells. J. Proteome Res. 2013, 12, 3679–3688. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.; Chang, C.; Vitek, O. MSstats: Protein Significance Analysis in DDA, SRM and DIA for Label-Free or Label-Based Proteomics Experiments. Available online: http://bioconductor.jp/packages/3.14/bioc/manuals/MSstats/man/MSstats.pdf (accessed on 31 May 2021).
- Sørensen, J.G.; Kristensen, T.N.; Loeschcke, V. The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 2003, 6, 1025–1037. [Google Scholar] [CrossRef]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).