Utilizing Remote Sensing Data for Species Distribution Modeling of Birds in Croatia
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Elith, J.; Leathwick, J.R. Species distribution models: Ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- Gallien, L.; Münkemüller, T.; Albert, C.H.; Boulangeat, I.; Thuiller, W. Predicting potential distributions of invasive species: Where to go from here? Divers. Distrib. 2010, 16, 331–342. [Google Scholar] [CrossRef]
- Bannari, A.; Huete, A.R.; Morin, D.; Bonn, F. A review of vegetation indices. Remote Sens. Rev. 1995, 13, 95–120. [Google Scholar] [CrossRef]
- Peterson, A.T.; Soberón, J.; Pearson, R.G.; Anderson, R.P.; Martínez-Meyer, E.; Nakamura, M.; Araújo, M.B. Ecological Niches and Geographic Distributions; Princeton University Press: Princeton, NJ, USA, 2011. [Google Scholar] [CrossRef]
- Guisan, A.; Thuiller, W. Predicting species distribution: Offering more than simple habitat models. Ecol. Lett. 2005, 8, 993–1009. [Google Scholar] [CrossRef]
- Broennimann, O.; Treier, U.A.; Müller-Schärer, H.; Thuiller, W.; Peterson, A.T.; Guisan, A. Analyzing niche dynamics during biological invasions. Ecol. Lett. 2007, 10, 701–719. [Google Scholar] [CrossRef]
- Stein, A.; Gerstner, K.; Kreft, H. Environmental heterogeneity as a universal driver of species richness across taxa, biomes and spatial scales. Ecol. Lett. 2014, 17, 866–880. [Google Scholar] [CrossRef]
- Petersen, C. Integrating remote sensing data into species distribution models. Ecography 2013, 36, 789–800. [Google Scholar]
- Merow, C.; Smith, M.J.; Silander, J.A., Jr. A practical guide to MaxEnt for modeling species’ distributions: What it does, and why inputs and settings matter. Ecography 2013, 36, 1058–1069. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.R.; Hastie, T. A working guide to boosted regression trees. J. Anim. Ecol. 2011, 77, 802–813. [Google Scholar] [CrossRef]
- He, K.S.; Bradley, B.A.; Cord, A.F.; Rocchini, D.; Tuanmu, M.N.; Schmidtlein, S.; Turner, W.; Wegmann, M.; Pettorelli, N. Integrating remote sensing data into species distribution models. Ecography 2013, 36, 789–800. [Google Scholar]
- Franklin, J. Mapping Species Distributions: Spatial Inference and Prediction; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar] [CrossRef]
- Thomas, C.D.; Cameron, A.; Green, R.E.; Bakkenes, M.; Beaumont, L.J.; Collingham, Y.C.; Erasmus, B.F.N.; Ferreira de Siqueira, M.; Grainger, A.; Hannah, L.; et al. Extinction risk from climate change. Nature 2004, 427, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Pearson, R.G.; Dawson, T.P. Predicting the impacts of climate change on the distribution of species: Are bioclimate envelope models useful? Glob. Ecol. Biogeogr. 2003, 12, 361–371. [Google Scholar] [CrossRef]
- Bueno de Mesquita, C.P.; King, A.J.; Schmidt, S.K.; Farrer, E.C.; Suding, K.N. Incorporating biotic factors in species distribution modelling: Are interactions with soil microbes important? Ecography 2016, 39, 970–980. [Google Scholar] [CrossRef]
- Zhang, L. Use of remote sensing in biodiversity monitoring and ecosystem management. Biodivers. Conserv. 2019, 28, 2225–2237. [Google Scholar]
- Hirzel, A.H.; Le Lay, G. Habitat suitability modelling and niche theory. J. Appl. Ecol. 2008, 45, 1372–1381. [Google Scholar] [CrossRef]
- GBIF. Global Biodiversity Information Facility. Available online: https://www.gbif.org/ (accessed on 15 December 2024).
- Kapelj, S.; Radović, A.; Zec, M.; Mihelič, T.; Mikac, S.; Maslać Mikulec, M.; Patčev, E.; Dender, D.; Taylor, L.; Mikuška, T.; et al. Završno Izvješće Usluge Definiranja SMART Ciljeva Očuvanja i Osnovnih Mjera Očuvanja Ciljnih Vrsta i Stanišnih Tipova—Grupa 5: Definiranje Ciljeva i Mjera Očuvanja za Nedovoljno Poznate Prste Ptica; Udruga BIOM, Geonatura, DOPPS: Zagreb, Croatia, 2023; p. 36. [Google Scholar]
- European Environmental Agency. Environmental Datasets. Available online: https://www.eea.europa.eu/en/datahub/datahubitem-view/3c362237-daa4-45e2-8c16-aaadfb1a003b (accessed on 15 May 2023).
- EEA, EU_DEM Ver 1.1., 2016. Available online: https://www.eea.europa.eu/en/datahub/datahubitem-view/d08852bc-7b5f-4835-a776-08362e2fbf4b (accessed on 10 July 2023).
- Habitat Map of Croatia. Available online: https://www.haop.hr/hr/baze-i-portali/karta-stanista-rh-2004 (accessed on 10 July 2023).
- Non-Forest Habitat Map of Croatia. Available online: https://www.haop.hr/hr/baze-i-portali/karta-kopnenih-nesumskih-stanista-republike-hrvatske-2016 (accessed on 16 June 2023).
- Zenodo. Copernicus Land Cover Data. Available online: https://zenodo.org/communities/copernicus-land-cover/ (accessed on 10 June 2023).
- Riitters, K.H.; O’Neill, R.V.; Hunsaker, C.T.; Wickham, J.D.; Yankee, D.H.; Timmins, S.P.; Jones, K.B.; Jackson, B.L. A factor analysis of landscape pattern and structure metrics. Landsc. Ecol. 1995, 10, 23–40. [Google Scholar] [CrossRef]
- Hesselbarth, M.H.K.; Sciaini, M.; With, K.A.; Wiegand, K.; Nowosad, J. landscapemetrics: An open-source R tool to calculate landscape metrics. Ecography 2019, 42, 1648–1657. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Barbosa, M.; Ghosh, A.; Mandel, A. _geodata: Download Geographic Data_. R package version 0.5–9. 2023. Available online: https://CRAN.R-project.org/package=geodata (accessed on 10 June 2023).
- R Core Team. _R: A Language and Environment for Statistical Computing_; R Foundation for Statistical Computing: Vienna, Austria; Available online: https://www.R-project.org/ (accessed on 10 June 2023).
- Phillips, S.J.; Dudík, M. Understanding and applying MaxEnt, a machine-learning approach for species distribution modeling. J. Biogeogr. 2008, 35, 1–11. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2002, 2, 18–22. [Google Scholar]
- Schauberger, P.; Walker, A. _openxlsx: Read, Write and Edit xlsx Files_. R package version 4.2.5.2. Available online: https://CRAN.R-project.org/package=openxlsx (accessed on 10 June 2023).
- Pebesma, E. Simple features for R: Standardized support for spatial vector data. R J. 2018, 10, 1–8. [Google Scholar] [CrossRef]
- Pebesma, E.; Bivand, R. Spatial Data Science: With Applications in R, 1st ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 2023. [Google Scholar] [CrossRef]
- Aiello-Lammens, M.E.; Boria, R.A.; Radosavljevic, A.; Vilela, B.; Anderson, R.P. spThin: An R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 2015, 38, 541–545. Available online: https://onlinelibrary.wiley.com/doi/10.1111/ecog.01132 (accessed on 15 June 2023).
- Hijmans, R.; Phillips, S.; Leathwick, J.; Elith, J. _dismo: Species Distribution Modeling_. R Package Version 1.3–16, 2024. Available online: https://CRAN.R-project.org/package=dismo (accessed on 15 June 2023).
- Bivand, R.; Rundel, C. rgeos: Interface to Geometry Engine—Open Source (‘GEOS’); R Package Version 0.6-2. Available online: https://CRAN.R-project.org/package=rgeos (accessed on 15 June 2023).
- Kass, J.M.; Pinilla-Buitrago, G.E.; Paz, A.; Johnson, B.A.; Grisales-Betancur, V.; Meenan, S.I.; Attali, D.; Broennimann, O.; Galante, P.J.; Maitner, B.S.; et al. wallace 2: A shiny app for modeling species niches and distributions redesigned to facilitate expansion via module contributions. Ecography 2023, 2023, e06547. [Google Scholar] [CrossRef]
- Hijmans, R. Terra: Spatial Data Analysis; R Package Version 1.7–29. Available online: https://CRAN.R-project.org/package=terra (accessed on 15 June 2023).
- Hijmans, R. _raster: Geographic Data Analysis and Modeling_. R Package Version 3.6–26, 2023. Available online: https://CRAN.R-project.org/package=raster (accessed on 15 June 2023).
- Kass, J.M.; Muscarella, R.; Galante, P.J.; Bohl, C.L.; Pinilla-Buitrago, G.E.; Boria, R.A.; Soley-Guardia, M.; Anderson, R.P. ENMeval 2.0: Redesigned for customizable and reproducible modeling of species’ niches and distributions. Methods Ecol. Evol. 2021, 12, 1602–1608. [Google Scholar] [CrossRef]
- Long, A.M.; Pierce, B.L.; Anderson, A.D.; Skow, K.L.; Smith, A.; Lopez, R.R. Integrating citizen science and remotely sensed data to help inform time-sensitive policy decisions for species of conservation concern. Biol. Conserv. 2019, 237, 463–469. [Google Scholar] [CrossRef]
- Valavi, R.; Guillera-Arroita, G.; Lahoz-Monfort, J.J.; Elith, J. Predictive performance of presence-only species distribution models: A benchmark study with reproducible code. Ecol. Monogr. 2022, 92, 1. [Google Scholar] [CrossRef]
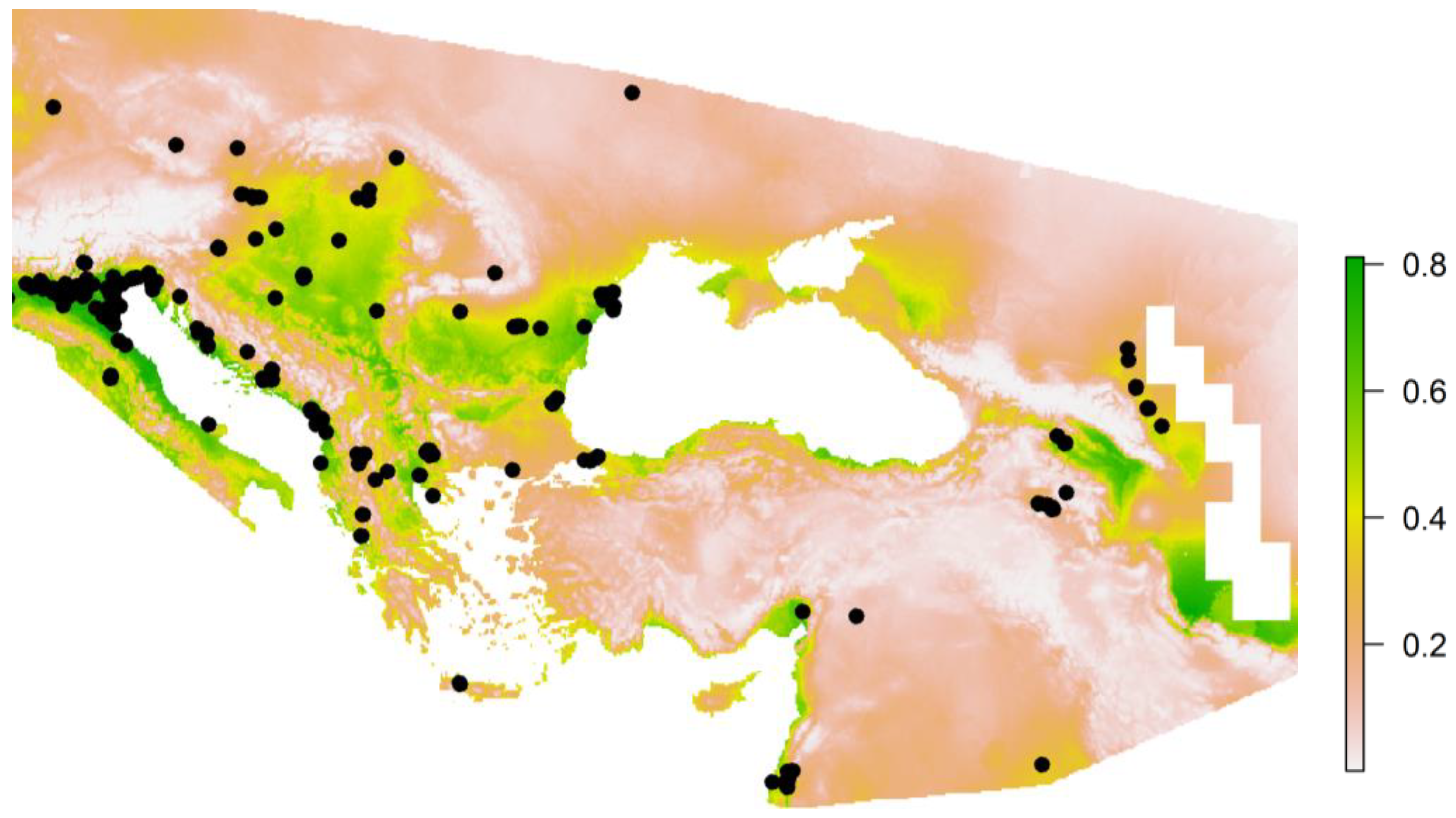
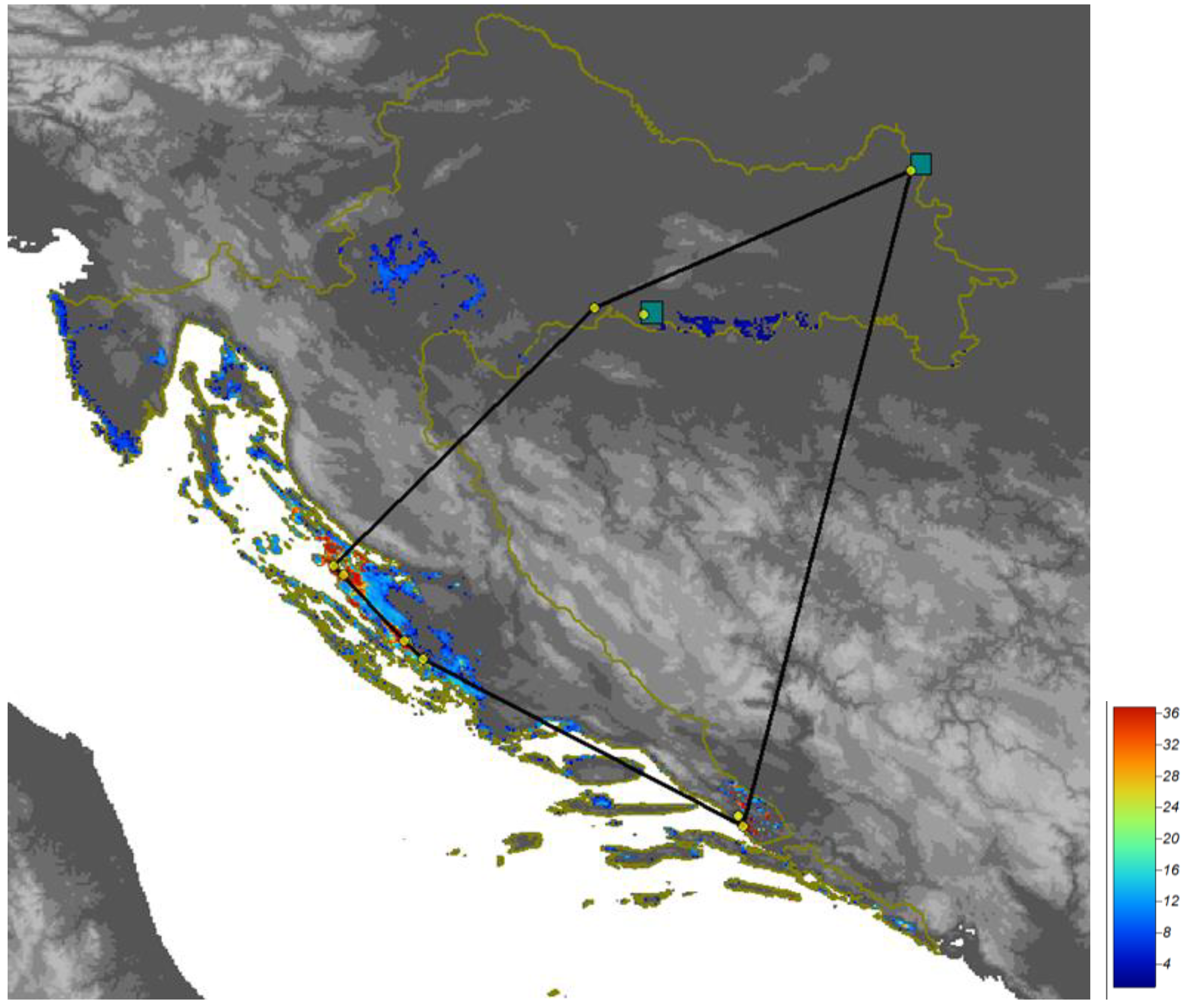
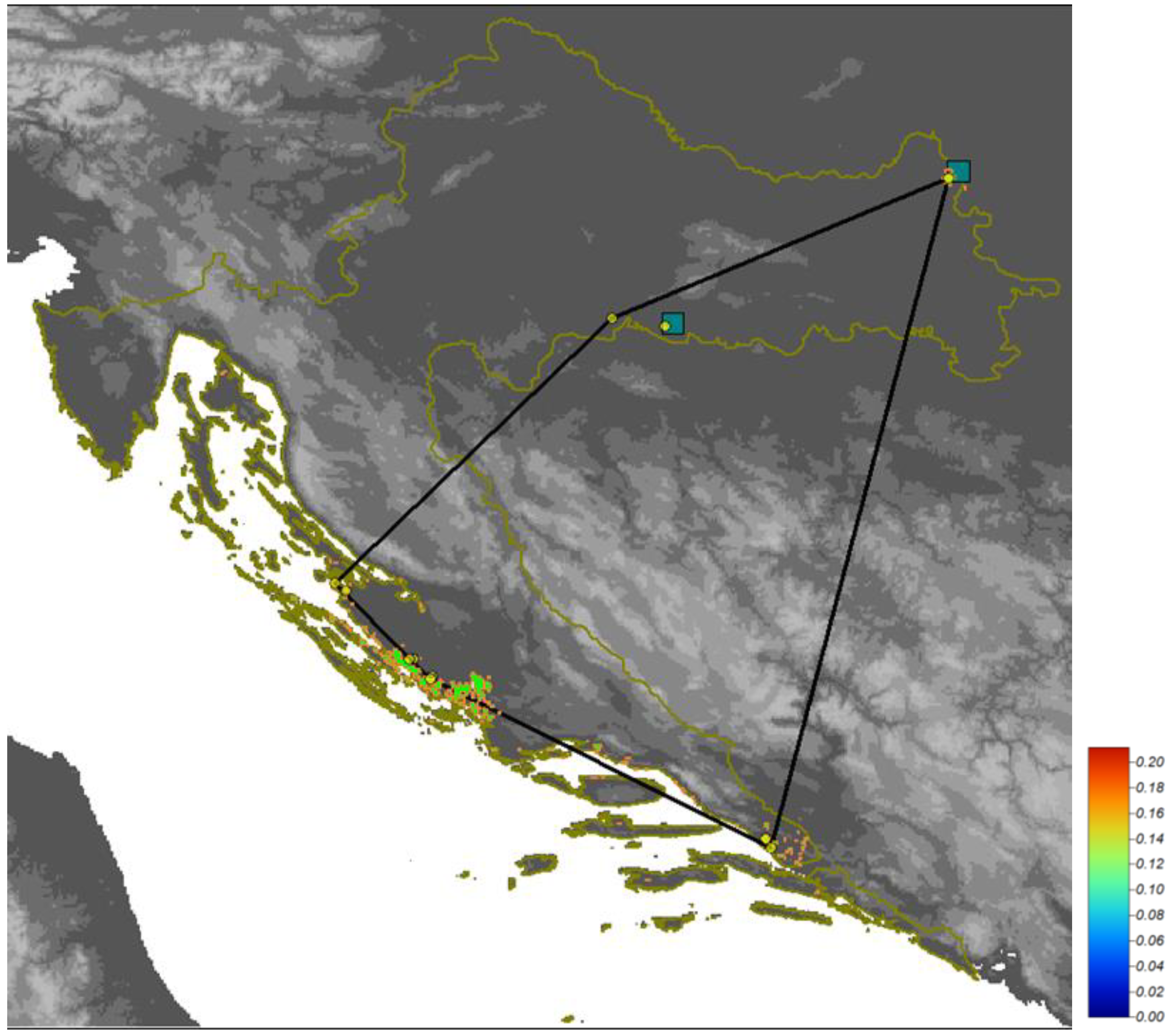
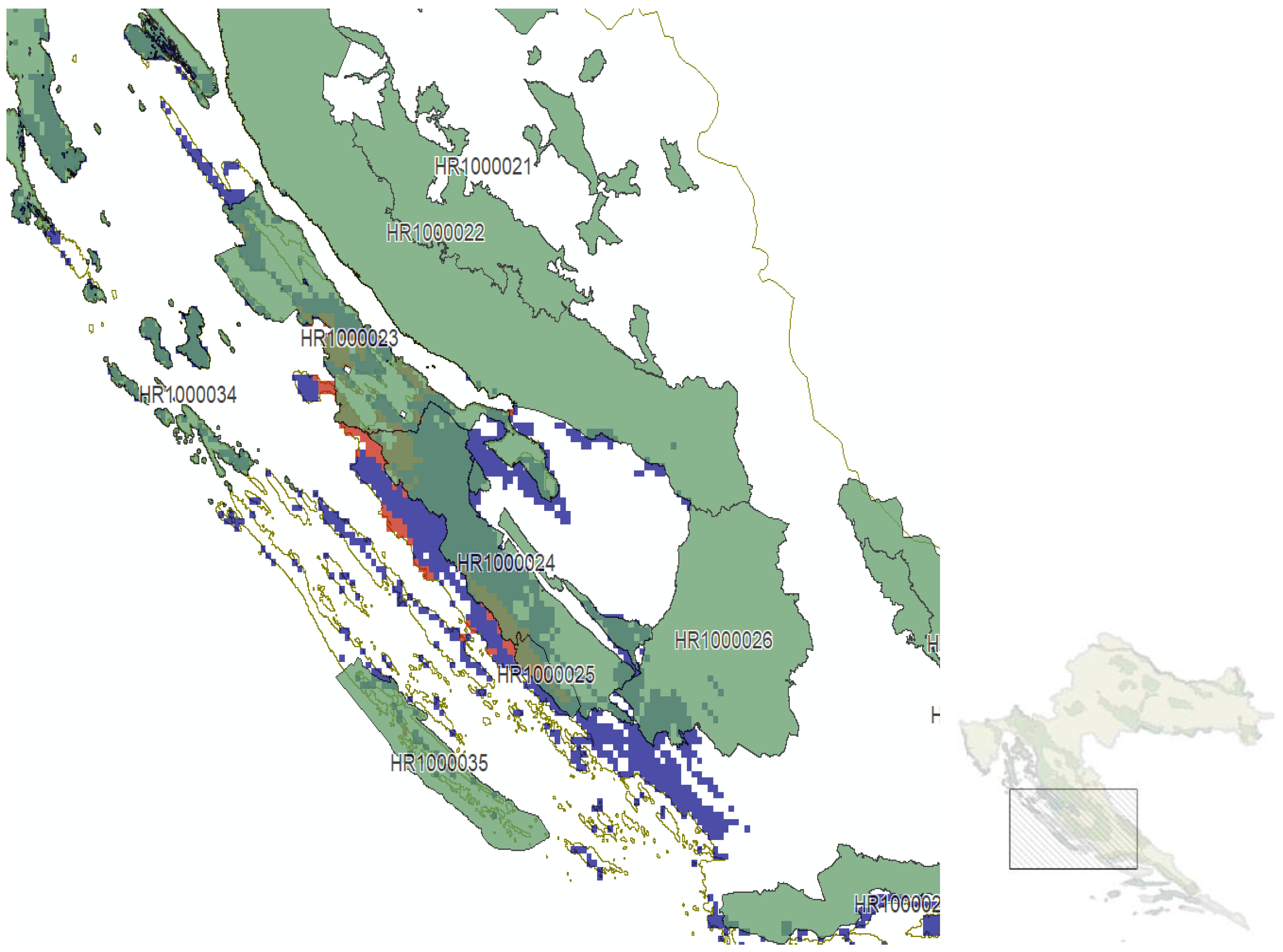
| Ecological Group | Species |
|---|---|
| Woodpeckers and Picids | Dendrocopos leucotos, Dendrocopos major, Dendrocopos syriacus, Dryobates minor, Dryocopus martius, Leiopicus medius, Picoides tridactylus, Picus canus, Picus viridis |
| Birds of Prey (Raptors) | Circaetus gallicus, Accipiter brevipes, Accipiter gentilis, Tachyspiza nisus, Aquila fasciata, Clanga pomarina, Buteo buteo, Falco subbuteo, Falco vespertinus, Falco biarmicus, Falco columbarius, Hieraaetus pennatus, Pernis apivorus, Circus cyaneus, Milvus migrans |
| Game Birds | Alectoris graeca, Tetrao urogallus, Bonasa bonasia |
| Nocturnal Birds | Caprimulgus europaeus, Aegolius funereus, Glaucidium passerinum |
| Herons, Egrets, and Allies | Ardea alba, Ardea cinerea, Ardea purpurea, Ardeola ralloides, Egretta garzetta, Nycticorax nycticorax |
| Spoonbills and Ibises | Platalea leucorodia, Plegadis falcinellus |
| Cormorants | Microcarbo pygmaeus, Phalacrocorax carbo sinensis |
| Terns and Gulls | Chlidonias hybrida, Chlidonias niger, Thalasseus sandvicensis, Hydrocoloeus minutus, Larus melanocephalus, Larus ridibundus |
| Rails and Crakes | Fulica atra, Gallinula chloropus, Porzana porzana, Rallus aquaticus, Zapornia parva, Zapornia pusilla |
| Ducks, Geese, and Swans | Anas acuta, Anas crecca, Anas platyrhynchos, Anser albifrons albifrons, Anser anser, Anser fabalis rossicus, Aythya ferina, Aythya fuligula, Aythya nyroca, Bucephala clangula, Cygnus olor, Mareca penelope, Mareca strepera, Netta rufina, Spatula clypeata, Spatula querquedula |
| Grebes | Podiceps cristatus, Podiceps grisegena, Podiceps nigricollis, Tachybaptus ruficollis |
| Passerines | Alcedo atthis, Riparia riparia, Acrocephalus arundinaceus, Acrocephalus melanopogon, Acrocephalus schoenobaenus, Acrocephalus scirpaceus, Cettia cetti, Cisticola juncidis, Emberiza schoeniclus, Locustella fluviatilis, Locustella luscinioides, Panurus biarmicus, Remiz pendulinus, Hippolais olivetorum |
| Shorebirds and Waders | Actitis hypoleucos, Calidris alpina, Calidris pugnax, Charadrius alexandrinus, Charadrius dubius, Grus grus, Haematopus ostralegus, Himantopus himantopus, Limosa limosa, Numenius arquata arquata, Numenius phaeopus, Pluvialis squatarola, Recurvirostra avosetta, Tringa erythropus, Tringa glareola, Tringa nebularia |
| Large Waterbirds | Botaurus stellaris, Ixobrychus minutus |
| Variable | Description | Abs Significance |
|---|---|---|
| DEM | Digital elevation model | 0.01715 |
| BIO_5 | Max Temperature of Warmest Month | 0.01437 |
| AGG_A4 | Area of habitat A1 (element of inland surface water and wetlands) | 0.01173 |
| BIO_9 | Mean Temperature of Driest Quarter | 0.00879 |
| BIO_18 | Precipitation of Warmest Quarter | 0.00845 |
| BIO_1 | Annual Mean Temperature | 0.00683 |
| BIO_3 | Isothermality | 0.00609 |
| BIO_10 | Mean Temperature of Warmest Quarter | 0.00554 |
| AGG_A1 | Area of habitat A4 (element of inland surface water and wetlands) | 0.00548 |
| Category | Variable Name | Description |
|---|---|---|
| Morphometric Variables | Digital Elevation Model (DEM) | Surface elevation model |
| Wetness Index (WI) | Potential water accumulation index | |
| Slope | Terrain slope | |
| Habitat Variables | Habitat Type Presence | Presence (1) or absence (0) of habitat type at reference grid |
| Habitat Type Area | Area of each habitat type at 1 km reference grid | |
| Aggregated Habitat Types | Spatial aggregation of habitat types up to 2nd level of national classification scheme (e.g., agg_A1, agg_A2, agg_B12, etc.) | |
| Habitat Heterogeneity Variables | Averaged Connectivity | Average connectivity between habitat fragments |
| Connectivity | Connectivity between habitat fragments | |
| Diversity | Degree of habitat heterogeneity | |
| Number of Categories | Number of different land cover categories per grid | |
| Bioclimatic Variables (WorldClim BIOCLIM) | BIO1—Annual Mean Temperature | Average annual temperature |
| BIO2—Mean Diurnal Range | Mean difference between daily max and min temperatures | |
| BIO3—Isothermality | Ratio of mean diurnal range to the annual temperature range | |
| BIO4—Temperature Seasonality | Variation in temperature throughout the year | |
| BIO5—Max Temperature of Warmest Month | Maximum temperature of the hottest month | |
| BIO6—Min Temperature of Coldest Month | Minimum temperature of the coldest month | |
| BIO7—Temperature Annual Range | Difference between BIO5 and BIO6 | |
| BIO8—Mean Temperature of Wettest Quarter | Mean temperature during the wettest 3-month period | |
| BIO9—Mean Temperature of Driest Quarter | Mean temperature during the driest 3-month period | |
| BIO10—Mean Temperature of Warmest Quarter | Mean temperature during the warmest 3-month period | |
| BIO11—Mean Temperature of Coldest Quarter | Mean temperature during the coldest 3-month period | |
| BIO12—Annual Precipitation | Total annual precipitation | |
| BIO13—Precipitation of Wettest Month | Precipitation in the wettest month | |
| BIO14—Precipitation of Driest Month | Precipitation in the driest month | |
| BIO15—Precipitation Seasonality | Variation in monthly precipitation levels | |
| BIO16—Precipitation of Wettest Quarter | Total precipitation in the wettest 3-month period | |
| BIO17—Precipitation of Driest Quarter | Total precipitation in the driest 3-month period | |
| BIO18—Precipitation of Warmest Quarter | Total precipitation in the warmest 3-month period | |
| BIO19—Precipitation of Coldest Quarter | Total precipitation in the coldest 3-month period | |
| Land Cover Categories (ESA LC 2021) | Tree Cover | Percentage of tree cover |
| Shrubland | Percentage of shrub cover | |
| Grassland | Percentage of grassland cover | |
| Cropland | Percentage of agricultural land | |
| Built-up Areas | Percentage of artificial surfaces | |
| Bare/Sparse Vegetation | Percentage of barren land | |
| Snow and Ice | Percentage of snow and ice cover | |
| Permanent Water Bodies | Percentage of surface covered by water | |
| Herbaceous Wetland | Percentage of wetland cover | |
| Mangrove | Percentage of mangrove cover | |
| Moss and Lichen | Percentage of moss and lichen cover |
| Sort Site ID | Count/Area of the Site | N | MIN | MAX | MEAN |
|---|---|---|---|---|---|
| 0 | 40,139,618 | 177,536 | 1 | 62 | 10.32 |
| 1 | 228,484 | 0 | |||
| 2 | 46,923 | 0 | |||
| 3 | 31,817 | 2242 | 7 | 12 | 10.73 |
| 4 | 204,609 | 1641 | 2 | 31 | 11.38 |
| 5 | 97,578 | 0 | |||
| 6 | 14,486 | 0 | |||
| 7 | 45,377 | 0 | |||
| 8 | 24,664 | 0 | |||
| 9 | 5883 | 5126 | 1 | 51 | 29.41 |
| 10 | 21,768 | 0 | |||
| 11 | 61,252 | 27,452 | 1 | 62 | 24.56 |
| 12 | 10,752 | 0 | |||
| 13 | 23,405 | 0 | |||
| 14 | 88,764 | 7891 | 1 | 29 | 10.12 |
| 15 | 67,109 | 47,896 | 1 | 62 | 17.57 |
| 16 | 69,104 | 0 | |||
| 17 | 87,799 | 0 | |||
| 18 | 30,239 | 0 | |||
| 19 | 38,423 | 235 | 4 | 59 | 9.95 |
| 20 | 47,334 | 1403 | 1 | 12 | 6.92 |
| 21 | 14,550 | 0 | |||
| 22 | 20,203 | 600 | 3 | 9 | 5.66 |
| 23 | 20,571 | 0 | |||
| 24 | 39 | 0 | |||
| 25 | 38,410 | 0 | |||
| 26 | 85,758 | 8455 | 1 | 29 | 8.25 |
| 27 | 14,594 | 7076 | 1 | 19 | 10.72 |
| 28 | 13,464 | 0 | |||
| 29 | 18,267 | 3990 | 1 | 9 | 5.96 |
| 30 | 1928 | 196 | 1 | 11 | 7.98 |
| 31 | 39,973 | 14,091 | 1 | 4 | 1.97 |
| 32 | 35,839 | 17,417 | 1 | 10 | 6.04 |
| 33 | 123,454 | 1198 | 1 | 4 | 2.41 |
| 34 | 1668 | 0 | |||
| 35 | 23,790 | 0 | |||
| 36 | 29,287 | 2842 | 3 | 34 | 9.33 |
| 37 | 24,891 | 5971 | 1 | 62 | 32.36 |
| 38 | 120,927 | 33,542 | 1 | 32 | 8.69 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radović, A.; Kapelj, S.; Taylor, L.T. Utilizing Remote Sensing Data for Species Distribution Modeling of Birds in Croatia. Diversity 2025, 17, 399. https://doi.org/10.3390/d17060399
Radović A, Kapelj S, Taylor LT. Utilizing Remote Sensing Data for Species Distribution Modeling of Birds in Croatia. Diversity. 2025; 17(6):399. https://doi.org/10.3390/d17060399
Chicago/Turabian StyleRadović, Andreja, Sven Kapelj, and Louie Thomas Taylor. 2025. "Utilizing Remote Sensing Data for Species Distribution Modeling of Birds in Croatia" Diversity 17, no. 6: 399. https://doi.org/10.3390/d17060399
APA StyleRadović, A., Kapelj, S., & Taylor, L. T. (2025). Utilizing Remote Sensing Data for Species Distribution Modeling of Birds in Croatia. Diversity, 17(6), 399. https://doi.org/10.3390/d17060399





