IT Challenges in Designing and Implementing Online Natural History Collection Systems
Abstract
1. Introduction
2. Materials and Methods
3. Challenges Categorisation
4. Requirements
4.1. Target Groups
4.1.1. Scientists
4.1.2. State and Local Government Administration
4.1.3. Services and State Officials
4.1.4. Non-Governmental Organisations and Society
4.1.5. Education
4.2. Interdisciplinarity
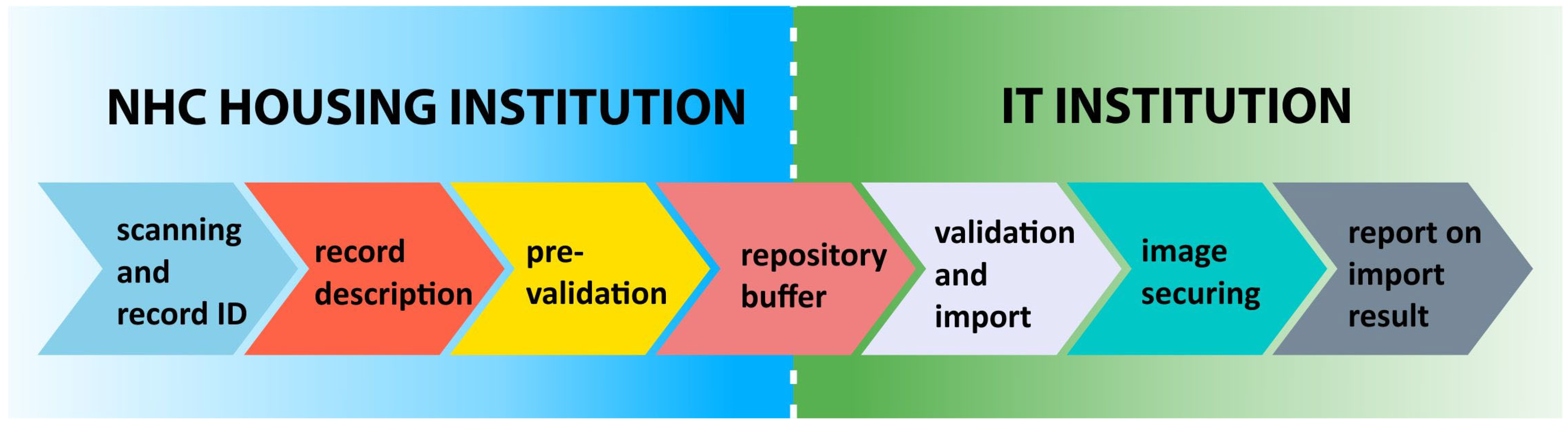
5. Digitisation
5.1. Digitisation Process
5.2. Data Quality—Date Uncertainty and Ambiguity
5.3. Iconographic Data Formats
5.4. Automation of Digitalisation Procedures
6. Design
6.1. Metadata Definition
6.2. Data Access Restrictions
6.2.1. Specimen Data Protection Levels
6.2.2. Record Field Protection Levels
6.2.3. User Roles
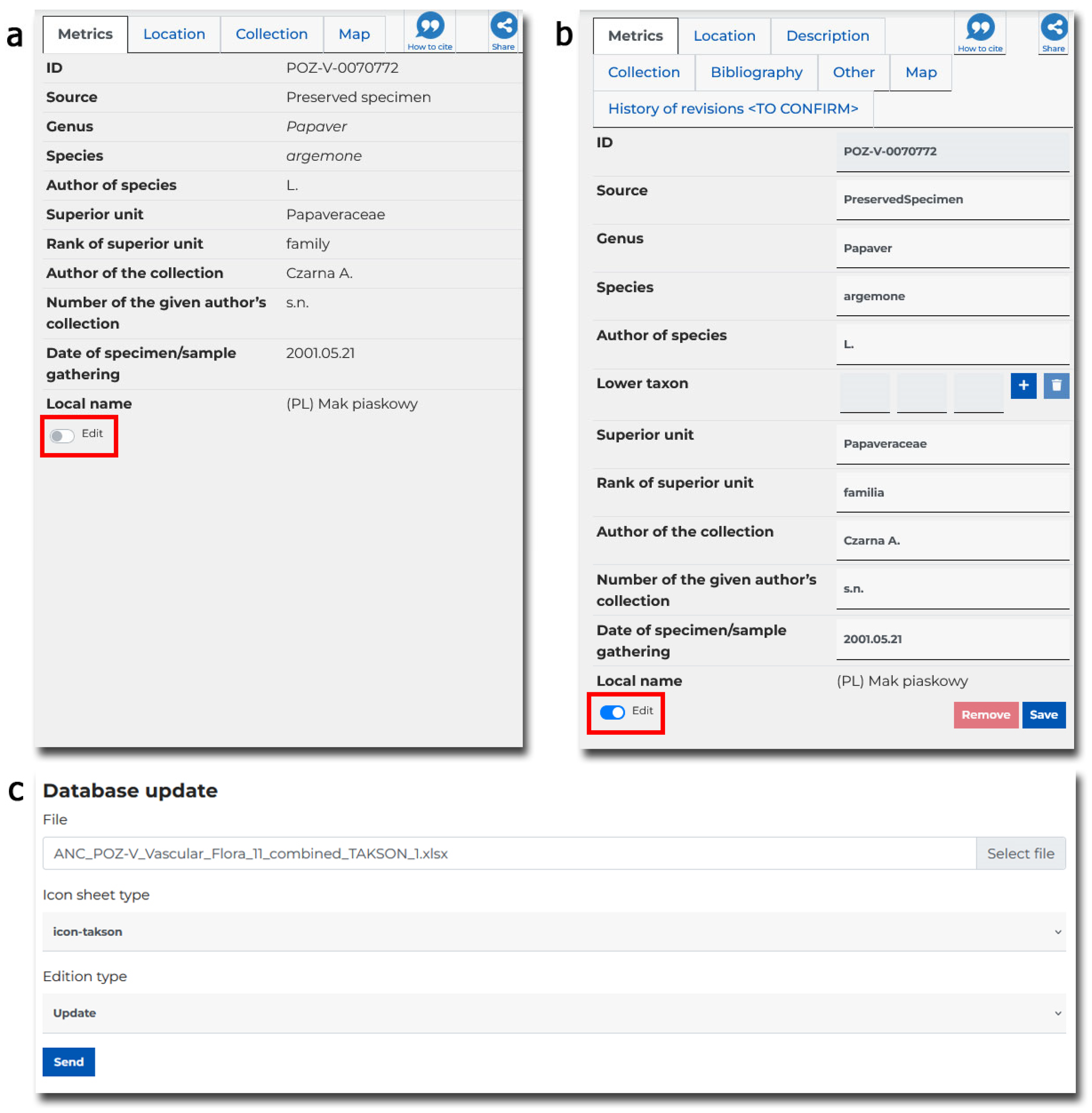
6.3. Data Correction
6.4. Graphic Design
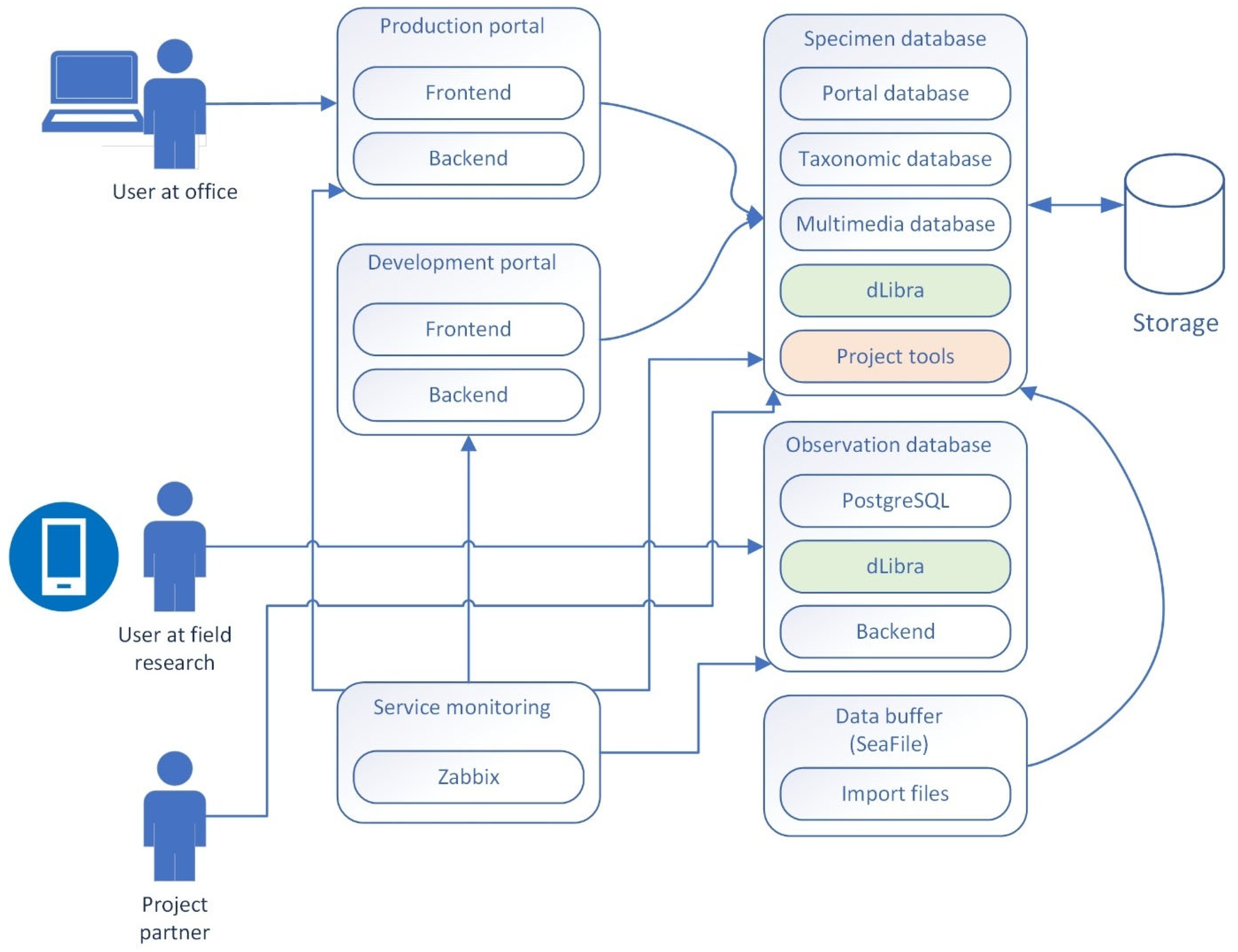
7. Technology
7.1. Infrastructure
7.1.1. Understanding the Specimen Quantity Factor
7.1.2. Storage Space
7.1.3. Computing and Service Resources
7.2. Interoperability
7.3. Security
7.3.1. Security Programming Practices
7.3.2. Security System Audit
7.3.3. Securing Iconographic Data
Removing the Outer Pixels Around the Image
Image Metadata
Watermarks
Digital Signature
8. Main Aspects and Efforts of the System Implementation
8.1. Aspects of Consideration Under the NHC System Implementation
8.1.1. Selected Digitisation Aspects
- exact coordinates—the precise location of the object was determined in the geotagging process;
- approximate coordinates—the location of the object is approximately determined, and the geographic coordinates indicate the centroid of the area that could be assigned to the specimen;
- approximate coordinates due to legal protection—the specimen is legally protected in accordance with national law, and the user does not have sufficient permissions to view the exact coordinates of such specimens;
- unspecified coordinates—the specimen has not been geotagged.
8.1.2. Selected Design Aspects
- (a)
- editing a single record—in the details view of a given record (specimen, samples, bibliography, iconography, and multimedia), an authorised person can switch the details view to the editing mode. It is then possible to correct all fields of a given record and save it. Changes are made and are visible immediately.
- (b)
- group editing of many taxonomic records—often, the attribute value for records is corrected simultaneously. Such a change is possible from the level of statistical tools—reports. Search results can be grouped according to a selected field, and then the value of the chosen field can be changed for all records from this group. For more, see Section 6.3.
8.1.3. Selected Technology Aspects
8.2. Estimated Efforts of Implementation
9. Discussion
10. Summary
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABCD | Access to Biological Collection Data |
| ACID | Atomicity, Consistency, Isolation, Durability |
| AMUNATCOLL | Adam Mickiewicz University Nature Collections |
| API | Application Programming Interface |
| BPS | BioCASe Provider Software |
| BioGIS | BioGeographic Information System |
| CGH | Computer Generated Hologram |
| CI/CD | Continuous Integration/Continuous Delivery |
| DMP | Data Management Plan |
| EBV | Essential Biodiversity Variables |
| EXIF | Exchangeable Image File Format |
| FAIR | Findable, Accessible, Interoperable and Reusable |
| GBIF | Global Biodiversity Information Facility |
| GDPR | General Data Protection Regulation |
| GIS | Geographic Information System |
| HDD | Hard Disk Drive |
| IPR | Intellectual Property Rights |
| ISO | International Organisation for Standardisation |
| IT | Information Technology |
| JWT | JSON Web Token |
| NHC | Natural History Collection |
| OWASP | Open Web Application Security Project |
| PCI DSS | PCI Data Security Standard |
| RAID | Redundant Array of Independent Disks |
| REST | REpresentational State Transfer |
| SDLC | Software Development LifeCycle |
| SDL | Security Development Lifecycle |
| SEO | Search Engine Optimisation |
| SSD | Solid State Drive |
| SSRF | Server-Side Request Forgery |
| TIFF | Tag Image File Format |
| UX | User Experience |
| UI | User Interface |
| WCAG | Web Content Accessibility Guidelines |
References
- Page, L.M.; MacFadden, B.J.; Fortes, J.A.; Soltis, P.S.; Riccardi, G. Digitization of biodiversity collections reveals biggest data on biodiversity. BioScience 2015, 65, 841–842. [Google Scholar] [CrossRef]
- Soltis, P.S.; Nelson, G.; James, S.A. Green digitization: Online botanical collections data answering real-world questions. Appl. Plant Sci. 2018, 6, e1028. [Google Scholar] [CrossRef]
- Corlett, R.T. Achieving zero extinction for land plants. Trends Plant Sci. 2023, 28, 913–923. [Google Scholar] [CrossRef]
- Pereira, H.M.; Ferrier, S.; Walters, M.; Geller, G.N.; Jongman, R.H.; Scholes, R.J.; Bruford, M.W.; Brummitt, N.; Butchart, S.H.; Cardoso, A.C.; et al. Essential Biodiversity Variables. Science 2013, 339, 277–278. [Google Scholar] [CrossRef]
- Jackowiak, B.; Błoszyk, J.; Celka, Z.; Konwerski, S.; Szkudlarz, P.; Wiland-Szymańska, J. Digitization and online access to data on natural history collections of Adam Mickiewicz University in Poznan: Assumptions and implementation of the AMUNATCOLL project. Biodivers. Res. Conserv. 2022, 65, 23–34. [Google Scholar] [CrossRef]
- KEW. KEW Data Portal. 2025. Available online: https://data.kew.org/?lang=en-US (accessed on 25 May 2025).
- MNP. Muséum National d’Histoire Naturelle in Paris—Collections. 2025. Available online: https://www.mnhn.fr/en/databases (accessed on 25 May 2025).
- PVM. Plantes Vasculaires at Muséum National d’Histoire Naturelle. 2025. Available online: https://www.mnhn.fr/fr/collections/ensembles-collections/botanique/plantes-vasculaires (accessed on 25 May 2025).
- TRO. Tropicos Database. 2025. Available online: http://www.tropicos.org/ (accessed on 25 May 2025).
- NDP. Natural History Museum Data Portal. 2025. Available online: https://data.nhm.ac.uk/about (accessed on 25 May 2025).
- SMI. Smithsonian National Museum of Natural History 2025. Available online: https://collections.nmnh.si.edu/search/ (accessed on 25 May 2025).
- Nowak, M.M.; Lawenda, M.; Wolniewicz, P.; Urbaniak, M.; Jackowiak, B. The Adam Mickiewicz University Nature Collections IT system (AMUNATCOLL): Portal, mobile application and graphical interface. Biodivers. Res. Conserv. 2022, 65, 49–67. [Google Scholar] [CrossRef]
- Schmeller, D.S.; Weatherdon, L.V.; Loyau, A.; Bondeau, A.; Brotons, L.; Brummitt, N.; Geijzendorffer, I.R.; Haase, P.; Kuemmerlen, M.; Martin, C.S.; et al. A suite of essential biodiversity variables for detecting critical biodiversity change. Biol. Rev. 2018, 93, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Jetz, W.; McGeoch, M.A.; Guralnick, R.; Ferrier, S.; Beck, J.; Costello, M.J.; Fernandez, M.; Geller, G.N.; Keil, P.; Merow, C.; et al. Essential biodiversity variables for mapping and monitoring species populations. Nat. Ecol. Evol. 2019, 3, 539–551. [Google Scholar] [CrossRef]
- Kissling, W.D.; Ahumada, J.A.; Bowser, A.; Fernandez, M.; Fernández, N.; García, E.A.; Guralnick, R.P.; Isaac, N.J.; Kelling, S.; Los, W.; et al. Building essential biodiversity variables (EBVs) of species distribution and abundance at a global scale. Biol. Rev. 2018, 93, 600–625. [Google Scholar] [CrossRef]
- Apache Cassandra. 2025. Available online: https://cassandra.apache.org/_/index.html (accessed on 25 May 2025).
- Apache Hadoop. 2025. Available online: https://hadoop.apache.org/ (accessed on 25 May 2025).
- DwC. Darwin Core Standard 2025. Available online: https://dwc.tdwg.org/ (accessed on 25 May 2025).
- Open Geospatial Consortium 2025. Available online: https://www.ogc.org/standards/ (accessed on 25 May 2025).
- Hardisty, A.R.; Michener, W.K.; Agosti, D.; García, E.A.; Bastin, L.; Belbin, L.; Bowser, A.; Buttigieg, P.L.; Canhos, D.A.; Egloff, W.; et al. The Bari Manifesto: An interoperability framework for essential biodiversity variables. Ecol. Inform. 2019, 49, 22–31. [Google Scholar] [CrossRef]
- Lawenda, M.; Wiland-Szymańska, J.; Nowak, M.M.; Jędrasiak, D.; Jackowiak, B. The Adam Mickiewicz University Nature Collections IT system (AMUNATCOLL): Metadata structure, database and operational procedures. Biodivers. Res. Conserv. 2022, 65, 35–48. [Google Scholar] [CrossRef]
- Gadelha, L.M.R., Jr.; de Siracusa, P.C.; Dalcin, E.C. A survey of biodiversity informatics: Concepts, practices, and challenges. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2021, 11, e1394. [Google Scholar] [CrossRef]
- Feest, A.; Van Swaay, C.; Aldred, T.D.; Jedamzik, K. The biodiversity quality of butterfly sites: A metadata assessment. Ecol. Indic. 2011, 11, 669–675. [Google Scholar] [CrossRef]
- Walls, R.L.; Deck, J.; Guralnick, R.; Baskauf, S.; Beaman, R.; Blum, S.; Bowers, S.; Buttigieg, P.L.; Davies, N.; Endresen, D.; et al. Semantics in Support of Biodiversity Knowledge Discovery: An Introduction to the Biological Collections Ontology and Related Ontologies. PLoS ONE 2014, 9, e89606. [Google Scholar] [CrossRef]
- da Silva, J.R.; Castro, J.A.; Ribeiro, C.; Honrado, J.; Lomba, Â.; Gonçalves, J. Beyond INSPIRE: An Ontology for Biodiversity Metadata Records. OTM Confederated International Conferences “On the Move to Meaningful Internet Systems”; Springer: Berlin, Germany, 2014. [Google Scholar]
- ANC. AMUNatColl Project. 2025. Available online: http://anc.amu.edu.pl/eng/index.php (accessed on 25 May 2025).
- FBAMU. Faculty of Biology of the Adam Mickiewicz University in Poznań. 2025. Available online: http://biologia.amu.edu.pl/ (accessed on 25 May 2025).
- PSNC. Poznan Supercomputing and Networking Center. 2025. Available online: https://www.psnc.pl/ (accessed on 25 May 2025).
- ANCPortal. AMUNATCOLL Portal. 2025. Available online: https://amunatcoll.pl/ (accessed on 25 May 2025).
- AMUNATCOLL, Android 2025; Faculty of Biology of Adam Mickiewicz University in Poznan: Poznan, Poland.
- AMUNATCOLL, iOS 2021; Faculty of Biology of Adam Mickiewicz University in Poznan: Poznan, Poland.
- Seafile. Open-Source File Sync and Share Software; Seafile Ltd.: Beijing, China; Available online: https://www.seafile.com/en/home/ (accessed on 25 May 2025).
- ISO. International Organization for Standardization. 2017. Available online: https://www.iso.org/iso-8601-date-and-time-format.html (accessed on 25 May 2025).
- RFC 3339: Date and Time on the Internet: Timestamps. 2002. Available online: https://www.rfc-editor.org/rfc/rfc3339.html (accessed on 25 May 2025).
- Guo, N.; Xiong, W.; Wu, Q.; Jing, N. An Efficient Tile-Pyramids Building Method for Fast Visualization of Massive Geospatial Raster Datasets. Adv. Electr. Comput. Eng. 2016, 16, 3–8. [Google Scholar] [CrossRef]
- BIS. Biodiversity Information Standards. 2025. Available online: https://www.tdwg.org/ (accessed on 25 May 2025).
- DCMI. The Dublin Core™ Metadata Initiative. 2025. Available online: https://www.dublincore.org/ (accessed on 25 May 2025).
- Michener, W.K.; Jones, M.B. Ecoinformatics: Supporting ecology as a data-intensive science. Trends Ecol. Evol. 2012, 27, 85–93. [Google Scholar] [CrossRef]
- Kacprzak, E.; Koesten, L.; Ibáñez, L.D.; Blount, T.; Tennison, J.; Simperl, E. Characterising dataset search—An analysis of search logs and data requests. J. Web Semant. 2018, 55, 37–55. [Google Scholar] [CrossRef]
- Löffler, F.; Wesp, V.; König-Ries, B.; Klan, F. Dataset search in biodiversity research: Do metadata in data repositories reflect scholarly information needs? PLoS ONE 2021, 16, e0246099. [Google Scholar] [CrossRef]
- ABCD. Access to Biological Collections Data Standard. 2025. Available online: https://www.tdwg.org/standards/abcd/ (accessed on 25 May 2025).
- Krug, S. Don’t Make Me Think! Web & Mobile Usability; MITP-Verlags GmbH & Co. KG: Frechen, Germany, 2018. [Google Scholar]
- Web Content Accessibility Guidelines (WCAG) 2.1. 2025. Available online: https://www.w3.org/TR/WCAG21/ (accessed on 25 May 2025).
- EUDirective2019-882. European Accessibility Act (EAA)—Directive (EU) 2019/882. 2019. Available online: https://eur-lex.europa.eu/eli/dir/2019/882/oj/eng (accessed on 25 May 2025).
- EUDirective2016-2102. Web Accessibility Directive—Directive (EU) 2016/2102. 2016. Available online: https://eur-lex.europa.eu/eli/dir/2016/2102/oj/eng (accessed on 25 May 2025).
- EN301549. European Standard EN 301 549. 2021. Available online: https://www.etsi.org/deliver/etsi_en/301500_301599/301549/03.02.01_60/en_301549v030201p.pdf (accessed on 25 May 2025).
- GBIF. Global Biodiversity Information Facility. 2025. Available online: https://www.gbif.org/en/ (accessed on 25 May 2025).
- Liu, Q.; Xing, L. Reliability Modeling of Cloud-RAID-6 Storage System. Int. J. Future Comput. Commun. 2015, 4, 415–420. [Google Scholar] [CrossRef]
- ISOUNITS. Standards by ISO/TC 12 Quantities and Units. 2025. Available online: https://www.iso.org/committee/46202/x/catalogue/ (accessed on 25 May 2025).
- ByteUnits. Wikipedia—Multiple-Byte Units. 2025. Available online: https://en.wikipedia.org/w/index.php?title=Byte&utm_campaign=the-difference-between-kilobytes-and-kibibytes&utm_medium=newsletter&utm_source=danielmiessler.com#Multiple-byte_units (accessed on 25 May 2025).
- Seagate. Why Does my Hard Drive Report Less Capacity than Indicated on the Drive’s Label? 2025. Available online: https://www.seagate.com/gb/en/support/kb/why-does-my-hard-drive-report-less-capacity-than-indicated-on-the-drives-label-172191en/ (accessed on 25 May 2025).
- Mazrae, P.R.; Mens, T.; Golzadeh, M.; Decan, A. On the usage, co-usage and migration of CI/CD tools: A qualitative analysis. Empir. Softw. Eng. 2023, 28, 52. [Google Scholar] [CrossRef]
- PYT. Python Language. 2025. Available online: https://www.python.org/ (accessed on 25 May 2025).
- DJA. Django Framework. 2025. Available online: https://www.djangoproject.com/ (accessed on 25 May 2025).
- PAN. Pandas—Data Analysis Tool. 2025. Available online: https://pandas.pydata.org/ (accessed on 25 May 2025).
- GUN. Gunicorn—Python Web Server. 2025. Available online: https://gunicorn.org/ (accessed on 25 May 2025).
- REA. React—JavaScript Library. 2025. Available online: https://reactjs.org/ (accessed on 25 May 2025).
- GEO. GeoJSON Geographic Data Structures Encoding Format. 2025. Available online: https://geojson.org/ (accessed on 25 May 2025).
- LEA. Leaflet—Library for Mobile-Friendly Interactive Maps. 2025. Available online: https://leafletjs.com/ (accessed on 25 May 2025).
- PIX. PixiJS—Advanced Text Rendering Graphic. 2025. Available online: https://pixijs.com/ (accessed on 25 May 2025).
- JWT. JSON Web Tokens. 2025. Available online: https://jwt.io/ (accessed on 25 May 2025).
- Guo, F.; Chen, C.; Li, K. Research on Zabbix Monitoring System for Large-scale Smart Campus Network from a Distributed Perspective. J. Electr. Syst. 2024, 20, 631–648. [Google Scholar]
- PostgreSQL. PostgreSQL Database Web Site. 2025. Available online: https://www.postgresql.org/ (accessed on 25 May 2025).
- dLibra. Digital Library Framework. 2025. Available online: https://www.psnc.pl/digital-libraries-dlibra-the-most-popular-in-poland/ (accessed on 25 May 2025).
- Wilde, E.; Pautasso, C. REST: From Research to Practice; Springer: New York, NY, USA, 2011. [Google Scholar]
- BioCASe. Biological Collection Access Service. 2025. Available online: http://www.biocase.org/ (accessed on 25 May 2025).
- GBIF Data Standards. 2025. Available online: https://www.gbif.org/standards (accessed on 25 May 2025).
- Saeed, H.; Shafi, I.; Ahmad, J.; Khan, A.A.; Khurshaid, T.; Ashraf, I. Review of Techniques for Integrating Security in Software Development Lifecycle. Comput. Mater. Contin. 2025, 82, 139–172. [Google Scholar] [CrossRef]
- Ryan Dewhurst. Static Code Analysis. 2025. Available online: https://owasp.org/www-community/controls/Static_Code_Analysis (accessed on 25 May 2025).
- ISO 27001 Information Security Management Systems. Available online: https://www.iso.org/standard/27001 (accessed on 25 May 2025).
- National Institute of Standards and Technology. Available online: https://www.nist.gov/standards (accessed on 25 May 2025).
- PCI DSS—Payment Card Industry Data Security Standard. Available online: https://www.pcisecuritystandards.org/document_library/ (accessed on 25 May 2025).
- OWASP. Open Web Application Security Project. 2025. Available online: https://owasp.org/ (accessed on 25 May 2025).
- EXIF. Exchangeable Image File Format. 2025. Available online: https://en.wikipedia.org/wiki/Exif (accessed on 25 May 2025).
- Pi, D.; Wang, J.; Li, J.; Wu, J.; Zhao, W.; Wang, Y.; Liu, J. High-security holographic display with content and copyright protection based on complex amplitude modulation. Opt. Express 2024, 32, 30555–30564. [Google Scholar] [CrossRef] [PubMed]
- Software Development Life Cycle—SDLC. Available online: https://www.ibm.com/think/topics/sdlc (accessed on 25 May 2025).
- Hardisty, A.; Roberts, D. A decadal view of biodiversity informatics: Challenges and priorities. BMC Ecol. 2013, 2013, 16. [Google Scholar] [CrossRef] [PubMed]
- Convention on Biological Diversity. 2025. Available online: https://www.cbd.int/ (accessed on 25 May 2025).





| Action | Species Search Language | Search | Browsing | |
|---|---|---|---|---|
| View | ||||
| Scientists | Latin | Simple—a few selected fields Extended—adding multiple fields to search conditions Advanced—any definition of complex conditions, including logical expressions | Browse as a list, a map, and aggregated reports. Full information about specimens | |
| State and local government administration | National, Latin | Search among species | Aggregated reports for selected species | |
| Services and state officials | National, Latin | Mainly from protected and similar species | Graphic information is essential | |
| Non-governmental organisations and society | National (e.g., English, Polish) | Simplified—one field whose value is matched to many fields from the specimen description | Browse in list, map and aggregated reports. Key information about specimens | |
| Education | National, including everyday language | Simplified—mainly searching from available educational materials | Educational materials about the specimen | |
| Read Value | Comment |
|---|---|
| 19.VI | Lack of year |
| 20.7.1889, 3.X.1890 | The month given in Arabic or Roman numerals |
| 7.1876 ü 1882 | Date given (probably) without day and with double year |
| mid July (in Polish “połowa lipca”) 1919 | Imprecise harvest day |
| end of May (in Polish “koniec maja”) 1916 | Imprecise harvest day |
| flowers (in Polish “kwiaty”) 16.04, leaves (in Polish “liście”) 15.07.1893 | Providing the date (without the year), and two specimens on one sheet |
| 19?5 | One of the year digits is not legible |
| Code | Format Description | Extension | Volume | Creation Method | Typical Applications | Remarks |
|---|---|---|---|---|---|---|
| RAW | Output files obtained directly from the digitising device, in a device-specific format (e.g., recording format) | depending on the device, e.g., .CRW or .CR2 for Canon cameras | big | It is created manually or semi-automatically (depending on the equipment), in the digitisation process. | long-term data archiving | The format is often manufacturer-dependent and may be closed, patented, etc., so it is not recommended as the only form of long-term archiving. Some devices (e.g., scanners) can generate TIFF files immediately without the RAW option. |
| MASTER | Lossless transformation of a RAW file to an open format, without any other changes. | most often TIFF | big | It can be automated for some RAW formats, if batch processing tools exist for these formats. If they do not exist, manual actions, e.g., using the manufacturer’s software supplied with the camera, are necessary. | long-term data archiving | The basic format for long-term data archiving is preserved in parallel to the RAW format. In case an error occurs during the transformation from RAW to MASTER, MASTER files can be restored from RAW. |
| MASTER CORRECTED | MASTER file subjected to necessary corrections (e.g., straightening, cropping), still saved in lossless form. | most often TIFF | big | Depending on the type of corrections, this may be a manual or partially automated operation. | long-term data archiving limited sharing of sample files | This is the base form for generating further formats for sharing purposes. It contains the file in a usable form (after processing) in the highest possible quality. If an error occurs during the corrections of the MASTER file, you can always go back to the clean MASTER form and repeat the corrections. |
| PRESENTATIONHI-RES | MASTER CORRECTED files converted to a format dedicated to online sharing in high resolution. Low-loss compression may be used here but does not have to be. | e.g., TIFF (pyramidal) or JPEG2000 | big | It can be created fully automatically based on the MASTER CORRECTED file. | online sharing | Sharing such files online usually requires the use of a special protocol that allows for gradual loading of details—the standard here is the IIIF protocol. |
| PRESENTATIONLOW-RES | MASTER CORRECTED files converted to a format dedicated for online sharing in medium/low resolution—there is a loss of quality. | e.g., JPG, PDF, PNG | small | It can be created fully automatically based on the MASTER CORRECTED file. | online and offline sharing (download) | Sharing by simply displaying on web pages or downloading files from the site’s pages. |
| Name | Type | Description |
|---|---|---|
| Converter | console or internet application | A tool for automatic formatting of Excel files as a web application. Its existence is based on the assumption that previously digitised data require adaptation to the new format. Its task is to convert input files into files compliant with the metadata specification using specific configurations (sets of rules), allowing for optional editing of input data. |
| Form | spreadsheet | The basis for proper preparation of input data is compliance with the metadata specification. This is also intended to automate the processes used by the developed applications. In order to avoid inconsistencies between the data filled in (often by different people), a form in the form of a spreadsheet file was prepared. It contains all the columns in accordance with the metadata specification, divided into sheets for taxa, samples, iconography, and bibliography. |
| Validator | console or internet application | A tool for validating spreadsheet files available as a web application. Its task is to check the presence and correctness of filling in the appropriate sheets in the file. The tool returns a report with errors and details of their occurrence, divided into each detected sheet. |
| Aggregator | console application | A program used to combine spreadsheet files that comply with metadata descriptions. It is used in the digitisation process, mainly at the stage of describing records by the georeferencing or translation teams. |
| Reporter | console application | A program that summarises the number of AMINATCOLL-compliant records in a spreadsheet. Used by project coordinators primarily to monitor progress. |
| Field content description: | Contains brief information about the type of information that should be entered into a given field. |
| Field format: | Specifies whether the field is a field of a specific format: Integer field, Float field, Text field, Date field in ABCD format. |
| Allowed values: | This field contains only values from the allowed list. Each list item is entered on a separate line. |
| Required field: | The word YES in this field description element indicates that the field is mandatory. The word NO in this field description element indicates that the field is optional. |
| Example values: | This description element provides example values for the field. |
| Comments: | Space for any additional information related to the field. |
| Level | Name | Description |
|---|---|---|
| 0 | specimen made public | information about the specimen (record) is not protected; full information is available to all logged-in and non-logged-in users |
| 1 | specimen made public with restrictions | information about the specimen (record) is partially protected—sensitive data (e.g., geographical coordinates, habitat) is protected |
| 2 | specimen is restricted | information about the specimen (record) is made available only to external and internal users verified in terms of competence |
| 3 | non-public specimen | information about the specimen (record) is made available only to authorised internal users (e.g., selected from among the employees of the hosting institution) |
| Level | Description |
|---|---|
| 0 | the field is not protected, everyone can see it, even if they are not logged in, e.g., genus, species; |
| 1 | the field is only available to logged-in users (e.g., ATPOL coordinates), |
| 2 | the field is only available to trusted collaborators, e.g., exact coordinates, exact habitat; |
| 3 | the field is only available to authorised internal users, e.g., suitability for target groups, technical information regarding file import. |
| Classification of Specimens | Number of Specimens | ||
|---|---|---|---|
| Total number of specimens | 2.25 million | ||
| Botanical collections (algae and plants) | approx. 500 thousand | ||
| Included nomenclatural types | approx. 350 | ||
| Mycological collections (fungi and lichens) | approx. 50 thousand | ||
| Zoological collections | approx. 1.7 million | ||
| Included nomenclatural types | over 1000 | ||
| Regular | Economical | ||
|---|---|---|---|
| Operation Name or Location | Factor | TB | TB |
| Source material | 90 | 90 | |
| Processed and protected material | 1.6 | 144 | 144 |
| RAID 6 | 0.2 | 46.8 | 46.8 |
| Conversion to ISO | 0.1 | 28.08 | 28.08 |
| Total in one location | 308.88 | 308.88 | |
| Geographic copy | 308.88 | 308.88 | |
| Local copy | 308.88 | 118.8 | |
| Buffer for the digitisation process | 120 | 120 | |
| Total | 1046.64 | 856.56 | |
| Conversion rate | ~11.63 | ~9.52 | |
| Dataset-Related Fields | ||
|---|---|---|
| ABCD | AMUNATCOLL | |
| Property Name | Link to Specification | Property Name/Comment |
| /DataSets/DataSet/ContentContacts/ContentContact/Name | https://terms.tdwg.org/wiki/abcd2:ContentContact-Name (accessed on 26 May 2025) | Information generated automatically during export depending on the custodian of a given collection |
| TechnicalContact/Name | https://terms.tdwg.org/wiki/abcd2:TechnicalContact-Name (accessed on 26 May 2025) | Information generated automatically during export |
| /DataSets/DataSet/ContentContacts/ContentContact/Organization/Name/Representation/@language | https://terms.tdwg.org/wiki/abcd2:DataSet-Representation-@language (accessed on 26 May 2025) | Information generated automatically during export |
| /DataSets/DataSet/Metadata/Description/Representation/Title | https://terms.tdwg.org/wiki/abcd2:DataSet-Title (accessed on 26 May 2025) | Information generated automatically during export |
| /DataSets/DataSet/Metadata/RevisionData/DateModified | https://terms.tdwg.org/wiki/abcd2:DataSet-DateModified (accessed on 26 May 2025) | Date of last import, date generated automatically by the system during export |
| Specimen Fields | ||
| /DataSets/DataSet/Units/Unit/SourceInstitutionID | https://terms.tdwg.org/wiki/abcd2:SourceInstitutionID (accessed on 26 May 2025) | Institution |
| /DataSets/DataSet/Units/Unit/SourceID | https://terms.tdwg.org/wiki/abcd2:SourceID (accessed on 26 May 2025) | Botany/Zoology |
| /DataSets/DataSet/Units/Unit/UnitID | https://terms.tdwg.org/wiki/abcd2:UnitID (accessed on 26 May 2025) | Collection/Specimen number |
| /DataSets/DataSet/Units/Unit/RecordBasis | https://terms.tdwg.org/wiki/abcd2:RecordBasis (accessed on 26 May 2025) | Source |
| /DataSets/DataSet/Units/Unit/SpecimenUnit/NomenclaturalTypeDesignations/NomenclaturalTypeDesignation/TypifiedName/NameAtomised/Botanical/GenusOrMonomial | https://terms.tdwg.org/wiki/abcd2:TaxonIdentified-Botanical-GenusOrMonomial (accessed on 26 May 2025) | Genus |
| /DataSets/DataSet/Units/Unit/Identifications/Identification/Result/TaxonIdentified/ScientificName/NameAtomised/Zoological/GenusOrMonomial | https://terms.tdwg.org/wiki/abcd2:TaxonIdentified-Zoological-GenusOrMonomial (accessed on 26 May 2025) | Genus |
| /DataSets/DataSet/Units/Unit/Identifications/Identification/Result/TaxonIdentified/ScientificName/NameAtomised/Botanical/FirstEpithet | https://terms.tdwg.org/wiki/abcd2:TaxonIdentified-FirstEpithet (accessed on 26 May 2025) | Species |
| /DataSets/DataSet/Units/Unit/Identifications/Identification/Result/TaxonIdentified/ScientificName/NameAtomised/Zoological/SpeciesEpithet | https://terms.tdwg.org/wiki/abcd2:TaxonIdentified-Zoological-SpeciesEpithet (accessed on 26 May 2025) | Species |
| /DataSets/DataSet/Units/Unit/Gathering/Agents/GatheringAgent/Person/FullName | https://terms.tdwg.org/wiki/abcd2:GatheringAgent-FullName (accessed on 26 May 2025) | Author of the collection |
| /DataSets/DataSet/Units/Unit/Identifications/Identification/Identifiers/Identifier/PersonName/FullName | https://terms.tdwg.org/wiki/abcd2:Identifier-FullName (accessed on 26 May 2025) | Author of designation |
| /DataSets/DataSet/Units/Unit/Gathering/DateTime/ISODateTimeBegin | https://terms.tdwg.org/wiki/abcd2:Gathering-DateTime-ISODateTimeBegin (accessed on 26 May 2025) | Date of specimen/sample collection |
| /DataSets/DataSet/Units/Unit/SpecimenUnit/Preparations/Preparation/PreparationType | https://terms.tdwg.org/wiki/abcd2:SpecimenUnit-PreparationType (accessed on 26 May 2025) | Storage method |
| /DataSets/DataSet/Units/Unit/Gathering/SiteCoordinateSets/SiteCoordinates/CoordinatesLatLong/LatitudeDecimal | https://terms.tdwg.org/wiki/abcd2:Gathering-LatitudeDecimal (accessed on 26 May 2025) | Latitude |
| /DataSets/DataSet/Units/Unit/Gathering/SiteCoordinateSets/SiteCoordinates/CoordinatesLatLong/LongitudeDecimal | https://terms.tdwg.org/wiki/abcd2:Gathering-LongitudeDecimal (accessed on 26 May 2025) | Longitude |
| /DataSets/DataSet/Units/Unit/Identifications/Identification/Result/TaxonIdentified/ScientificName/FullScientificNameString | https://terms.tdwg.org/wiki/abcd2:TaxonIdentified-FullScientificNameString (accessed on 26 May 2025) | Required by Darwin Core. Merging of several fields: Genus + Species + SpeciesAuthor + YearOfCollection |
| Name | Description |
|---|---|
| Broken Access Control | Access controls implement policies designed to restrict users from operating beyond their designated permissions. When these controls fail, they often lead to unauthorised information disclosure, data alteration or destruction, or the execution of business functions that exceed the user’s authorised limits. |
| Cryptographic Failures | It emphasises the importance of safeguarding data both during transmission and while stored. Sensitive information, including passwords, credit card details, personal data, and proprietary information, necessitates enhanced security measures, particularly when such data falls under privacy laws like the EU General Data Protection Regulation (GDPR) or financial data protection standards such as the PCI Data Security Standard (PCI DSS). |
| Injection | This vulnerability arises from the absence of verification for user-provided data, specifically the lack of filtering or sanitisation measures. It pertains to the use of non-parameterised, context-sensitive calls that are executed directly within the interpreter. Additionally, it encompasses the utilisation of malicious data, such as that found in SQL queries, dynamic queries, commands, or stored procedures. |
| Insecure Design | A broad category representing various weaknesses, expressed as “missing or ineffective control design.” Design flaws and implementation flaws must be distinguished for some reason, and have different root causes and remedies. A secure design may still have implementation flaws that lead to exploitable vulnerabilities. An insecure design cannot be fixed by a perfect implementation, because by definition, the necessary security controls were never designed to defend against the specific attacks. |
| Security Misconfiguration | The application stack may be susceptible to attacks due to insufficient hardening or improper configuration of various services, such as enabling unnecessary ports, services, pages, accounts, or permissions. Additionally, error handling mechanisms may disclose excessive information, including stack traces or other overly detailed error messages. Furthermore, the most recent security features may either be disabled or incorrectly configured. The server might fail to transmit security headers or directives, or it may not be configured with secure values. |
| Vulnerable and Outdated Components | Lastly, the software could be outdated or contain vulnerabilities. The scope encompasses the operating system, web or application server, database management system, applications, APIs, and all associated components, runtimes, and libraries. It is essential that the underlying platform is consistently patched and updated, and software developers must verify the compatibility of any updated, enhanced, or patched libraries. |
| Identification and Authentication Failures | To protect against authentication attacks, user identity confirmation, authentication, and session management are key. Frequently, vulnerabilities arise from automated attacks wherein the perpetrator possesses a compilation of legitimate usernames and passwords. Additionally, breaches can occur due to inadequate or ineffective credential recovery methods, forgotten passwords, and the transmission of passwords in plaintext or through poorly hashed password storage systems. The absence or ineffectiveness of multi-factor authentication further exacerbates these risks. Moreover, there may be instances of improper invalidation of session identifiers or the exposure of session identifiers within URLs. |
| Software and Data Integrity Failures | Software and data integrity failures pertain to code and infrastructure that do not adequately safeguard against breaches of integrity. It is impermissible for an application to depend on plugins, libraries, or modules sourced from unverified origins. A frequent method for exploiting vulnerabilities is through the auto-update feature, which allows updates to be downloaded and implemented on a previously trusted application without adequate integrity checks. |
| Security Logging and Monitoring Failures | Logging and monitoring service activity helps detect, escalate, and respond to active breaches. Events such as logins, failed logins, and high-value transactions should be audited. Warnings and errors should generate clear log messages when they occur, and they themselves should be monitored for suspicious activity. Alert thresholds and response escalation processes should be defined at the appropriate level, and information about their exceedance should be provided in real or near real time. |
| Server-Side Request Forgery (SSRF) | An SSRF vulnerability arises when a web application retrieves a remote resource without validating the URL supplied by the user. This allows an attacker to manipulate the server-side application into directing the request to an unintended destination. Such vulnerabilities can impact services that are exclusively internal to the organisation’s infrastructure, in addition to any external systems. Consequently, this may lead to the exposure of sensitive information, including authorisation credentials. |
| Tag | Exemplary Value |
|---|---|
| ID | POZ-V-0000001 |
| Image description | The image depicts a scan from the Natural History Collections of Housing Institution Name. |
| Copyright | Copyright © 2025 Housing Institution Name, City. All rights reserved. |
| Copyright note | This image or any part of it cannot be reproduced without the prior written permission of Housing Institution Name, City. |
| Additional information | Deleting or changing the image metadata is strictly prohibited. For more information on restrictions on the use of photos, please visit: https://www.domain.com/ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lawenda, M.; Wolniewicz, P. IT Challenges in Designing and Implementing Online Natural History Collection Systems. Diversity 2025, 17, 388. https://doi.org/10.3390/d17060388
Lawenda M, Wolniewicz P. IT Challenges in Designing and Implementing Online Natural History Collection Systems. Diversity. 2025; 17(6):388. https://doi.org/10.3390/d17060388
Chicago/Turabian StyleLawenda, Marcin, and Paweł Wolniewicz. 2025. "IT Challenges in Designing and Implementing Online Natural History Collection Systems" Diversity 17, no. 6: 388. https://doi.org/10.3390/d17060388
APA StyleLawenda, M., & Wolniewicz, P. (2025). IT Challenges in Designing and Implementing Online Natural History Collection Systems. Diversity, 17(6), 388. https://doi.org/10.3390/d17060388








