Genetic Diversity in Sporophytic Apomictic Neotropical Savanna Trees: Insights from Eriotheca and Handroanthus Agamic Complexes
Abstract
1. Introduction
2. Methods
2.1. Biological Material
2.2. DNA Extraction and ISSR-PCR
2.3. Data Analyses
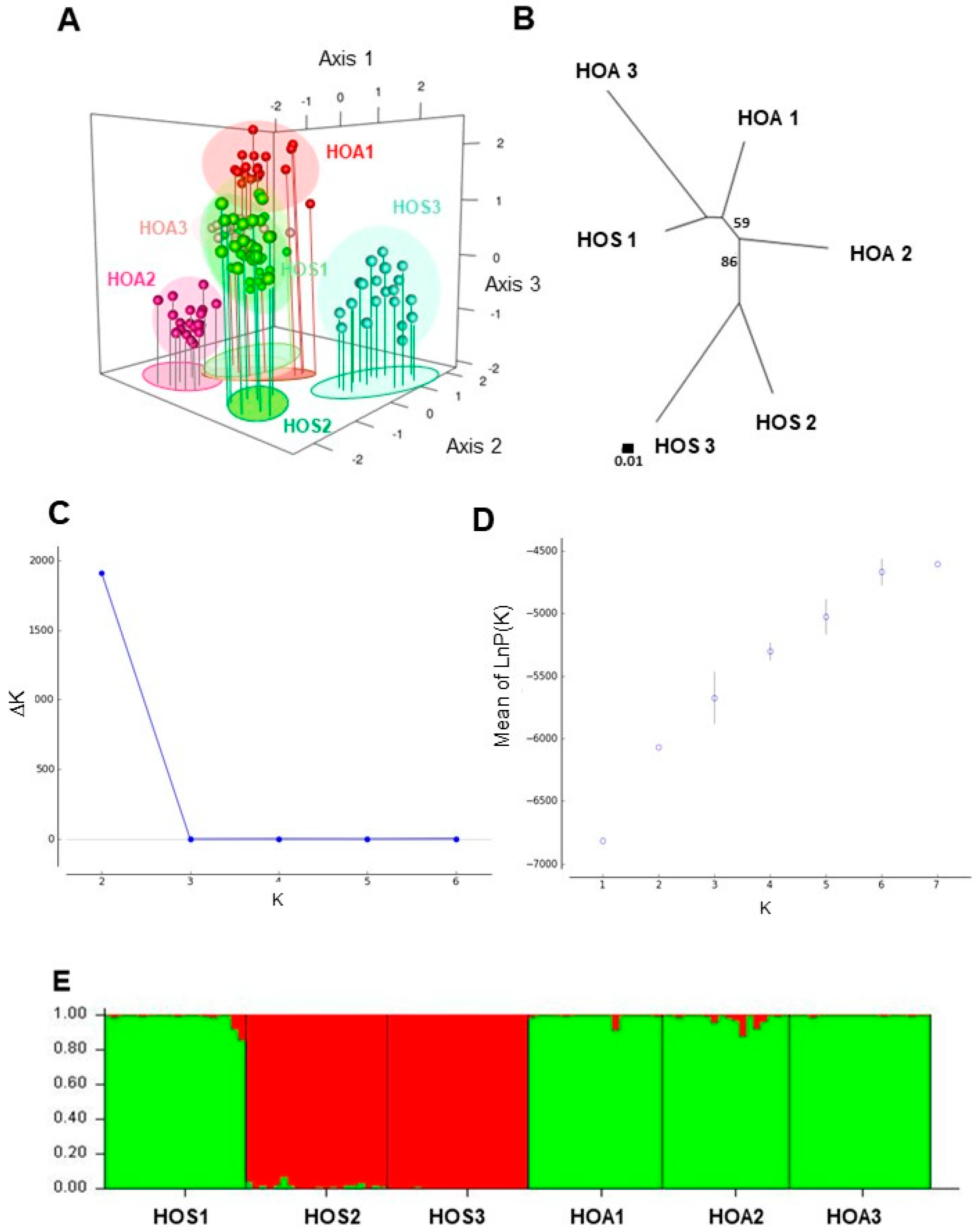
3. Results
4. Discussion
4.1. Genetic Diversity in Apomictic vs. Sexual Populations
4.2. Evolutionary and Ecological Implications of Facultative Apomixis
4.3. General Considerations
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Koltunow, A.M. Apomixis: Embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 1993, 5, 1425. [Google Scholar] [CrossRef]
- Richards, A.J. Apomixis in flowering plants: An overview. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2003, 358, 1085–1093. [Google Scholar] [CrossRef]
- Kaushal, P.; Dwivedi, K.K.; Radhakrishna, A.; Srivastava, M.K.; Kumar, V.; Roy, A.K.; Malaviya, D.R. Partitioning apomixis components to understand and utilize gametophytic apomixis. Front. Plant Sci. 2019, 10, 256. [Google Scholar] [CrossRef] [PubMed]
- Hörandl, E. Geographical Parthenogenesis in Alpine and Arctic Plants. Plants 2023, 12, 844. [Google Scholar] [CrossRef] [PubMed]
- Asker, S.E.; Jerling, L. Apomixis in Plants; CRC Press: Boca Raton, FL, USA, 1992. [Google Scholar]
- Hojsgaard, D.; Klatt, S.; Baier, R.; Carman, J.G.; Hörandl, E. Taxonomy and biogeography of apomixis in angiosperms and associated biodiversity characteristics. Crit. Rev. Plant Sci. 2014, 33, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Koltunow, A.M.; Grossniklaus, U. Apomixis: A developmental perspective. Annu. Rev. Plant Biol. 2003, 54, 547–574. [Google Scholar] [CrossRef]
- Bicknell, R.A.; Koltunow, A.M. Understanding apomixis: Recent advances and remaining conundrums. Plant Cell 2004, 16, 228–245. [Google Scholar] [CrossRef]
- Paun, O.; Greilhuber, J.; Temsch, E.M.; Hörandl, E. Patterns, sources and ecological implications of clonal diversity in apomictic Ranunculus carpaticola (Ranunculus auricomus complex, Ranunculaceae). Mol. Ecol. 2006, 15, 897–910. [Google Scholar] [CrossRef]
- Voigt-Zielinski, M.L.; Piwczyński, M.; Sharbel, T.F. Differential effects of polyploidy and diploidy on fitness of apomictic Boechera. Sex. Plant Reprod. 2012, 25, 97–109. [Google Scholar] [CrossRef]
- Mendes-Rodrigues, C.; Marinho, R.C.; Balao, F.; Arista, M.; Ortiz, P.L.; Carmo-Oliveira, R.; Oliveira, P.E. Reproductive diversity, polyploidy, and geographical parthenogenesis in two Eriotheca (Malvaceae) species from Brazilian Cerrado. Perspect. Plant Ecol. Evol. Syst. 2019, 36, 1–12. [Google Scholar] [CrossRef]
- Hörandl, E.; Paun, O. Patterns and sources of genetic diversity in apomictic plants: Implications for evolutionary potentials. In Apomixis: Evolution, Mechanisms and Perspectives; ARG Gantner Verlag KG: Vaduz, Lichtenstein, 2007; pp. 169–194. [Google Scholar]
- Dias, A.C.C.; Serra, A.C.; Sampaio, D.S.; Borba, E.L.; Bonetti, A.M.; Oliveira, P.E. Unexpectedly high genetic diversity and divergence among populations of the apomictic Neotropical tree Miconia albicans. Plant Biol. 2018, 20, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Dias, A.C.C.; Marinho, R.C.; Sampaio, D.S.; Bonetti, A.M.; Oliveira, P.E. Clone worth? Genetic diversity in obligate apomictic Miconia albicans (Melastomataceae). Plant Biol. 2021, 23, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Hojsgaard, D.; Hörandl, E. The rise of apomixis in natural plant populations. Front. Plant Sci. 2019, 10, 358. [Google Scholar] [CrossRef]
- Hörandl, E. Novel approaches for species concepts and delimitation in polyploids and hybrids. Plants 2022, 11, 204. [Google Scholar] [CrossRef] [PubMed]
- Mendes-Rodrigues, C.; Carmo-Oliveira, R.; Talavera, S.; Arista, M.; Ortiz, P.L.; Oliveira, P.E. Polyembryony and apomixis in Eriotheca pubescens (Malvaceae-Bombacoideae). Plant Biol. 2005, 7, 533–540. [Google Scholar] [CrossRef]
- Hojsgaard, D.; Hörandl, E. Apomixis as a facilitator of range expansion and diversification in plants. In Evolutionary Biology: Biodiversification from Genotype to Phenotype; Springer: Cham, Switzerland, 2015; pp. 305–327. [Google Scholar] [CrossRef]
- Firetti-Leggieri, F.; Lohmann, L.G.; Alcantara, S.; da Costa, I.R.; Semir, J. Polyploidy and polyembryony in Anemopaegma (Bignonieae, Bignoniaceae). Plant Reprod. 2013, 26, 43–53. [Google Scholar] [CrossRef]
- Mendes, M.G.; de Oliveira, A.P.; Oliveira, P.E.; Bonetti, A.M.; Sampaio, D.S. Sexual, apomictic and mixed populations in Handroanthus ochraceus (Bignoniaceae) polyploid complex. Plant Syst. Evol. 2018, 304, 817–829. [Google Scholar] [CrossRef]
- Sampaio, D.S.; Bittencourt Júnior, N.S.; Oliveira, P.E. Sporophytic apomixis in polyploid Anemopaegma species (Bignoniaceae) from Central Brazil. Bot. J. Linn. Soc. 2013, 173, 77–91. [Google Scholar] [CrossRef]
- Gibbs, P.E. Late-acting self-incompatibility–the pariah breeding system in flowering plants. New Phytol. 2014, 203, 717–734. [Google Scholar] [CrossRef]
- Sampaio, D.S.; Bittencourt Jr, N.S.; Oliveira, P.E. Mating in the pseudogamic apomictic Anemopaegma acutifolium DC: Another case of pseudo-self-compatibility in Bignoniaceae? Plant Biol. 2013, 15, 919–924. [Google Scholar] [CrossRef]
- Marinho, R.C.; Mendes-Rodrigues, C.; Balao, F.; Ortiz, P.L.; Yamagishi-Costa, J.; Bonetti, A.M.; Oliveira, P.E. Do chromosome numbers reflect phylogeny? New counts for Bombacoideae and a review of Malvaceae sl. Am. J. Bot. 2014, 101, 1456–1465. [Google Scholar] [CrossRef] [PubMed]
- Dufresne, F.; Stift, M.; Vergilino, R.; Mable, B.K. Recent progress and challenges in population genetics of polyploid organisms: An overview of current state-of-the-art molecular and statistical tools. Mol. Ecol. 2014, 23, 40–69. [Google Scholar] [CrossRef] [PubMed]
- Tokgöz, H.B.; Çetin, Ö.; Kaya, H.B.; Akkale, C.; Yildirim, H.; Pirhan, A.F.; Pirhan, A.F.; Kaya, E.; Altan, F. Genetic diversity of Lilium candidum natural populations in Türkiye evaluated with ISSR and M13-tailed SSR markers. Plant Syst. Evol. 2024, 310, 5. [Google Scholar] [CrossRef]
- Sampaio, D.S. Biologia Reprodutiva de Espécies de Bignoniaceae Ocorrentes no Cerrado e Variações no Sistema de Autoincompatibilidade. Ph.D. Thesis, Federal University of Uberlândia, Uberlândia, Brazil, 2010. [Google Scholar]
- Amiteye, S. Basic concepts and methodologies of DNA marker systems in plant molecular breeding. Heliyon 2021, 7, e08093. [Google Scholar] [CrossRef]
- Peakall, R.O.D.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Adler, D.; Murdoch, D.; Nenadic, O.; Urbanek, S.; Chen, M.; Gebhardt, A. rgl: 3D Visualization Device System (OpenGL), R package version 0.93; Semantic Scholar: Seattle, WA, USA, 2013; Available online: http://rgl.neoscientists.org (accessed on 15 July 2023).
- Soetaert, K. Plot3D: Plotting Multi-Dimensional Data, R package version 1.0; NIOZ: Yerseke, The Netherlands, 2013. [Google Scholar]
- Vekemans, X.; Beauwens, T.; Lemaire, M.; Roldán-Ruiz, I. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol. Ecol. 2002, 11, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. PHYLIP: Phylogeny Inference Package, version 3.66; Universidade de Washington: Seattle, WA, USA, 2006. [Google Scholar]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Hörandl, E. The complex causality of geographical parthenogenesis. New Phytol. 2006, 171, 525–538. [Google Scholar] [CrossRef]
- Martins, R.L.; Oliveira, P.E. RAPD evidence for apomixis and clonal populations in Eriotheca (Bombacaceae). Plant Biol. 2003, 5, 338–340. [Google Scholar] [CrossRef]
- Karunarathne, P.; Hojsgaard, D. Single independent autopolyploidization events from distinct diploid gene pools and residual sexuality support range expansion of locally adapted tetraploid genotypes in a South American grass. Front. Genet. 2021, 12, 736088. [Google Scholar] [CrossRef]
- Moura, Y.A.; Alves-Pereira, A.; Da Silva, C.C.; Souza, L.M.; de Souza, A.P.; Koehler, S. Secondary origin, hybridization and sexual reproduction in a diploid–tetraploid contact zone of the facultatively apomictic orchid Zygopetalum mackayi. Plant Biol. 2020, 22, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Viana, M.L.; Oliveira, P.E.; Romero, R.; Caetano, A.P.S. The best of both worlds: Apomixis and sexuality co-occur in species of Microlicia, Melastomataceae. Plant Species Biol. 2021, 36, 476–488. [Google Scholar] [CrossRef]
- Venter, S.M.; Glennon, K.L.; Witkowski, E.T.F.; Baum, D.; Cron, G.V.; Tivakudze, R.; Karimi, N. Baobabs (Adansonia digitata L.) are self-incompatible and ‘male’ trees can produce fruit if hand-pollinated. S. Afr. J. Bot. 2017, 109, 263–268. [Google Scholar] [CrossRef]
- Lobo, J.A.; Quesada, M.; Stoner, K.E. Effects of pollination by bats on the mating system of Ceiba pentandra (Bombacaceae) populations in two tropical life zones in Costa Rica. Am. J. Bot. 2005, 92, 370–376. [Google Scholar] [CrossRef]
- Figueredo, A.; de Loliveira, Á.W.; Carvalho-Sobrinho, J.G.; Souza, G. Karyotypic stability in the paleopolyploid genus Ceiba Mill. (Bombacoideae, Malvaceae). Braz. J. Bot. 2016, 39, 1087–1093. [Google Scholar] [CrossRef]
- Munthali, C.R.Y.; Chirwa, P.W.; Changadeya, W.J.; Akinnifesi, F.K. Genetic differentiation and diversity of Adansonia digitata L (baobab) in Malawi using microsatellite markers. Agrofor. Syst. 2013, 87, 117–130. [Google Scholar] [CrossRef][Green Version]
- Brondani, R.P.V.; Gaiotto, F.A.; Missiaggia, A.A.; Kirst, M.; Gribel, R.; Grattapaglia, D. Microsatellite markers for Ceiba pentandra (Bombacaceae), an endangered tree species of the Amazon Forest. Mol. Ecol. Notes 2003, 3, 177–179. [Google Scholar] [CrossRef]
- Bertoni, B.W.; Telles, M.P.d.C.; Malosso, M.G.; Torres, S.C.; Pereira, J.O.; Lourenço, M.V.; França, S.d.C.; Pereira, A.M. Genetic diversity in natural populations of Jacaranda decurrens Cham. determined using RAPD and AFLP markers. Genet. Mol. Biol. 2010, 33, 532–538. [Google Scholar] [CrossRef]
- Batistini, A.P.; Telles, M.P.D.C.; Bertoni, B.W.; Coppede, J.D.S.; Môro, F.V.; Pereira, A.M.S.; França, S.D.C. Genetic diversity of natural populations of Anemopaegma arvense (Bignoniaceae) in the Cerrado of São Paulo State, Brazil. Genet. Mol. Res. 2009, 8, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Geng, Y.; Tersing, T.; Liu, N.; Wang, Q.; Zhong, Y. High genetic differentiation and low genetic diversity in Incarvillea younghusbandii, an endemic plant of Qinghai-Tibetan Plateau, revealed by AFLP markers. Biochem. Syst. Ecol. 2009, 37, 589–596. [Google Scholar] [CrossRef]
- Petit, C.; Bretagnolle, F.; Felber, F. Evolutionary consequences of diploid–polyploid hybrid zones in wild species. Trends Ecol. Evol. 1999, 14, 306–311. [Google Scholar] [CrossRef]
- Marinho, R.C.; Mendes-Rodrigues, C.; Resende-Moreira, L.C.; Lovato, M.B.; Bonetti, A.M.; Oliveira, P.E. Phylogeography of Eriotheca species complex: Insights into the origin and range expansion of apomictic and polyploid trees in Neotropical Savannas. Plant Biol. 2023, 25, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, L.C.; Reis MN, O.; Melo, W.A.; Collevatti, R.G. Comparative phylogeography shows congruent co-divergence in Neotropical seasonally dry forest and savanna tree species. J. Biogeogr. 2024, 51, 1064–1078. [Google Scholar] [CrossRef]
- Strassburg, B.B.; Brooks, T.; Feltran-Barbieri, R.; Iribarrem, A.; Crouzeilles, R.; Loyola, R.; Latawiec, A.E.; Oliveira Filho, F.J.; Scaramuzza, C.A.D.M.; Scarano, F.R.; et al. Moment of truth for the Cerrado hotspot. Nat. Ecol. Evol. 2017, 1, 0099. [Google Scholar] [CrossRef]


| Species Municipality-State | Code | Geographical Coordinates | Sample Size | Embryonic Pattern | Ploidy Level and Chromosome Number | Breeding System | Voucher Number |
|---|---|---|---|---|---|---|---|
| E. crenulata (comb. n. E. gracilipes) | |||||||
| Uberlândia-MG | ECS | 19°18′34″ S; 48°26′48″ W | 17 | Mono 3,4 | 2n = 2x = 92 1 | Sexual | HUFU-15008 |
| Caldas Novas-GO | ECA | 17°47′08″ S; 48°40′09″ W | 19 | Poly 4 | 2n = 6x = 276 1 | Apomitic | HUFU-15122 |
| E. pubescens | |||||||
| Cristalina-GO | EPS | 16°52′32″ S; 47°40′44″ W | 19 | Mono 3,4 | 2n = 4x = 184 1 | Sexual | HUFU-15073 |
| Catalão-GO | EPA | 18°08′42″ S; 47°54′24″ W | 19 | Poly 3,4 | 2n = 6x = 276 1 | Apomitic | HUFU-25854 * |
| H. ochraceus | |||||||
| Uberlândia-MG | HOS1 | 19°18′34″ S; 48°26′48″ W | 20 | Mono 2 | 2n = 2x = 40 2 | Sexual | HUFU-52585 |
| Pires do Rio-GO | HOS2 | 17°11′37″ S; 47°45′03″ W | 20 | Mono 2 | 2n = 2x = 40 2 | Sexual | HUFU-55507 |
| Biribiri-MG | HOS3 | 18°07′54″ S; 43°36′56″ W | 20 | Mono 5 | 2n = 2x = 40 5 | Sexual | HUFU-52678 |
| Córrego Danta-MG | HOA1 | 19°41′12″ S; 46°02′20″ W | 19 | Poly 2 | 2n = 2x = 40; 2n = 4x = 80 2 | Apomitic | HUFU-52593 |
| Fidalgo-MG | HOA2 | 19°32′50″ S; 43°59′13″ W | 18 | Poly 5 | 2n = 4x = 80 5 | Apomitic | BHCB-68100 |
| São José do Rio Preto-SP | HOA3 | 20°47′10″ S; 49°21′32″ W | 20 | Poly 2 | 2n = 4x = 80 2 | Apomitic | SJRP-29235 |
| Populations (Breeding Systems) | P (%) | I | He | NDL | Total Loci |
|---|---|---|---|---|---|
| Eriotheca crenulata | |||||
| ECS (sexual) | 50.51 | 0.294 ± 0.031 a | 0.201 ± 0.021 a | 3–25 loci | 93 |
| ECA (apomictic) | 59.60 | 0.363 ± 0.031 a | 0.251 ± 0.022 a | 2–27 loci | 88 |
| Species mean | 55.05 | 0.328 ± 0.022 | 0.226 ± 0.015 | ||
| Eriotheca pubescens | |||||
| EPS (sexual) | 66.25 | 0.396 ± 0.034 a | 0.273 ± 0.024 a | 6–25 loci | 71 |
| EPA (apomictic) | 68.75 | 0.396 ± 0.033 a | 0.271 ± 0.023 a | 0–26 loci | 73 |
| Species mean | 67.50 | 0.396 ± 0.023 | 0.272 ± 0.017 | ||
| Handroanthus ochraceus | |||||
| HOS1 (sexual) | 78.85 | 0.447 ± 0.267 a | 0.306 ± 0.193 a | 17–46 loci | 89 |
| HOS2 (sexual) | 74.04 | 0.355 ± 0.267 ab | 0.234 ± 0.190 ab | 14–44 loci | 101 |
| HOS3 (sexual) | 66.35 | 0.318 ± 0.268 b | 0.208 ± 0.187 b | 13–41 loci | 87 |
| Sexual populations mean | 73.08 | 0.373 ± 0.066 | 0.249 ± 0.051 | ||
| HOA1 (apomictic) | 53.85 | 0.316 ± 0.307 b | 0.217 ± 0.215 b | 10–34 loci | 82 |
| HOA2 (apomictic) | 72.12 | 0.342 ± 0.250 b | 0.221 ± 0.174 b | 14–42 loci | 87 |
| HOA3 (apomictic) | 52.88 | 0.267 ± 0.280 b | 0.177 ± 0.194 b | 12–37 loci | 88 |
| Apomictic populations mean | 59.62 | 0.308 ± 0.038 | 0.205 ± 0.024 | ||
| Species mean | 66.35 | 0.341 ± 0.060 | 0.227 ± 0.043 |
| Species | Source of Variation | Degrees of Freedom | Sum of Squares | Variance Components | Total Variance Percentage | p-Value |
|---|---|---|---|---|---|---|
| E. crenulata | ||||||
| Among populations | 1 | 172.676 | 9.232 | 53% | <0.001 | |
| Within populations | 34 | 238.445 | 7.013 | 47% | <0.001 | |
| Total | 35 | 411.121 | 16.245 | 100% | <0.001 | |
| E. pubescens | ||||||
| Among populations | 1 | 169.479 | 8.418 | 47% | <0.001 | |
| Within populations | 36 | 343.127 | 9.531 | 53% | <0.001 | |
| Total | 37 | 512.605 | 17.950 | 100% | <0.001 | |
| H. ochraceus | Among populations | 5 | 875.138 | 8.322 | 39.00 | <0.001 |
| Within populations | 11 | 1420.766 | 12.800 | 61.00 | <0.001 | |
| Grouped by breeding system | ||||||
| Among groups/breeding system | 1 | 183.914 | 0.199 | 1.00 | <0.046 | |
| Among populations | 4 | 688.760 | 8.168 | 38.00 | <0.001 | |
| Within populations | 111 | 1441.104 | 12.983 | 61.00 | <0.001 | |
| Grouped by Bayesian analysis | ||||||
| Among groups (HOS1, HOA1, HOA2, HOA3 and HOS1, HOS2) | 1 | 290.046 | 290.046 | 12.00 | <0.001 | |
| Among populations | 4 | 582.777 | 145.694 | 30.00 | <0.001 | |
| Within populations | 111 | 1440.954 | 12.982 | 58.00 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marinho, R.C.; Mendes, M.G.; Mendes-Rodrigues, C.; Bonetti, A.M.; Borba, E.L.; Oliveira, P.E.; Sampaio, D.S. Genetic Diversity in Sporophytic Apomictic Neotropical Savanna Trees: Insights from Eriotheca and Handroanthus Agamic Complexes. Diversity 2025, 17, 254. https://doi.org/10.3390/d17040254
Marinho RC, Mendes MG, Mendes-Rodrigues C, Bonetti AM, Borba EL, Oliveira PE, Sampaio DS. Genetic Diversity in Sporophytic Apomictic Neotropical Savanna Trees: Insights from Eriotheca and Handroanthus Agamic Complexes. Diversity. 2025; 17(4):254. https://doi.org/10.3390/d17040254
Chicago/Turabian StyleMarinho, Rafaela Cabral, Mariana Gonçalves Mendes, Clesnan Mendes-Rodrigues, Ana Maria Bonetti, Eduardo Leite Borba, Paulo Eugênio Oliveira, and Diana Salles Sampaio. 2025. "Genetic Diversity in Sporophytic Apomictic Neotropical Savanna Trees: Insights from Eriotheca and Handroanthus Agamic Complexes" Diversity 17, no. 4: 254. https://doi.org/10.3390/d17040254
APA StyleMarinho, R. C., Mendes, M. G., Mendes-Rodrigues, C., Bonetti, A. M., Borba, E. L., Oliveira, P. E., & Sampaio, D. S. (2025). Genetic Diversity in Sporophytic Apomictic Neotropical Savanna Trees: Insights from Eriotheca and Handroanthus Agamic Complexes. Diversity, 17(4), 254. https://doi.org/10.3390/d17040254






