Abstract
Rhesus macaque (Macaca mulatta), the iconic species of genus Macaca, is characterized by the greatest geographical distribution of all nonhuman primates and is an important resource in many wildlife-related tourism areas, especially in China. In the current study, the genetic diversity was assessed by ten microsatellite loci with DNA obtained from muscle tissue of deceased individuals of free-ranging but tourist-habituated rhesus population naturally inhabiting the Wulongkou Scenic Area, Jiyuan, China, where they have been exploited for tourism since the early 1980s. The results showed that the genetic diversity for the studied rhesus population was relatively lower compared with its wild and captive counterparts, and the samples collected from the population subdivision in the studied area could mask the finding. Therefore, we proposed that a group-based study of the genetic diversity would help to clarify the genetic structure/diversity of rhesus macaques in this area, and then reasonable management recommendations could be provided for the sustainable development of local wildlife-dominated tourism.
1. Introduction
Wildlife tourism, undertaken predominantly to view and/or encounter non-domesticated vertebrates, directly/indirectly connects humans with wild animals, which could satisfy human being’s thirst for knowing the unknown of wild animals [1,2,3]. This sector naturally attracts people of all ages from around the world, which promotes the blossoming of wildlife tourism [1,2]. However, the emerging infectious disease pandemic (i.e., COVID-19) could negatively affect the tourism industry, including the wildlife tourism sector [4,5,6]. Nowadays, the recovery of wildlife tourism post the COVID-19 pandemic, especially from the tourism-dependent countries/areas [7,8,9], is gaining momentum driven by the pent-up demand for nature-based experiences [10,11,12,13].
Though wildlife tourism plays a significant role in both international and regional economies, it could result in potential and actual impacts across a range of wildlife, habitats, and interactive situations, such as behavioral changes, stresses induced by human disturbance, and lower dispersal due to food provisioning, especially in nonhuman primates [1,13]. Nonhuman primates are our closest evolutionary relatives, and both their morphological and behavioral features attract the attention of people of all age groups [9]. For instance, a tourist presence and interactions could increase the anxiety levels of wild male Barbary macaque (Macaca sylvanus), and tourists aggressive interactions would significantly trigger the physiological stress estimated by fecal glucocorticoid concentrations [14]. The violation of the 7-m distance rule by observers can result in elevated physiological stress in wild western lowland gorilla (Gorilla gorilla gorilla) [15]. Implementing food-provisioning sites for wildlife-related tourism could not only increase the risk of parasite transmission [16] but also lead to lower genetic diversity and higher relatedness in the endangered Yunnan snub-nosed monkey (Rhinopithecus bieti) [17].
The rhesus macaque (M. mulatta), iconic species of the genus Macaca, is characterized by the greatest geographical distribution of all nonhuman primates and is commonly present in many zoos around the world [18,19,20], an important resource in many wildlife-related tourism areas in Asia, especially in China [20,21]. Though a large number of studies of rhesus macaques have been conducted to estimate the genetic diversity of wild, free-ranging, and captive populations, there is a lack of a genetic assessment of rhesus populations inhabiting tourist-habituated locations, especially in indigenous populations [22,23,24,25,26,27]. Therefore, the wildness of rhesus macaques in the aspect of genetic diversity might be underestimated due to the high prevalence of rhesus macaque-involved tourism [20].
The Taihangshan macaque (M. m. tcheliensis) is an endemic subspecies and currently occupies the northernmost range of rhesus macaque, the southern end of Mountain Taihang, and the northern bank of the Yellow River, in China [28,29]. Its current distribution range spreads from 111°51′ E to 113°45′ E in longitude and from 34°54′ N to 35°42′ N in latitude, demonstrating a very narrow but relatively long-range shape (Figure 1A) [29]. A previous study has shown that the overall level of genetic diversity in Taihangshan macaques is lower compared with other wild rhesus populations in China, which could be due to natural limits and historical human disturbances [27,30,31]. However, it has been severely exploited for tourism in the past several decades [20], which might deteriorate the genetic diversity of the regional population of Taihangshan macaques that has been noticed in wild primates [17]. Therefore, we investigated the genetic diversity based on deceased individuals of tourist-habituated rhesus macaques in the Wulongkou area, Jiyuan City, China.
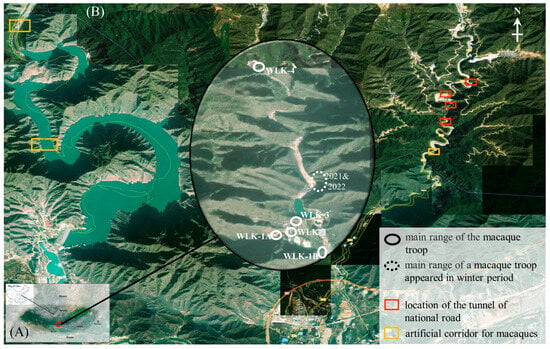
Figure 1.
The sketch map of the Wulongkou area, Jiyuan City, China. (A) showing the current distribution range of Taihangshan macaques in China, (B) showing the location of Wulongkou area, Jiyuan, China.
2. Materials and Methods
2.1. Subject and Sampling
From 2012 to 2021, deceased macaque individuals of Taihangshan macaques were collected ad libitum in Wulongkou area (35°12′49″ N, 112°41′25″ E) (Figure 1B), Jiyuan, China, and then were stored in −20 °C freezers (BC/BD-208DT, Changhong Meiling Co., Ltd., Hefei, China) for further processing [32]. This study area is located in the eastern end of Taihangshan Macaque National Nature Reserve (Jiyuan section) (34°54′–35°42′ N, 112°02′–113°45′ E), Henan Province, China. This area has been exploited for tourism since the early 1980s when the initial population of Taihangshan macaques included ca. 50 individuals [33].
During the past four decades, the macaque population in the core sightseeing area increased gradually in the first three decades, peaked by the end of 2012 with over 500 individuals divided into over 7 groups, then experienced a massive collapse during 2013–2016 and included ca. 350 individuals grouped into 5 groups since 2017 (Figure 2) [33]. One group (named WLK-4) mainly patrols in the northern backyard of this area, and the other four groups (WLK-1A, WLK-1B, WLK-2, and WLK-3) primarily wander in the core area for tourist sightseeing. The investigated samples might be collected mainly from all these named groups but could not be assigned to a specific group due to difficulties in identifying deceased macaque or body remains except for a few characterized individuals.
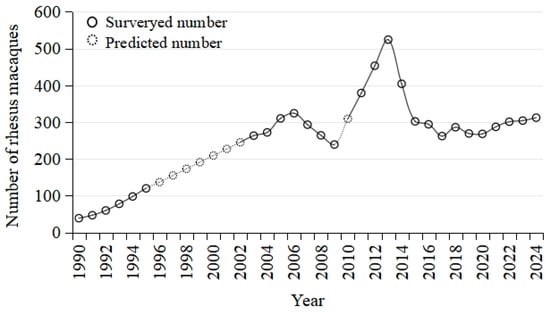
Figure 2.
Number of rhesus macaques inhabiting the Wulongkou area, Jiyuan City, China.
In total, 42 muscle samples, including 13 females and 29 males, were harvested from deceased individuals collected during 2012 and 2021. The age of the individuals ranged from a few months to ca. 15 years, and most of these individuals were adults (n = 31).
2.2. DNA Extraction and Genotyping
TIANamp Genomic DNA Kit (TIANGEN Biotech (Beijing) Co., Ltd., China) was employed in extracting total genomic DNA from tissue samples following the manual with few modifications. In total, 10 microsatellite markers were chosen as the target loci based on previous genetic studies of rhesus macaques (Table 1) [34].

Table 1.
Information of primers for the 10 microsatellite loci used in this study.
Three independent PCRs were performed for each locus of all samples to eliminate microsatellite locus genotyping errors. Polymerase chain reaction (PCR) amplification was performed with a 15 μL reaction volume containing 1 μL of template DNA, 0.4 μL of each forward and reverse primer (10 μM) (Tsingke Biotechnology Co., Ltd., Beijing, China), 7.5 μL of red mix (Beibei Biotechnology Co., Ltd., Zhengzhou, China) and the rest of the addition of ddH2O to 15 μL. The amplification conditions were 94 °C for 5 min, 35 cycles at 94 °C for 30 s, 53–58 °C (Ta for each locus referring to Table 1) for 30 s, 72 °C for 30 s, and final extension at 72 °C for 10 min. The PCR products were separated on 1.2% agarose gels by electrophoresis to visually assess the amplification efficiency, and then qualified products were genotyped by Shanghai Sangon Biological Engineering Technology and Service Co., Ltd. (Shanghai, China).
The allele sizes were scored against the GeneScanTM 500 LIZ® Size Standard (Thermo Fisher, Waltham, MA, USA), using GeneMapper v4.0 software (Applied Biosystems, Waltham, MA, USA). The length of allele fragments and the data were counted according to the integer multiples of 2 and 4 bp in the length of different alleles. These data were transposed into a spreadsheet of Office 2019 Excel (Microsoft, Redmond, WA, USA) for further analysis.
2.3. Statistical Analysis
Micro-Checker v2.2.3 [35] was used to check for possible null alleles, allelic dropout, and scoring errors due to stuttering. The significance of deviations from Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD) among loci was assessed with GENEPOP v4.7 [36], followed by the sequential Bonferroni test [37] to correct for multiple comparisons using Myriads v1.2 [38].
The variables assessing the genetic variation at the population level included the mean number of alleles per locus (Na), the effective number of alleles (Ne), the observed (Ho) and expected (He) heterozygosity, and fixation index (F, also called the inbreeding coefficient), which were calculated with GenAlEx 6.5 [39,40] plugged-in Office 2019 Excel (Microsoft, Redmond, WA, USA). The polymorphic information content (PIC) was calculated by Cervus v3.0 [41]. Furthermore, the allelic richness (AR) was computed using FSTAT v2.9.3 [42].
To test whether a past bottleneck was present or not, a method based on deviations of allele frequencies for calculations of heterozygosity was adopted within BOTTLENECK V1.2.02 [43]. In specific, the infinite alleles model (IAM), stepwise mutation model (SMM) with 1000 iterations, and two-phase model (TPM) with 95% single-step mutations and 10% multiple-step mutations [44] were performed. In specific, the theory of the BOTTLENECK program was generated by Cornuet and Luikart [43] and Luikart et al. [45]. The species or population that experienced a recent bottleneck simultaneously decreased the allele number and the expected levels of heterozygosity. Nevertheless, the allele number is reduced faster than the expected heterozygosity. Therefore, the value of the expected heterozygosity calculated through the allele number (Heq) is lower than the obtained expected heterozygosity (He). For neutral markers, in a population in gene mutation drift equilibrium, there is an equal probability that a given locus has a slight excess or deficit of heterozygosity regarding the heterozygosity calculated from the number of alleles. In contrast, in a bottlenecked population, a large fraction of the loci analyzed will exhibit a significant excess of the expected heterozygosity.
2.4. Ethical Statement of Ethics
This study adhered to the Ethical Regulations of the China Primatological Society. The study conformed to Chinese legal requirements and complied with protocols approved by the State Forestry Administration of China. The study was approved by the Administration Bureau of Taihangshan Macaque National Natural Reserve.
3. Results
3.1. Genetic Diversity
The average amplification success rate of the microsatellite loci was 94.8% (SD: 2.0%, n = 10), with a range of 83.3% to 100%. The characterization of the genotyped 10 microsatellite loci is summarized (Table 2). The mean values of Na, Ne, AR, and PIC were 6.9 (SE: 0.5), 3.464 (SE: 0.314), 6.745 (SE: 0.514), and 0.643 (SE: 0.033), respectively. The average values of Ho (0.693) and He (0.691) were statistically undifferentiable (Wilcox rank sum exact test with continuity correction, W = 49.5, p = 1.0), which meant that the population was basically in HWE. The average F ranged from −0.262 to 0.146, with a mean of −0.010. Nine out of the ten microsatellite loci showed no significant departure from HWE (all p values > 0.05), but the D7S513, both direct test and test corrected by Bonferroni method, significantly departed from the HWE (p < 0.05). Linkage disequilibrium was not detected between most paired loci within the population (all p values > 0.05), except D4S1626, and D12S1645 (p < 0.05). However, when the test was adjusted for multiple comparisons, all the paired loci showed no significant linkage disequilibrium (all p values > 0.05).

Table 2.
Summary of the 10 microsatellite loci in 42 Taihangshan macaques.
3.2. Bottleneck Effect Testing
Three models were adopted to diagnose the recent bottleneck effect, and the results were summarized (Table 3). None of the tests employed with the BOTTLENECK V1.2.02 software showed significant evidence of recent bottlenecks in the studied macaque population, except the D2S169. Under the IAM model, the He (0.828) of D2S169 was significantly higher than its Heq (0.676) (p = 0.023), which suggested a bottleneck effect. However, the other nine microsatellites under the IAM model did not show significant evidence of bottlenecks, suggesting the microsatellites fit better with SMM or TPM models, both of which did not show any significant trend correlated with bottleneck events. Contrarily, with the SMM model, three microsatellites (D2S2151, p = 0.030; D4S1626, p = 0.048; and D11S2002, p = 0.028) showed population expansion or subdivision in the studied area rather than recent bottlenecks.

Table 3.
Summary of bottleneck probability values under three models.
4. Discussion
Assessing genetic diversity is of great importance to wildlife conservation management [46]. In the present study, based on tissues collected from deceased individuals, we investigated the population genetics of tourist-habituated free-ranging Taihangshan macaques in Jiyuan, China.
Rhesus macaque is the most widely distributed nonhuman primate species ranging from western (Afghanistan) to eastern (China) Asia between ca. 15° and ca. 36°, north latitude [18,47], which may lead to a great variety of genetic diversity. The average PIC (mean ± SE: 0.643 ± 0.033) of the used 10 microsatellite loci could be classified into a high level of polymorphic information (≥0.5) [48]. Several studies of wild (estimated by He and Na, mean ± SE: 0.748 ± 0.065; 7.1 ± 1.7) [24,27,34,49] and captive (estimated by He and Na, mean ± SE: 0.760 ± 0.038; 10.28 ± 3.19) [50,51,52,53,54,55,56] rhesus populations have shown a relatively higher genetic diversity than that of this study (He: 0.691 ± 0.027, Na: 6.9 ± 0.5). Moreover, the genetic diversity (He) of the studied rhesus population in Wulongkou area was relatively lower than that of the genetic diversity of overall (He: 0.79) and western (He: overall 0.80, clade A 0.79, clade B 0.79, clade C 0.79) populations but relatively higher than that of the eastern population (He: 0.62) of Taihangshan macaques, and the average number of alleles per locus was relatively lower than the western populations (Na: 13) but similar to the eastern population (Na: 6) of Taihangshan macaques and wild Sichuan rhesus populations (Na: 5.3~6.7) [24,27,49]. All the above-discussed contents suggested that the population of Taihangshan macaques in the Wulongkou area might keep a normal or slightly lower level of genetic diversity.
Furthermore, the average F (−0.010 ± 0.041) of the studied population suggested a very low inbreeding coefficient compared with wild rhesus populations in China (0.090~0.323 for eight populations but −0.164 for one population) [24,27,34,49] but was similar to that of the Cayo Santiago rhesus populations (0.000) which was India-original but free-ranging rhesus macaques inhabiting an isolated island of Puerto Rico [25]. Moreover, the BOTTLENECK test showed that there was population expansion or subdivision rather than recent bottleneck evidence in the studied rhesus population. These findings showed that tourism could enhance or keep the genetic diversity of rhesus macaque. However, this could be due to the time lag for detecting a significant signal of the tourism-induced effects and/or mixed samples from several groups of rhesus macaques. The rhesus population in the studied area was composed of several groups during the past decades, and male dispersal could reduce the inbreeding coefficient for a period. Specifically, the initial group (original WLK-1, ca. 50 individuals) was attracted by food baits from the forest during the early 1980s, from which several groups were derived in the past decades. Besides, there was another group (original WLK-2, ca. 12 individuals) that was translocated from a neighboring area during the early 1990s, which could be supposed to have a very limited kinship with the initial group in this Wulongkou area. During the past three decades, the males in this area could freely disperse among groups, which could help, if not enhance, the slowing down of the decrease of the level of genetic diversity that normally reflects historical events with a relatively longer period. However, given the decrease in population size, observed more and more males stay in this area, and even in their natal group (Tian JD, field observations), we, therefore, propose that a group-based study is needed to detect the genetic structure/diversity of rhesus macaques in this area, and then reasonable management recommendation could be provided for sustainable development of local wildlife-dominated tourism.
Author Contributions
Conceptualization, J.T.; methodology, Y.W., Y.Z., J.T. and T.L.; software, Y.W., J.T. and Y.Z.; validation, J.T. and Y.W.; formal analysis, J.T., Y.W. and Y.Z.; investigation, Y.W., Y.Z., T.L. and J.T.; resources, J.T.; data curation, J.T. and Y.W.; writing—original draft preparation, Y.W. and J.T.; writing—review and editing, J.T. and J.L.; supervision, J.T. and J.L.; project administration, J.T. and J.L.; funding acquisition, J.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (No. 31600304) and the Cultivation Fund for Young Teachers in Natural Science Basic Research of Zhengzhou University (JC202043029) to J.T.
Institutional Review Board Statement
The study adhered to the Ethical Regulations of the China Primatological Society. The study conformed to Chinese legal requirements and complied with protocols approved by the State Forestry Administration of China. The study was permitted by the Administration Bureau of Taihangshan Macaque National Natural Reserve.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Acknowledgments
We are thankful to the Jiyuan Administration Bureau of Taihangshan Macaque National Nature Reserve for permission to carry out this study and to the Administration Bureau of Wulongkou Scenic Spot for the permission and logistic support. We are thankful to Qingjun Li, Leibo Guo, Pan Li, San’ao Kuang, Yali Kang, Xiaolei Miao, Zhengjing Kuang, and other staff for their help and kindness in the fieldwork. We are very grateful and thankful to the two reviewers for their critical but constructive and informative comments and corrections, which helped us to significantly improve and strengthen our manuscript.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| AR | allelic richness |
| bp | base pair |
| ca. | circa |
| COVID-19 | coronavirus disease 2019 |
| DNA | deoxyribonucleic acid |
| E | east longitude |
| F | fixation index |
| He | expected heterozygosity |
| Heq | expected equilibrium heterozygosity |
| Ho | observed heterozygosity |
| HWE | Hardy–Weinberg equilibrium |
| IAM | infinite alleles model |
| LD | linkage disequilibrium |
| n | sample size |
| N | northern latitude |
| Na | mean number of alleles per locus |
| Ne | effective number of alleles |
| m | meter |
| min | minute |
| PCR | polymerase chain reaction |
| PIC | polymorphic information content |
| s | second |
| SD | standard deviation of the mean |
| SE | standard error of the mean |
| SMM | stepwise mutation model |
| Ta | annealing temperature |
| TPM | two-phase model |
| WLK | Wulongkou |
| μL | microlitre |
| μM | micromole |
References
- Newsome, D.; Dowling, R.K.; Moore, S.A. Wildlife Tourism; Channel View Publications: Bristol, UK, 2005. [Google Scholar]
- Fatima, J.K. Wilderness of Wildlife Tourism; Apple Academic Press: Palm Bay, FL, USA, 2017. [Google Scholar] [CrossRef]
- Speiran, S.I.M.; Hovorka, A.J. Bringing animals in-to wildlife tourism. Sustainability 2024, 16, 7155. [Google Scholar] [CrossRef]
- Fotiadis, A.; Polyzos, S.; Huan, T.T.C. The good, the bad and the ugly on COVID-19 tourism recovery. Ann. Tour. Res. 2021, 87, 103117. [Google Scholar] [CrossRef] [PubMed]
- Newsome, D. The collapse of tourism and its impact on wildlife tourism destinations. J. Tour. Futures 2021, 7, 295–302. [Google Scholar] [CrossRef]
- Usui, R.; Sheeran, L.K.; Asbury, A.M.; Blackson, M. Impacts of the COVID-19 pandemic on mammals at tourism destinations: A systematic review. Mammal Rev. 2021, 51, 492–507. [Google Scholar] [CrossRef]
- UNWTO. COVID-19 Tourism Recovery Technical Assistance Package; World Tourism Organization: Madrid, Spain, 2020. [Google Scholar]
- Durant, I. We Urgently Need to Kickstart Tourism’s Recovery but Crisis Offers an Opportunity to Rethink it. World Economic Forum 2021. Available online: https://www.weforum.org/agenda/2021/08/tourism-still-in-deep-trouble/ (accessed on 17 February 2025).
- Usui, R.; Sheeran, L.K.; Asbury, A.M.; Pedersen, L. Building resilience in primate tourism: Insights from the COVID-19 pandemic and future directions. Primates 2024, 65, 191–201. [Google Scholar] [CrossRef]
- Aebli, A.; Volgger, M.; Taplin, R. A two-dimensional approach to travel motivation in the context of the COVID-19 pandemic. Curr. Issues Tour. 2021, 25, 60–75. [Google Scholar] [CrossRef]
- Buckley, R.C.; Cooper, M. Tourism as a tool in nature-based mental health: Progress and prospects post-pandemic. Int. J. Environ. Res. Public Health 2022, 19, 13112. [Google Scholar] [CrossRef]
- Yao, Y.B.; Zhao, X.X.; Ren, L.P.; Jia, G.M. Compensatory travel in the post COVID-19 pandemic era: How does boredom stimulate intentions? J. Hosp. Tour. Manag. 2023, 54, 56–64. [Google Scholar] [CrossRef]
- Stone, L.S.; Stone, M.T. Wildlife Tourism Dynamics in Southern Africa; Springer: Cham, Switzerland, 2024. [Google Scholar] [CrossRef]
- Maréchal, L.; Semple, S.; Majolo, B.; Qarro, M.; Heistermann, M.; MacLarnon, A. Impacts of tourism on anxiety and physiological stress levels in wild male Barbary macaques. Biol. Conserv. 2011, 144, 2188–2193. [Google Scholar] [CrossRef]
- Shutt, K.; Heistermann, M.; Kasim, A.; Todd, A.; Kalousova, B.; Profosouva, I.; Petrzelkova, K.; Fuh, T.; Dicky, J.; Bopalanzognako, J.; et al. Effects of habituation, research and ecotourism on faecal glucocorticoid metabolites in wild western lowland gorillas: Implications for conservation management. Biol. Conserv. 2014, 172, 72–79. [Google Scholar] [CrossRef]
- Afonso, E.; Fu, R.; Dupaix, A.; Goydadin, A.C.; Yu, Z.; Callou, C.; Villette, P.; Giraudoux, P.; Li, L. Feeding sites promoting wildlife-related tourism might highly expose the endangered Yunnan snub-nosed monkey (Rhinopithecus bieti) to parasite transmission. Sci. Rep. 2021, 11, 15817. [Google Scholar] [CrossRef] [PubMed]
- Afonso, E.; Fu, R.; Dupaix, A.; Goydadin, A.; Yu, Z.; Li, D.; Giraudoux, P.; Li, L. Creating small food-habituated groups might alter genetic diversity in the endangered Yunnan snub-nosed monkey. Glob. Ecol. Conserv. 2021, 26, e1422. [Google Scholar] [CrossRef]
- Maestripieri, D. Macachiavellian Intelligence: How Rhesus Macaques and Humans Have Conquered the World; University of Chicago Press: Chicago, IL, USA, 2007. [Google Scholar]
- Singh, M.; Kumar, A.; Kumara, H.N.; Macaca mulatta (Amended Version of 2020 Assessment). The IUCN Red List of Threatened Species 2024: E.T12554A256057746. Available online: https://www.iucnredlist.org/species/12554/17950825 (accessed on 17 February 2025).
- Wang, Y.W.; Lu, J.Q.; Tian, J.D. Survey on the status of rhesus macaque-involved tourism in China. Chi. J. Zool. 2022, 57, 514–520. [Google Scholar] [CrossRef]
- Bezanson, M.; McNamara, A. The what and where of primate field research may be failing primate conservation. Evol. Anthropol. 2019, 28, 166–178. [Google Scholar] [CrossRef]
- Andrade, M.C.R.; Penedo, M.C.T.; Ward, T.; Silva, V.F.; Cabello, P.H. Determination of genetic status in a closed colony of rhesus monkeys (Macaca mulatta). Primates 2004, 45, 183–186. [Google Scholar] [CrossRef]
- Kanthaswamy, S.; Kou, A.; Smith, D.G. Population genetic statistics from rhesus macaques (Macaca mulatta) in three different housing configurations at the California National Primate Research Center. J. Am. Assoc. Lab. Anim. 2010, 49, 598–609. [Google Scholar]
- Li, D.Y.; Xu, H.L.; Trask, J.S.; Zhu, Q.; Cheng, A.C.; Smith, D.G.; George, D.; Zhang, L. Genetic diversity and population structure in wild Sichuan rhesus macaques. Mol. Biol. Rep. 2013, 40, 3033–3041. [Google Scholar] [CrossRef] [PubMed]
- Kanthaswamy, S.; Ng, J.; Hernandez-Pacheco, R.; Ruiz-Lambides, A.; Maldonado, E.; Martinez, M.I.; Sariol, C.A. The population genetic composition of conventional and SPF colonies of rhesus macaques (Macaca mulatta) at the Caribbean Primate Research Center. J. Am. Assoc. Lab. Anim. 2016, 55, 147–151. [Google Scholar]
- Kanthaswamy, S.; Oldt, R.F.; Ng, J.; Ruiz-Lambides, A.V.; Maldonado, E.; Martinez, M.I.; Sariol, C.A. Population genetic structure of the Cayo Santiago colony of rhesus macaques (Macaca mulatta). J. Am. Assoc. Lab. Anim. 2017, 56, 396–401. [Google Scholar]
- Zhou, Y.Y.; Tian, J.D.; Lu, J.Q. Genetic structure and recent population demographic history of Taihangshan macaque (Macaca mulatta tcheliensis), North China. Integr. Zool. 2023, 18, 530–542. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Quan, G.Q.; Lin, Y.L.; Charles, S. Extinction of rhesus monkeys (Macaca mulatta) in Xinglung, North China. Int. J. Primatol. 1989, 10, 375–381. [Google Scholar] [CrossRef]
- Lu, J.Q.; Hou, J.H.; Wang, H.F.; Qu, W.Y. Current status of Macaca mulatta in Taihangshan Mountains Area, Jiyuan, Henan, China. Int. J. Primatol. 2007, 28, 1085–1091. [Google Scholar] [CrossRef]
- Wu, S.J.; Luo, J.; Li, Q.Q.; Wang, Y.Q.; Murphy, R.W.; Blair, C.; Wu, S.F.; Yue, B.S.; Zhang, Y.P. Ecological genetics of Chinese rhesus macaque in response to mountain building: All things are not equal. PLoS ONE 2013, 8, e55315. [Google Scholar] [CrossRef]
- Liu, Z.J.; Tan, X.; Orozco-terWengel, P.; Zhou, X.M.; Zhang, L.Y.; Tian, S.L.; Yan, Z.Z.; Xu, H.L.; Ren, B.P.; Zhang, P.; et al. Population genomics of wild Chinese rhesus macaques reveals a dynamic demographic history and local adaptation, with implications for biomedical research. Gigascience 2018, 7, giy106. [Google Scholar] [CrossRef]
- Pang, K.L.; Jin, Q.Q.; Yuan, Z.A.; Kuang, Z.J.; Lu, J.Q.; Tian, J.D. Spondyloarthritis in Taihangshan macaques (Macaca mulatta tcheliensis). Folia Primatol. 2021, 92, 203–210. [Google Scholar] [CrossRef]
- Kong, X.G.; Guo, W.D.; Kuang, S.A.; Yang, Q.; Lu, J.Q. Status and utilization of the rhesus macaques in Wulongkou Scenic Spot, Jiyuan, Henan, China. J. Henan Forest Sci. Tech. 2011, 31, 11–12. [Google Scholar]
- Wang, B.S.; Wang, Z.L.; Tian, J.D.; Cui, Z.W.; Lu, J.Q. Establishment of a microsatellite set for noninvasive paternity testing in free-ranging Macaca mulatta tcheliensis in Mount Taihangshan area, Jiyuan, China. Zool. Stud. 2015, 54, 8. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Rousset, F. GENEPOP’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Rice, W.R. Analyzing tables of statistical tests. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef]
- Carvajal-Rodriguez, A. Myriads: P-value-based multiple testing correction. Bioinformatics 2018, 34, 1043–1045. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Updated from Goudet (1995). J. Hered. 2001, 86, 485–486. [Google Scholar] [CrossRef]
- Cornuet, J.M.; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 2001–2014. [Google Scholar] [CrossRef]
- Cristescu, R.; Sherwin, W.B.; Handasyde, K.; Cahill, V.; Cooper, D.W. Detecting bottlenecks using BOTTLENECK 1.2.02 in wild populations: The importance of the microsatellite structure. Conserv. Genet. 2010, 11, 1043–1049. [Google Scholar] [CrossRef]
- Luikart, G.; Allendorf, F.W.; Cornuet, J.M.; Sherwin, W.B. Distortion of allele frequency distributions provides a test for recent population bottlenecks. J. Hered. 1998, 89, 238–247. [Google Scholar] [CrossRef]
- DeWoody, J.A.; Harder, A.M.; Mathur, S.; Willoughby, J.R. The long-standing significance of genetic diversity in conservation. Mol. Ecol. 2021, 30, 4147–4154. [Google Scholar] [CrossRef]
- Fooden, J. Systematic review of the rhesus macaque, Macaca mulatta (Zimmermann, 1780). Fieldiana Zool. 2000, 96, 1–180. [Google Scholar] [CrossRef]
- Serrote, C.; Reiniger, L.; Silva, K.B.; Rabaiolli, S.; Stefanel, C.M. Determining the polymorphism information content of a molecular marker. Gene 2020, 726, 144175. [Google Scholar] [CrossRef]
- Yao, Y.F.; Dai, Q.X.; Li, J.; Ni, Q.Y.; Zhang, M.W.; Xu, H.L. Genetic diversity and differentiation of the rhesus macaque (Macaca mulatta) population in western Sichuan, China, based on the second exon of the major histocompatibility complex class II DQB (MhcMamu-DQB1) alleles. BMC Evol. Biol. 2014, 14, 130. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.G.; George, D.; Kanthaswamy, S.; McDonough, J. Identification of country of origin and admixture between Indian and Chinese rhesus macaques. Int. J. Primatol. 2006, 27, 881–898. [Google Scholar] [CrossRef]
- Kanthaswamy, S.; von Dollen, A.; Kurushima, J.D.; Alminas, O.; Rogers, J.; Ferguson, B.; Lerche, N.W.; Allen, P.C.; Smith, D.G. Microsatellite markers for standardized genetic management of captive colonies of rhesus macaques (Macaca mulatta). Am. J. Primatol. 2006, 68, 73–95. [Google Scholar] [CrossRef] [PubMed]
- Satkoski, J.A.; Malhi, R.; Kanthaswamy, S.; Tito, R.; Malladi, V.; Smith, D. Pyrosequencing as a method for SNP identification in the rhesus macaque (Macaca mulatta). BMC Genom. 2008, 9, 256. [Google Scholar] [CrossRef]
- Bonhomme, M.; Cuartero, S.; Blancher, A.; Crouau-Roy, B. Assessing natural introgression in 2 biomedical model species, the rhesus macaque (Macaca mulatta) and the long-tailed macaque (Macaca fascicularis). J. Hered. 2009, 100, 158–169. [Google Scholar] [CrossRef]
- Xu, Y.R.; Li, J.H.; Zhu, Y.; Sun, B.H. Development of a microsatellite set for paternity assignment of captive rhesus macaques (Macaca mulatta) from Anhui province, China. Genetika 2013, 49, 838–845. [Google Scholar] [CrossRef] [PubMed]
- de Groot, N.; Doxiadis, G.G.; Otting, N.; de Vos-Rouweler, A.J.; Bontrop, R.E. Differential recombination dynamics within the MHC of macaque species. Immunogenetics 2014, 66, 535–544. [Google Scholar] [CrossRef]
- Xu, Y.T.; Hu, Z.X.; Li, W.J.; Zeng, T.; Zhang, X.Y.; Li, J.; Zhang, W.W.; Yue, B.S. Isolation and strategies of novel tetranucleotide microsatellites with polymorphisms from different chromosomes of the rhesus monkey (Macaca mulatta). Mol. Biol. Rep. 2019, 46, 3955–3966. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
