Effects of Temperature and Precipitation at Large Spatial Scales on Genetic Diversity, Genetic Structure, and Potential Distribution of Agropyron michnoi
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Extraction
2.3. Chloroplast DNA Amplification
2.4. Data Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hughes, A.R.; Inouye, B.D.; Johnson, M.T.; Underwood, N.; Vellend, M. Ecological Consequences of Genetic Diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef]
- Erickson, D.L.; Hamrick, J.; Kochert, G.D. Ecological Determinants of Genetic Diversity in an Expanding Population of the Shrub Myrica Cerifera. Mol. Ecol. 2004, 13, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Proksa, B.; Uhrin, D.; Adamcova, J.; Fuska, J. Ecological and Evolutionary responses to Recentclimate Change. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 637–669. [Google Scholar] [CrossRef]
- Aguirre-Liguori, J.A.; Ramírez-Barahona, S.; Tiffin, P.; Eguiarte, L.E. Climate Change Is Predicted to Disrupt Patterns of Local Adaptation in Wild and Cultivated Maize. Proc. R. Soc. B 2019, 286, 20190486. [Google Scholar] [CrossRef]
- Avolio, M.L.; Beaulieu, J.M.; Smith, M.D. Genetic Diversity of a Dominant C 4 Grass Is Altered with Increased Precipitation Variability. Oecologia 2013, 171, 571–581. [Google Scholar] [CrossRef]
- Zhang, C.; Sun, M.; Zhang, X.; Chen, S.; Nie, G.; Peng, Y.; Huang, L.; Ma, X. Aflp-Based Genetic Diversity of Wild Orchardgrass Germplasm Collections from Central Asia and Western China, and the Relation to Environmental Factors. PLoS ONE 2018, 13, e0195273. [Google Scholar] [CrossRef]
- Wang, T.; Wang, Z.; Xia, F.; Su, Y. Local Adaptation to Temperature and Precipitation in Naturally Fragmented Populations of Cephalotaxus Oliveri, an Endangered Conifer Endemic to China. Sci. Rep. 2016, 6, 25031. [Google Scholar] [CrossRef]
- Parmesan, C.; Yohe, G. A Globally Coherent Fingerprint of Climate Change Impacts across Natural Systems. Nature 2003, 421, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Han, X.; Zhang, Z.; Sun, O.J. Grassland Ecosystems in China: Review of Current Knowledge and Research Advancement. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.J. Grasses and Grassland Ecology; Oxford University Press: Oxford, UK, 2009. [Google Scholar] [CrossRef]
- Broadhurst, L.; Breed, M.; Lowe, A.; Bragg, J.; Catullo, R.; Coates, D.; Encinas-Viso, F.; Gellie, N.; James, E.; Krauss, S. Genetic Diversity and Structure of the Australian Flora. Divers. Distrib. 2017, 23, 41–52. [Google Scholar] [CrossRef]
- Hamann, E.; Kesselring, H.; Armbruster, G.F.; Scheepens, J.F.; Stöcklin, J. Evidence of Local Adaptation to Fine-and Coarse-Grained Environmental Variability in Poa Alpina in the Swiss Alps. J. Ecol. 2016, 104, 1627–1637. [Google Scholar] [CrossRef]
- Volis, S.; Ormanbekova, D.; Shulgina, I. Role of Selection and Gene Flow in Population Differentiation at the Edge Vs. Interior of the Species Range Differing in Climatic Conditions. Mol. Ecol. 2016, 25, 1449–1464. [Google Scholar] [CrossRef]
- Wu, Z.; Dijkstra, P.; Koch, G.W.; Hungate, B.A. Biogeochemical and Ecological Feedbacks in Grassland Responses to Warming. Nat. Clim. Change 2012, 2, 458–461. [Google Scholar] [CrossRef]
- Li, R.; Zhang, H.; Zhou, X.; Guan, Y.; Yao, F.; Song, G.; Wang, J.; Zhang, C. Genetic Diversity in Chinese Sorghum Landraces Revealed by Chloroplast Simple Sequence Repeats. Genet. Resour. Crop Evol. 2010, 57, 1–15. [Google Scholar] [CrossRef]
- Zhu, Z.-M.; Gao, X.-F.; Fougère-Danezan, M. Phylogeny of Rosa Sections Chinenses and Synstylae (Rosaceae) Based on Chloroplast and Nuclear Markers. Mol. Phylogenet. Evol. 2015, 87, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Yan, M.; Wang, J.; Zhao, L.; Zhou, F.; Hu, Z.; Jin, S.; Diao, Y. The Complete Chloroplast Genome Sequences of Five Miscanthus Species, and Comparative Analyses with Other Grass Plastomes. Ind. Crops Prod. 2021, 162, 113248. [Google Scholar] [CrossRef]
- Albert, V.A.; Backlund, A.; Bremer, K.; Chase, M.W.; Manhart, J.R.; Mishler, B.D.; Nixon, K.C. Functional Constraints and Rbcl Evidence for Land Plant Phylogeny. Ann. Mo. Bot. Gard. 1994, 81, 534–567. [Google Scholar] [CrossRef]
- Wu, Z.; Tian, C.; Yang, Y.; Li, Y.; Liu, Q.; Li, Z.; Jin, K. Comparative and Phylogenetic Analysis of Complete Chloroplast Genomes in Leymus (Triticodae, Poaceae). Genes 2022, 13, 1425. [Google Scholar] [CrossRef]
- Raggi, L.; Bitocchi, E.; Russi, L.; Marconi, G.; Sharbel, T.F.; Veronesi, F.; Albertini, E. Understanding Genetic Diversity and Population Structure of a Poa Pratensis Worldwide Collection through Morphological, Nuclear and Chloroplast Diversity Analysis. PLoS ONE 2015, 10, e0124709. [Google Scholar] [CrossRef]
- Tso, K.L.; Allan, G.J. Environmental Variation Shapes Genetic Variation in Bouteloua Gracilis: Implications for Restoration Management of Natural Populations and Cultivated Varieties in the Southwestern United States. Ecol. Evol. 2019, 9, 482–499. [Google Scholar] [CrossRef]
- Wani, G.A.; Shah, M.A.; Tekeu, H.; Reshi, Z.A.; Atangana, A.R.; Khasa, D.P. Phenotypic Variability and Genetic Diversity of Phragmites Australis in Quebec and Kashmir Reveal Contrasting Population Structure. Plants 2020, 9, 1392. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, C.; Yang, Y. Crecimiento de módulos clonales de Agropyron michnoi en la planicie Songnen del Noreste de China. Phyton 2015, 84, 417–422. [Google Scholar] [CrossRef]
- Yang, Y.-F.; Zheng, H.-Y.; Li, J.-D. The Effects of Grazing on Age Structure in Clonal Populations of Agropyron michnoi. Chin. J. Plant Ecol. 2001, 25, 71. [Google Scholar]
- Chen, S.; Ma, X.; Zhang, X.; Huang, L.; Zhou, J. Genetic Diversity and Relationships among Accessions of Five Crested Wheatgrass Species (Poaceae: Agropyron) Based on Gliadin Analysis. Genet. Mol. Res. 2013, 12, 5704–5713. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Shimizu, Y.; Ando, M.; Sakai, F. Clonal Structure of Natural Populations of Cryptomeria Japonica Growing at Different Positions on Slopes, Detected Using Rapd Markers. Biochem. Syst. Ecol. 2002, 30, 733–748. [Google Scholar] [CrossRef]
- Li, C.; Lu, S.; Yang, Q. Asian Origin for Polystichum (Dryopteridaceae) Based on Rbc L Sequences. Chin. Sci. Bull. 2004, 49, 1146–1150. [Google Scholar] [CrossRef]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal Primers for Amplification of Three Non-Coding Regions of Chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.; Harvester, B.V.S. A Website and Program for Visualizing Structure Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Rosenberg, N.A.; Pritchard, J.K.; Weber, J.L.; Cann, H.M.; Kidd, K.K.; Zhivotovsky, L.A.; Feldman, M.W. Genetic Structure of Human Populations. Science 2002, 298, 2381–2385. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Gao, Y.; Wang, J.; Ren, A.; Xu, H. Rapd Diversity of Stipa Grandis Populations and Its Relationship with Some Ecological Factors. Acta Ecol. Sin. 2006, 26, 1312–1318. [Google Scholar] [CrossRef]
- Huang, W.-D.; Zhao, X.-Y.; Zhao, X.; Li, Y.-L.; Pan, C.-C. Environmental Determinants of Genetic Diversity in Caragana microphylla (Fabaceae) in Northern China. Bot. J. Linn. Soc. 2016, 181, 269–278. [Google Scholar] [CrossRef]
- Hao, Z.; Xin, L.; Fenghe, J. Effects of Different Water Supply on the Reproduction of Stipa Krylovii and Artemisia Frigida Populations in Degraded Steppe. Acta Agrestia Sin. 2005, 13, 106–110. [Google Scholar] [CrossRef]
- Chen, S.; Chen, R.; Zeng, X.; Chen, X.; Qin, X.; Zhang, Z.; Sun, Y. Genetic Diversity, Population Structure, and Conservation Units of Castanopsis sclerophylla (Fagaceae). Forests 2022, 13, 1239. [Google Scholar] [CrossRef]
- Kabiel, H.; Hegazy, A.; Faisal, M.; Doma, E. Genetic Variations within and among Populations of Anastatica Heirochuntica at Macroscale Geographical Range. Appl. Ecol. Environ. Res. 2013, 11, 343–354. [Google Scholar] [CrossRef]
- Nybom, H. Comparison of Different Nuclear DNA Markers for Estimating Intraspecific Genetic Diversity in Plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef]
- Sexton, J.P.; Hangartner, S.B.; Hoffmann, A.A. Genetic Isolation by Environment or Distance: Which Pattern of Gene Flow Is Most Common? Evolution 2014, 68, 1–15. [Google Scholar] [CrossRef]
- Still, D.; Kim, D.-H.; Aoyama, N. Genetic Variation in Echinacea Angustifolia Along a Climatic Gradient. Ann. Bot. 2005, 96, 467–477. [Google Scholar] [CrossRef]
- Edelaar, P.; Bolnick, D.I. Non-Random Gene Flow: An Underappreciated Force in Evolution and Ecology. Trends Ecol. Evol. 2012, 27, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Hancock, A.M.; Witonsky, D.B.; Alkorta-Aranburu, G.; Beall, C.M.; Gebremedhin, A.; Sukernik, R.; Utermann, G.; Pritchard, J.K.; Coop, G.; Di Rienzo, A. Adaptations to Climate-Mediated Selective Pressures in Humans. PLoS Genet. 2011, 7, e1001375. [Google Scholar] [CrossRef] [PubMed]
- Al-Gharaibeh, M.; Hamasha, H.; Rosche, C.; Lachmuth, S.; Wesche, K.; Hensen, I. Environmental Gradients Shape the Genetic Structure of Two Medicinal Salvia Species in Jordan. Plant Biol. 2017, 19, 227–238. [Google Scholar] [CrossRef]
- Gray, M.M.; St Amand, P.; Bello, N.M.; Galliart, M.B.; Knapp, M.; Garrett, K.A.; Morgan, T.J.; Baer, S.G.; Maricle, B.R.; Akhunov, E.D. Ecotypes of an Ecologically Dominant Prairie Grass (Andropogon gerardii) Exhibit Genetic Divergence across the Us Midwest Grasslands’ Environmental Gradient. Mol. Ecol. 2014, 23, 6011–6028. [Google Scholar] [CrossRef]
- Song, R.; Zhang, X.; Zhang, Z.; Zhou, C. Climatic Factors, but Not Geographic Distance, Promote Genetic Structure and Differentiation of Cleistogenes squarrosa (Trin.) Keng Populations. Front. Bioinform. 2024, 4, 1454689. [Google Scholar] [CrossRef]
- Pacheco-Hernández, Y.; Villa-Ruano, N.; Lozoya-Gloria, E.; Barrales-Cortés, C.A.; Jiménez-Montejo, F.E.; Cruz-López, M.D.C. Influence of Environmental Factors on the Genetic and Chemical Diversity of Brickellia Veronicifolia Populations Growing in Fragmented Shrublands from Mexico. Plants 2021, 10, 325. [Google Scholar] [CrossRef] [PubMed]
- Brandt, J.S.; Haynes, M.A.; Kuemmerle, T.; Waller, D.M.; Radeloff, V.C. Regime Shift on the Roof of the World: Alpine Meadows Converting to Shrublands in the Southern Himalayas. Biol. Conserv. 2013, 158, 116–127. [Google Scholar] [CrossRef]
- Steinbauer, M.J.; Grytnes, J.-A.; Jurasinski, G.; Kulonen, A.; Lenoir, J.; Pauli, H.; Rixen, C.; Winkler, M.; Bardy-Durchhalter, M.; Barni, E. Accelerated Increase in Plant Species Richness on Mountain Summits Is Linked to Warming. Nature 2018, 556, 231–234. [Google Scholar] [CrossRef]

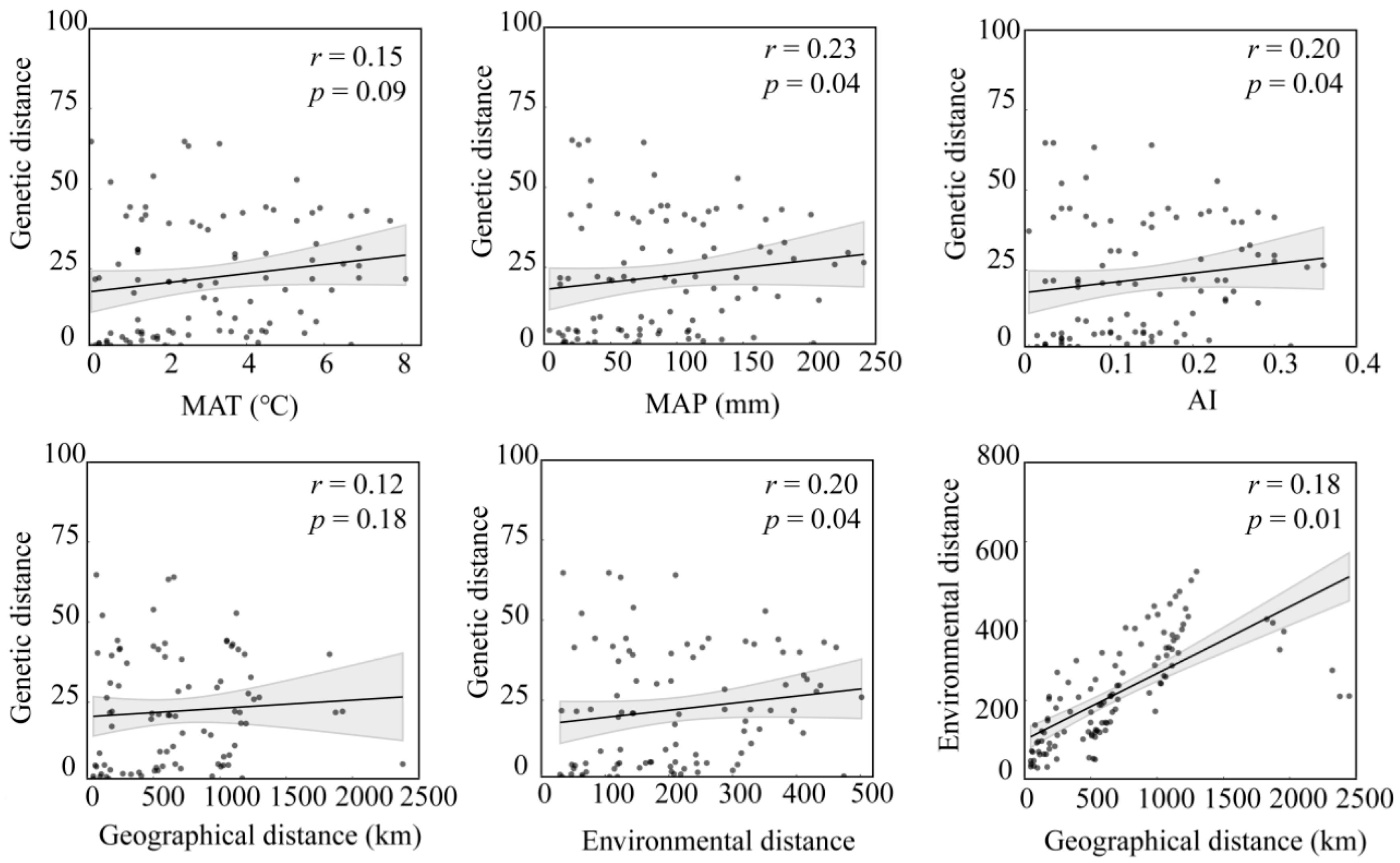
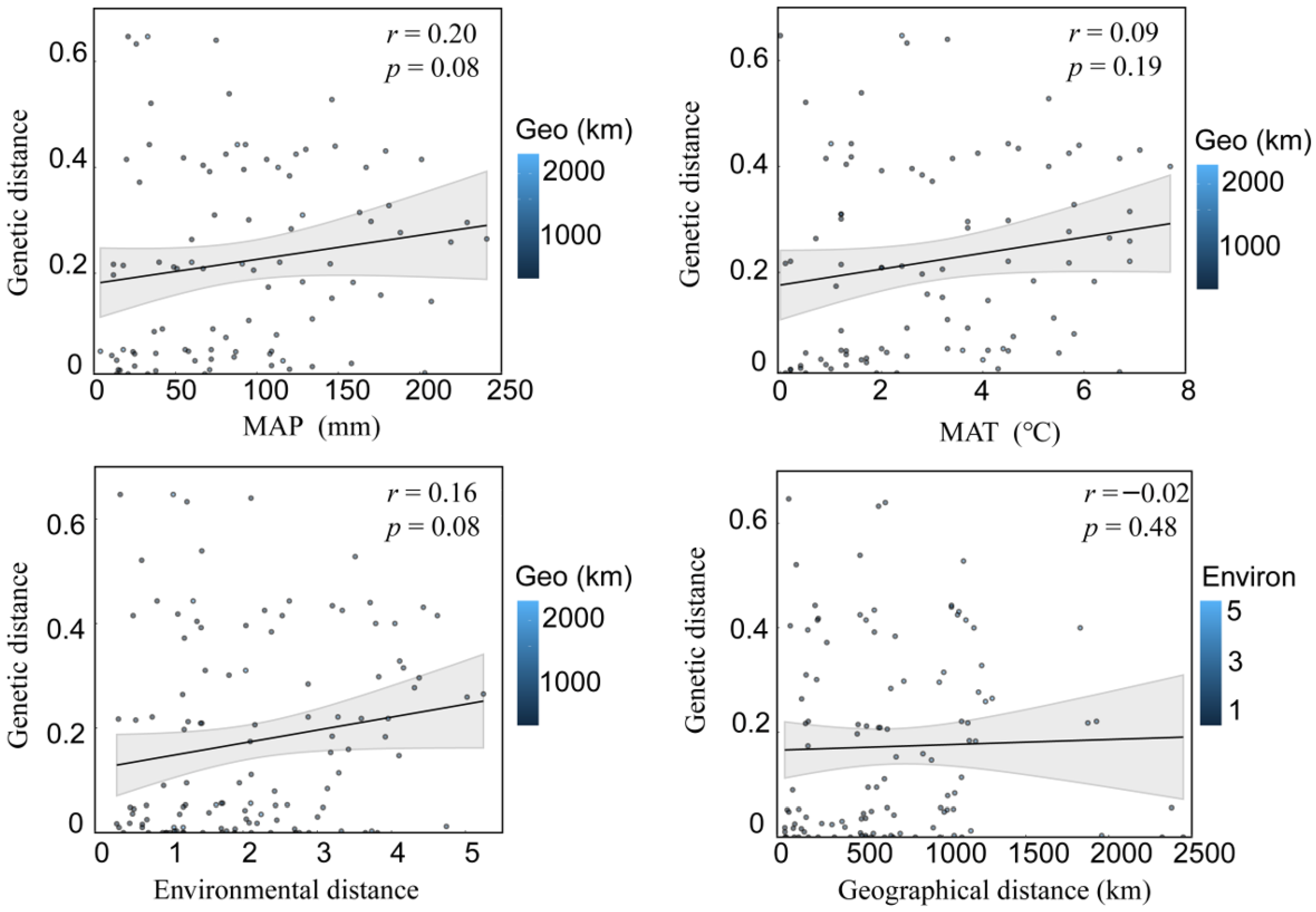
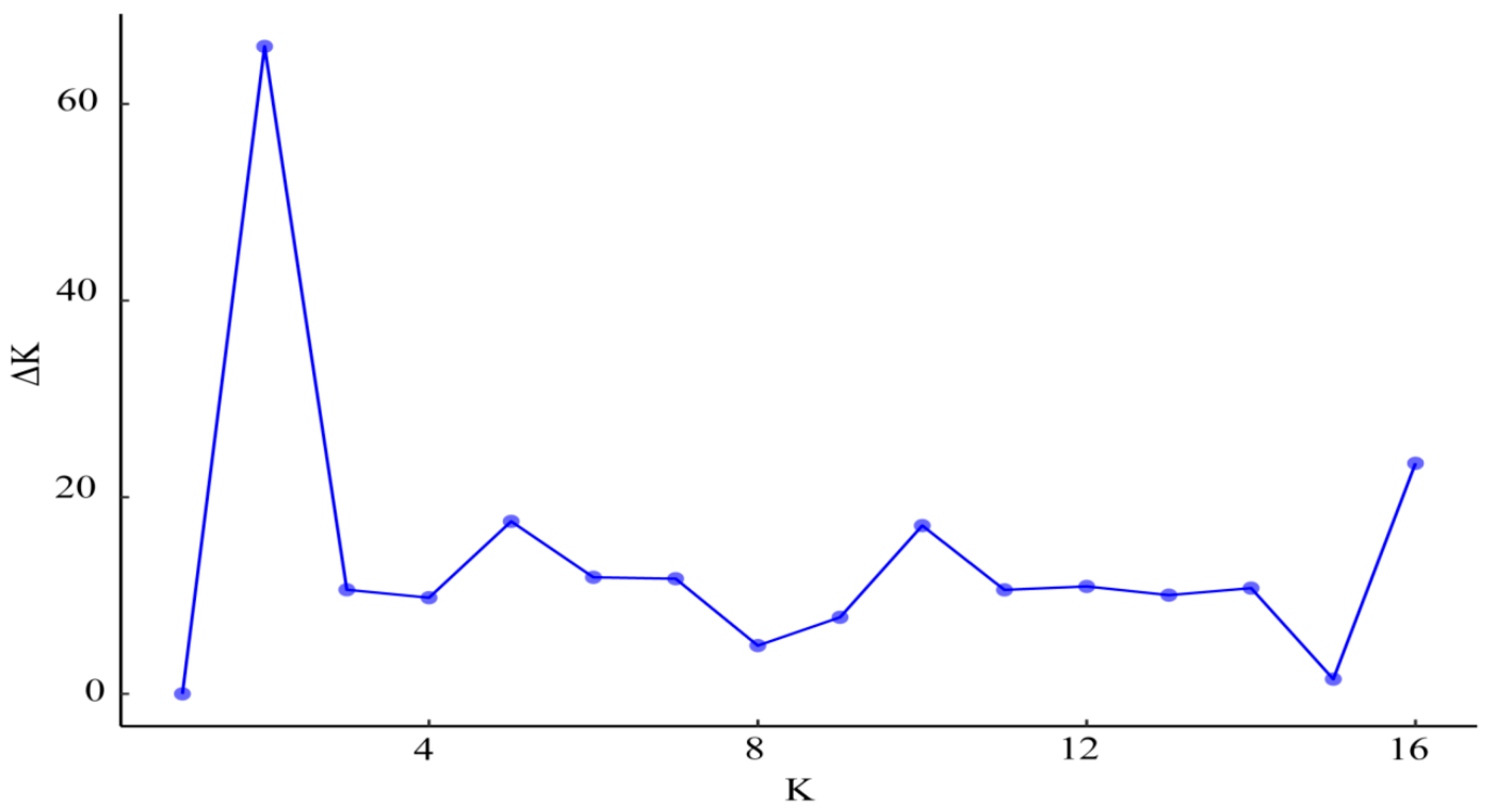
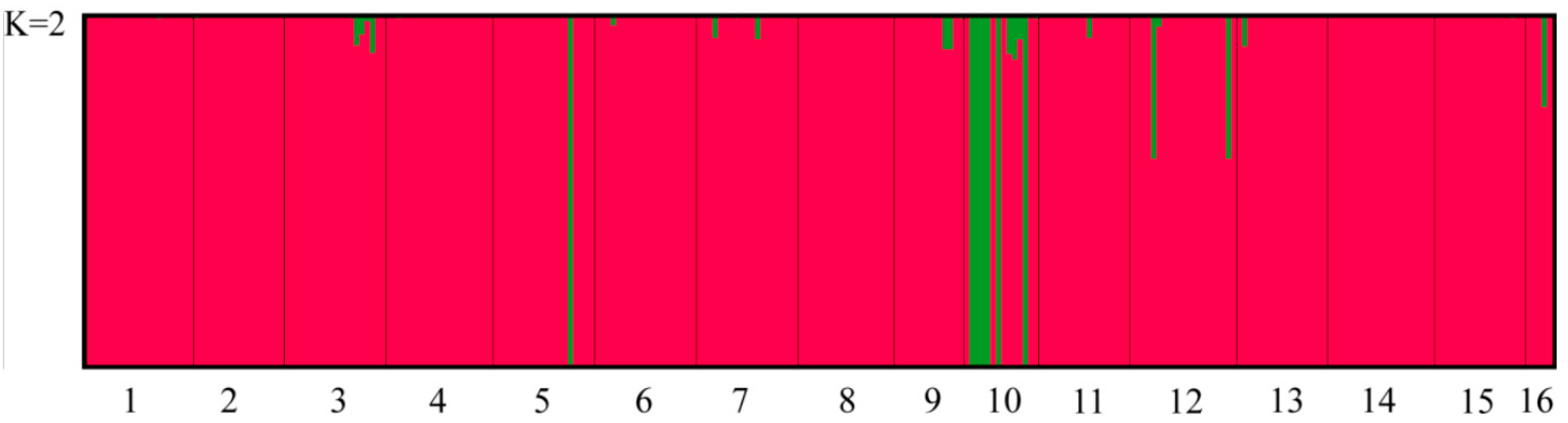
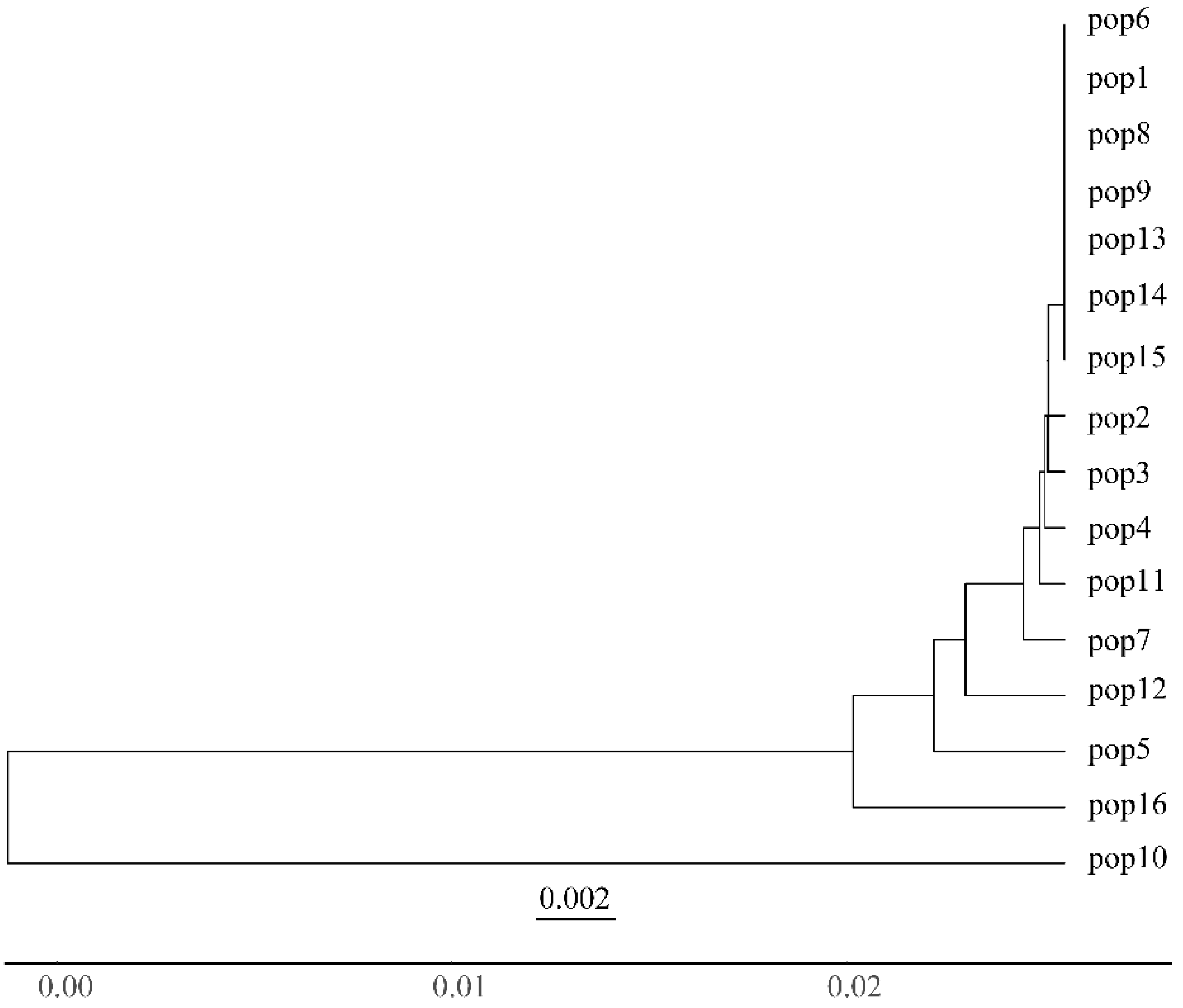
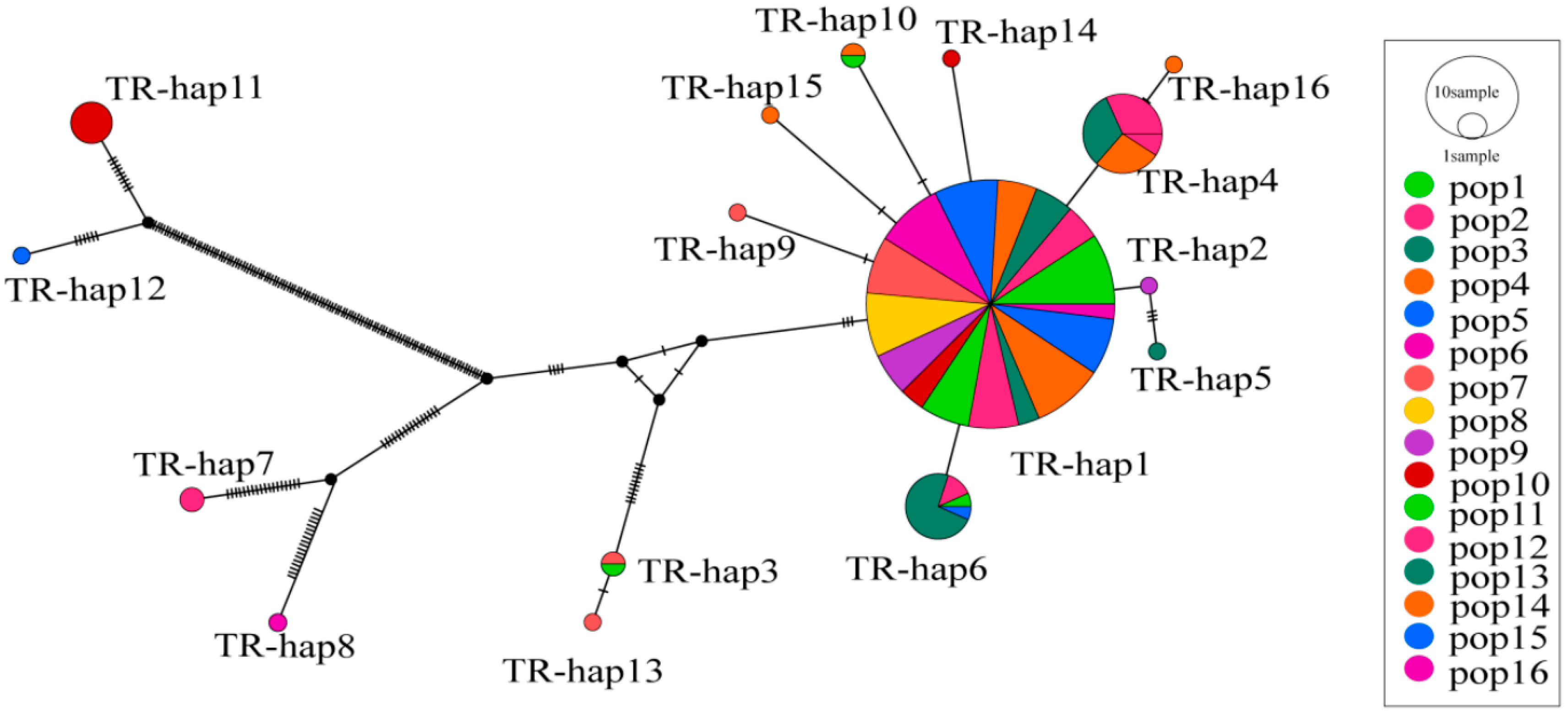
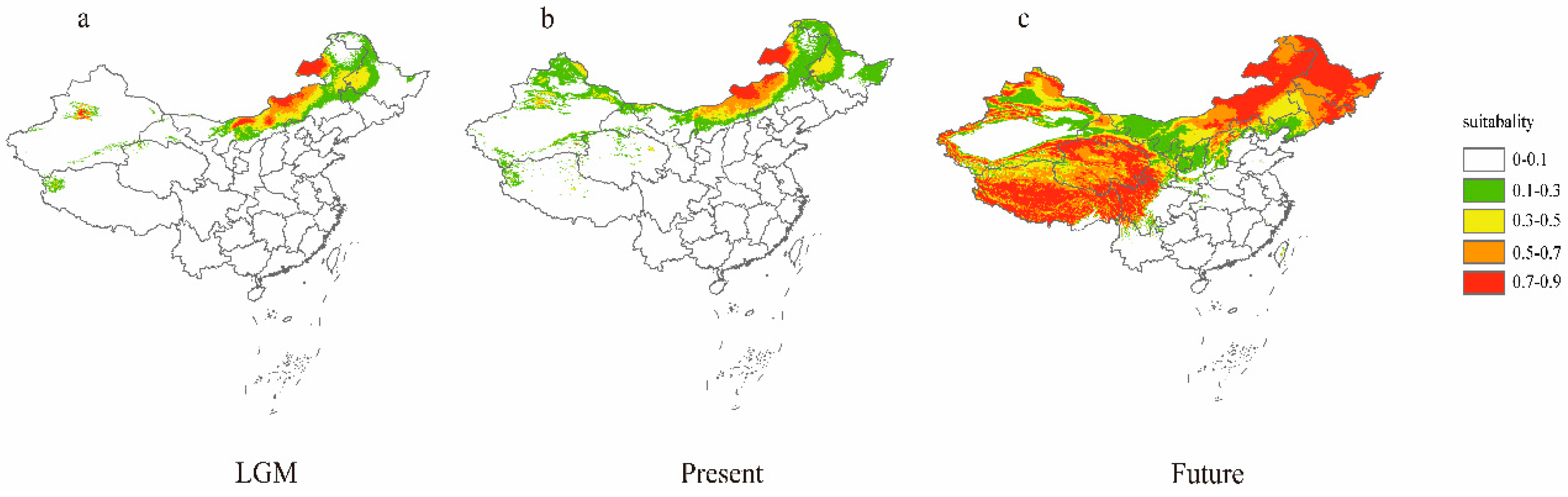
| Population | Sample Size | Latitude | Longitude | MAT | PET (mm) | MAP (mm) | AI = MAP/PET |
|---|---|---|---|---|---|---|---|
| 1 | 20 | 43.10 | 85.22 | −24.00 | 611.00 | 264.00 | 0.43 |
| 2 | 17 | 41.80 | 107.47 | 42.00 | 894.00 | 151.00 | 0.17 |
| 3 | 19 | 41.87 | 108.05 | 47.00 | 910.00 | 173.00 | 0.19 |
| 4 | 20 | 41.91 | 108.71 | 35.00 | 873.00 | 204.00 | 0.23 |
| 5 | 19 | 42.16 | 109.17 | 44.00 | 917.00 | 189.00 | 0.21 |
| 6 | 19 | 43.85 | 114.09 | 20.00 | 872.00 | 222.00 | 0.25 |
| 7 | 19 | 43.98 | 114.83 | 14.00 | 843.00 | 246.00 | 0.29 |
| 8 | 18 | 43.93 | 115.70 | 12.00 | 854.00 | 271.00 | 0.32 |
| 9 | 13 | 44.99 | 118.75 | 9.00 | 809.00 | 380.00 | 0.47 |
| 10 | 14 | 47.66 | 119.30 | −18.00 | 751.00 | 352.00 | 0.47 |
| 11 | 17 | 48.10 | 118.46 | −6.00 | 775.00 | 285.00 | 0.37 |
| 12 | 20 | 48.50 | 117.15 | 5.00 | 781.00 | 260.00 | 0.33 |
| 13 | 17 | 49.34 | 117.09 | −15.00 | 740.00 | 297.00 | 0.40 |
| 14 | 20 | 49.53 | 118.01 | −11.00 | 752.00 | 318.00 | 0.42 |
| 15 | 17 | 49.78 | 118.53 | −14.00 | 754.00 | 332.00 | 0.44 |
| 16 | 5 | 49.19 | 120.36 | −14.00 | 739.00 | 392.00 | 0.53 |
| Population | π | K | Hd | h | Haplotype Distribution |
|---|---|---|---|---|---|
| Pop1 | 0.00031 | 0.679 | 0483 | 1 | Hap1 (20) |
| Pop2 | 0.00047 | 1.074 | 0.728 | 2 | Hap1 (10); Hap4 (7) |
| Pop3 | 0.00066 | 1.368 | 0.754 | 3 | Hap1 (11); Hap4 (7); Hap5 (1) |
| Pop4 | 0.00047 | 1.068 | 0.732 | 5 | Hap1 (11); Hap4 (6); Hap10 (1); Hap15 (1); Hap16 (1) |
| Pop5 | 0.02426 | 53.860 | 0.743 | 2 | Hap1 (18); Hap12 (1) |
| Pop6 | 0.00036 | 0.807 | 0.450 | 1 | Hap1 (19) |
| Pop7 | 0.00314 | 7.088 | 0.544 | 4 | Hap1 (16); Hap3 (1); Hap9 (1); Hap13 (1) |
| Pop8 | 0.00026 | 0.582 | 0.542 | 1 | Hap1 (18) |
| Pop9 | 0.00007 | 0.154 | 0.154 | 2 | Hap1 (12); Hap2 (1) |
| Pop10 | 0.07762 | 124.363 | 0.714 | 3 | Hap1 (7); Hap11 (6); Hap14 (1) |
| Pop11 | 0.00209 | 4.721 | 0.735 | 4 | Hap1 (14); Hap3 (1); Hap6 (1); Hap10 (1) |
| Pop12 | 0.00674 | 13.605 | 0.795 | 4 | Hap1 (14); Hap4 (2); Hap6 (2); Hap7 (2) |
| Pop13 | 0.00028 | 0.603 | 0.522 | 2 | Hap1 (6); Hap6 (11) |
| Pop14 | 0.00004 | 0.100 | 0.100 | 1 | Hap1 (20) |
| Pop15 | 0.00026 | 0.588 | 0.426 | 2 | Hap1 (16); Hap6 (1) |
| Pop16 | 0.01632 | 36.400 | 0.400 | 2 | Hap1 (4); Hap8 (1) |
| Gene Fragment | π | K | Hd | h | Tajima’s D | Nst | Gst | HS | HT |
|---|---|---|---|---|---|---|---|---|---|
| rbcL | 0.00115 | 1.49721 | 0.268 | 11 | −2.41191 ** | 0.127 | 0.210 | 0.224 | 0.283 |
| trnL-F | 0.03500 | 8.39971 | 0.229 | 10 | −2.03966 * | 0.414 | 0.304 | 0.195 | 0.280 |
| Combined Sequence | 0.00536 | 8.22243 | 0.370 | 16 | −2.33804 ** | 0.000 | 0.035 | 0.172 | 0.178 |
| Source of Variation | df | Sum of Squares | Variance Components | Percentage of Variation |
|---|---|---|---|---|
| Within populations | 15 | 359.874 | 1.23227 Va | 29.43% |
| Among populations | 258 | 762.488 | 2.95538 Vb | 70.57% |
| Total | 273 | 1122.361 | 4.18765 | 100% |
| Fixation Index Fst = 0.29 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Song, R.; Yang, T.; Zhou, C. Effects of Temperature and Precipitation at Large Spatial Scales on Genetic Diversity, Genetic Structure, and Potential Distribution of Agropyron michnoi. Diversity 2025, 17, 798. https://doi.org/10.3390/d17110798
Zhang Z, Song R, Yang T, Zhou C. Effects of Temperature and Precipitation at Large Spatial Scales on Genetic Diversity, Genetic Structure, and Potential Distribution of Agropyron michnoi. Diversity. 2025; 17(11):798. https://doi.org/10.3390/d17110798
Chicago/Turabian StyleZhang, Zhuo, Ruyan Song, Tingting Yang, and Chan Zhou. 2025. "Effects of Temperature and Precipitation at Large Spatial Scales on Genetic Diversity, Genetic Structure, and Potential Distribution of Agropyron michnoi" Diversity 17, no. 11: 798. https://doi.org/10.3390/d17110798
APA StyleZhang, Z., Song, R., Yang, T., & Zhou, C. (2025). Effects of Temperature and Precipitation at Large Spatial Scales on Genetic Diversity, Genetic Structure, and Potential Distribution of Agropyron michnoi. Diversity, 17(11), 798. https://doi.org/10.3390/d17110798





