The Multipartite Mitogenome of Camellia sinensis cv. Xinyang10 Reveals Frequent Reorganization and Hints at Phylogeographic History
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Sequencing, Assembly, and Annotation
2.2. Mitogenome Collinearity Analysis
2.3. Repetitive Sequence Analysis
2.4. Intracellular Gene Transfer
2.5. RNA Editing Prediction
2.6. Codon Usage Analysis and Ka/Ks Estimation
2.7. Phylogenetic Analysis
3. Results
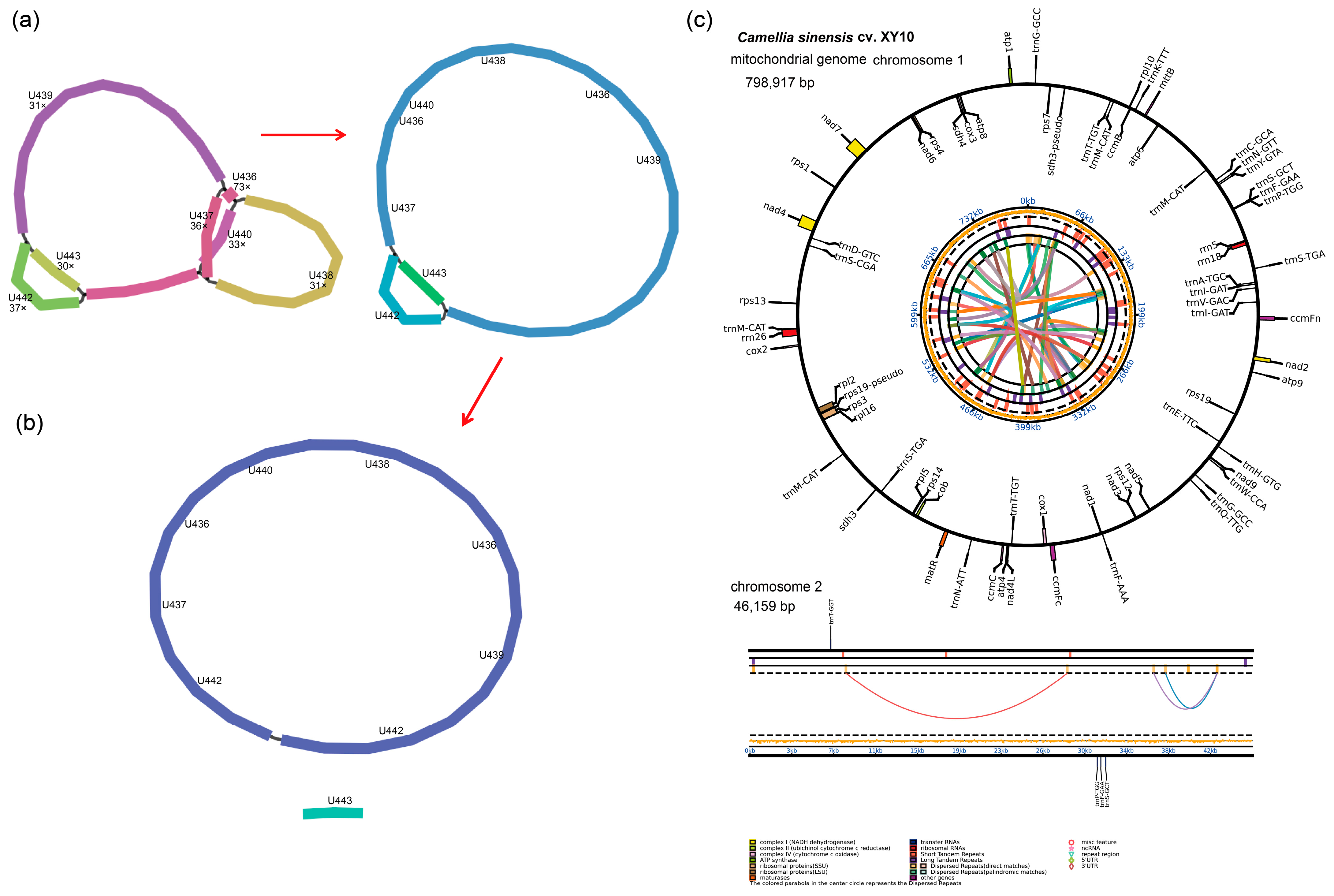
3.1. Mitogenome Assembly and Annotation
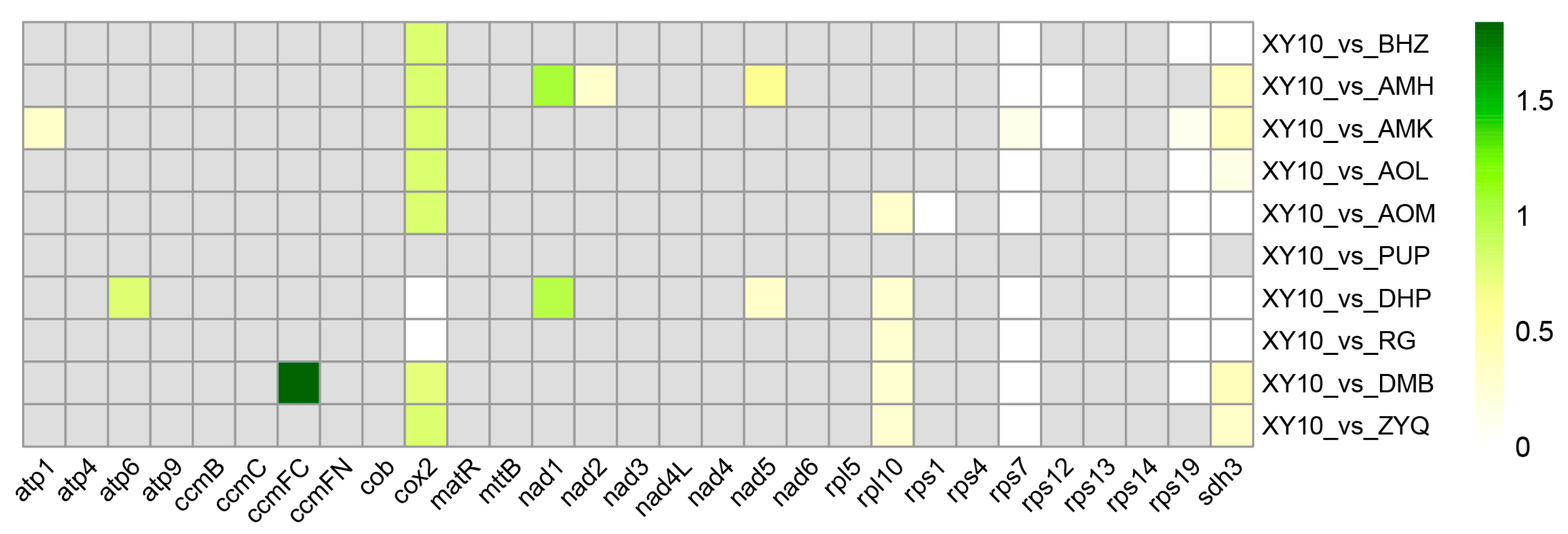
3.2. Mitochondrial Genome Collinearity Between Diverse Tea Plants
3.3. Repetitive Elements in the Mitochondrial Genome of C. sinensis cv. Xinyang10
3.4. Collinearity Analysis of the Mitochondrial GENOME and Chloroplast Genome of C. sinensis cv. Xinyang10
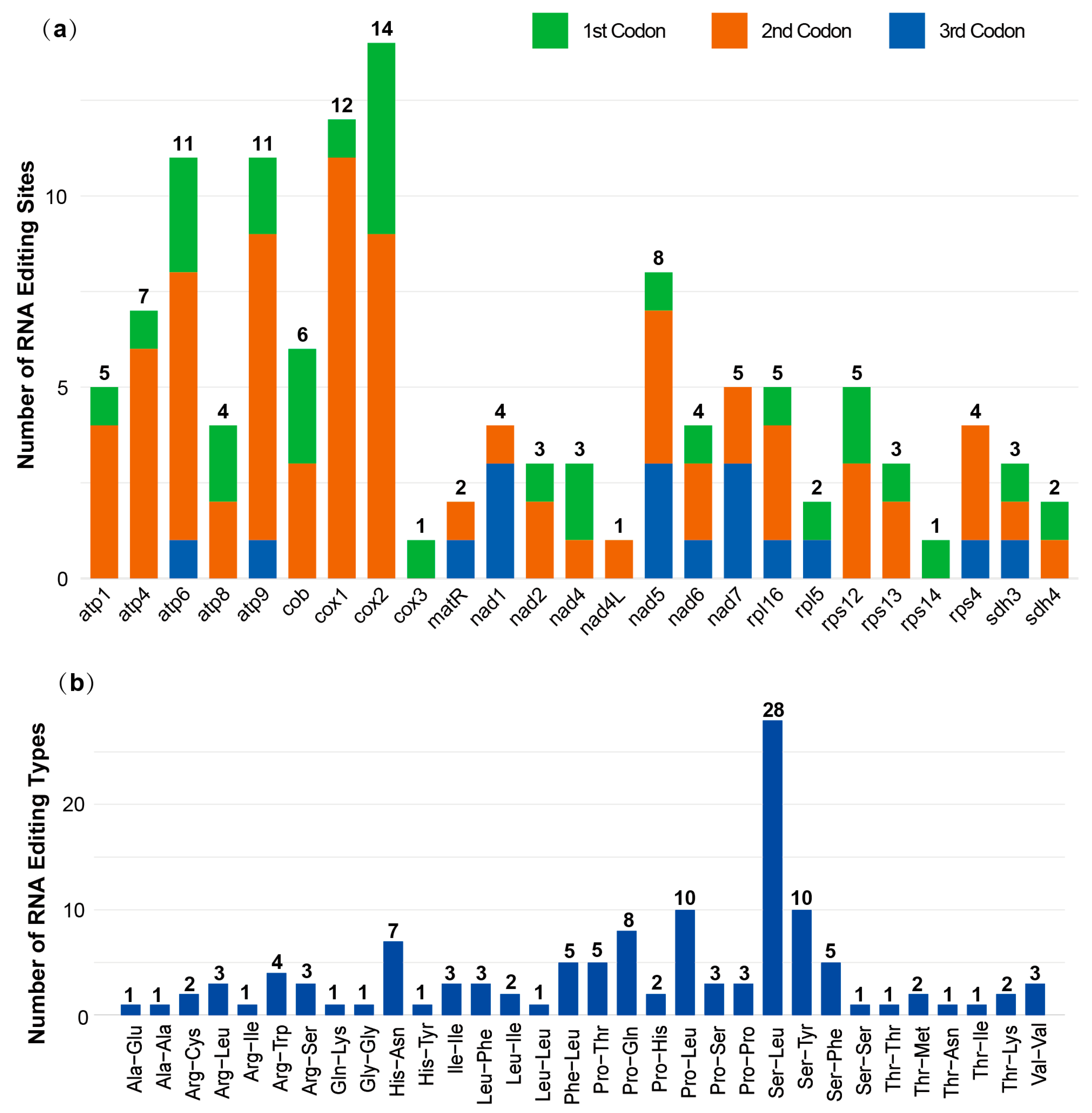
3.5. RNA Editing Serves to Modify the Physicochemical Properties of Encoded Amino Acids
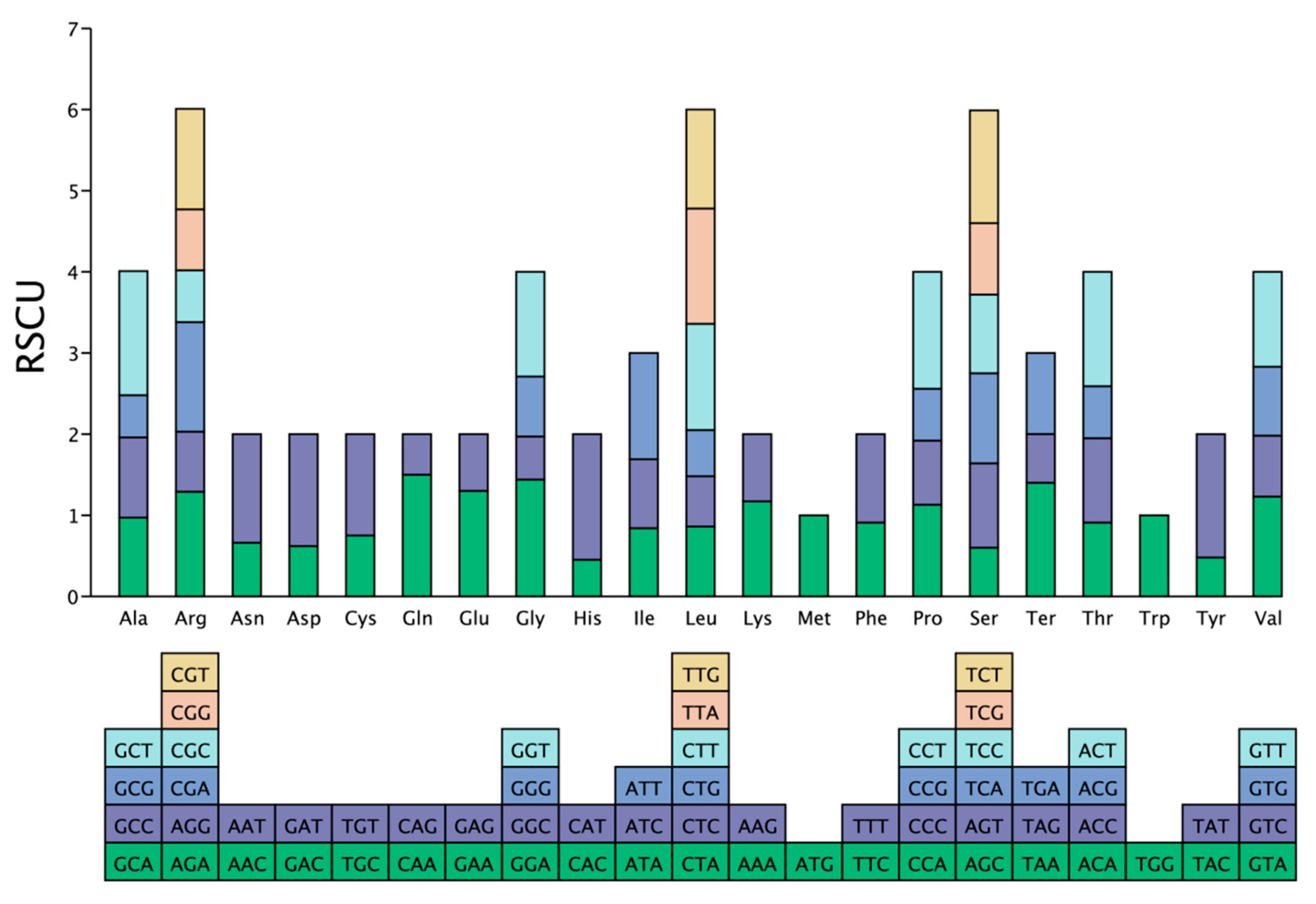
3.6. Codon Usage Bias in Mitochondrial Genome of C. sinensis cv. Xinyang10
3.7. Assessing Selective Pressure in Tea Plants
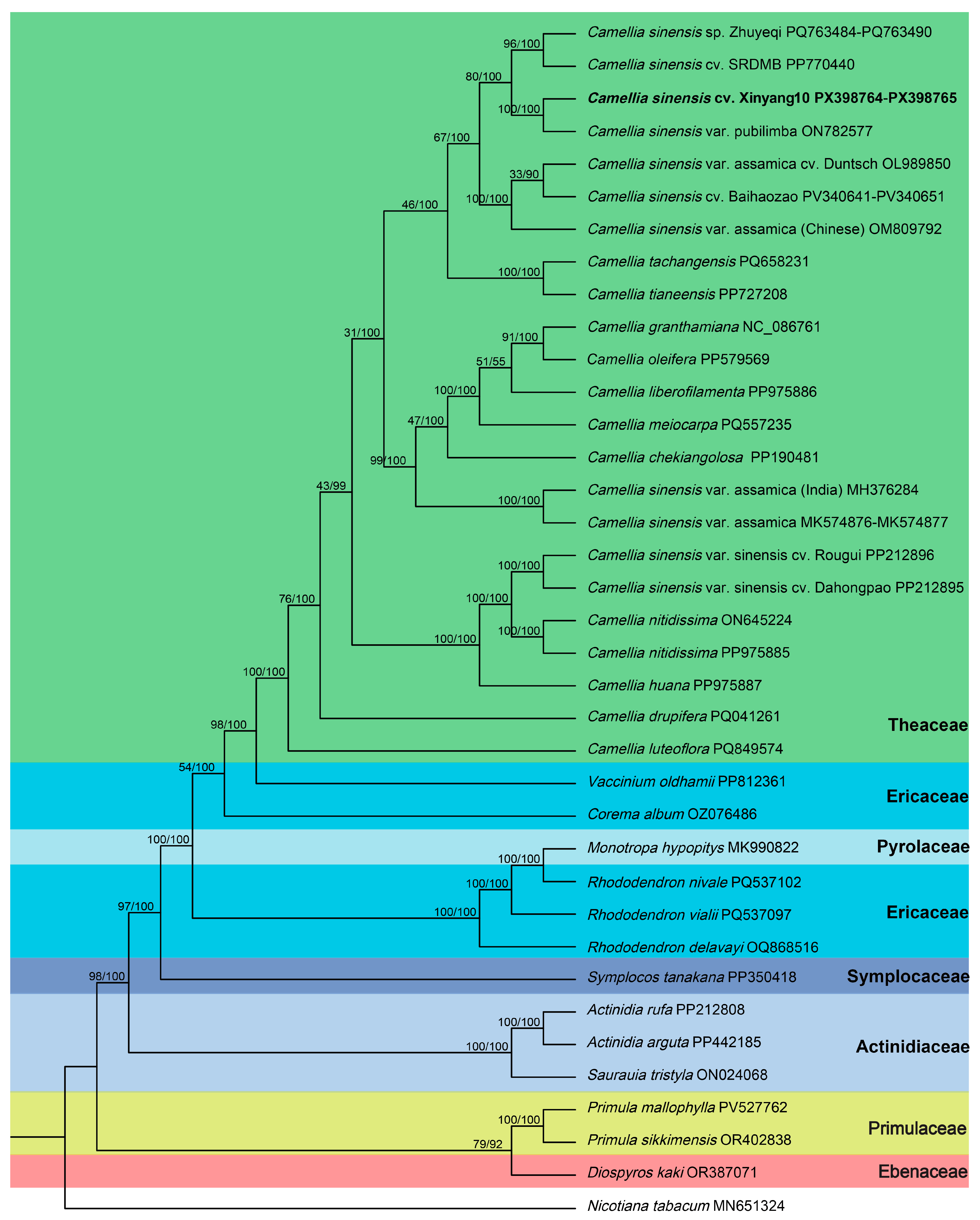
3.8. Phylogenetic Analysis
4. Discussion
4.1. Structure and Size of the C. sinensis cv. Xinyang10 Mitogenome
4.2. Codon Usage Bias and Its Evolutionary Implications in the Mitogenome of C. sinensis cv. Xinyang10
4.3. Phylogenetic Relationships Based on Mitochondrial Genomes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, Z.Q.; Liao, X.Z.; Zhang, X.N.; Tembrock, L.R.; Broz, A. Genomic architectural variation of plant mitochondria—A review of multichromosomal structuring. J. Syst. Evol. 2022, 60, 160–168. [Google Scholar] [CrossRef]
- Morley, S.A.; Nielsen, B.L. Plant mitochondrial DNA. Front. Biosci. (Landmark Ed.) 2017, 22, 1023–1032. [Google Scholar]
- Jethva, J.; Schmidt, R.R.; Sauter, M.; Selinski, J. Try or Die: Dynamics of Plant Respiration and How to Survive Low Oxygen Conditions. Plants 2022, 11, 205. [Google Scholar] [CrossRef] [PubMed]
- Darracq, A.; Varré, J.S.; Maréchal-Drouard, L.; Courseaux, A.; Castric, V.; Saumitou-Laprade, P.; Oztas, S.; Lenoble, P.; Vacherie, B.; Barbe, V.; et al. Structural and content diversity of mitochondrial genome in beet: A comparative genomic analysis. Genome Biol. Evol. 2011, 3, 723–736. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, N.; Li, S.; Grover, C.E.; Nie, H.; Wendel, J.F.; Hua, J. Plant Mitochondrial Genome Evolution and Cytoplasmic Male Sterility. Crit. Rev. Plant Sci. 2017, 36, 55–69. [Google Scholar] [CrossRef]
- Liberatore, K.L.; Dukowic-Schulze, S.; Miller, M.E.; Chen, C.; Kianian, S.F. The role of mitochondria in plant development and stress tolerance. Free Radic. Biol. Med. 2016, 100, 238–256. [Google Scholar] [CrossRef]
- Quesada, V. The roles of mitochondrial transcription termination factors (MTERFs) in plants. Physiol. Plant. 2016, 157, 389–399. [Google Scholar] [CrossRef]
- de Cássia Monteiro-Batista, R.; Siqueira, J.A.; da Fonseca Pereira, P.; Barreto, P.; Feitosa-Araujo, E.; Araújo, W.L.; Nunes-Nesi, A. Potential roles of mitochondrial carrier proteins in plant responses to abiotic stress. J. Exp. Bot. 2025, eraf032. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Jiao, Y.; Wang, Y.; He, D.; Liu, Q.; Linchen, R.; Gao, Y.; Wang, J.; Wang, X.; Huang, T.; et al. Complete mitochondrial genome assembly and comparative analysis of Fagopyrum dibotrys (Golden Buckwheat). BMC Plant Biol. 2025, 25, 985. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yan, R.R.; Yang, C.Y.; Ling, L.Z.; Bai, X.X.; Ren, Q.F.; Hu, G.X. Mitochondrial genome analysis of the endangered Oreocharis esquirolii: Insights into evolutionary adaptation and conservation. BMC Plant Biol. 2025, 25, 827. [Google Scholar] [CrossRef]
- Xu, C.; Bi, W.; Ma, R.Y.; Li, P.R.; Liu, F.; Liu, Z.W. Assembly and comparative analysis of the complete mitochondrial of Spodiopogon sagittifolius, an endemic and protective species from Yunnan, China. BMC Plant Biol. 2025, 25, 373. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Luo, X.; Zhang, T.; Zhou, G.; Li, J.; Zhang, B.; Li, P.; Huang, H. Assembly and comparative analysis of the complete mitochondrial genome of white towel gourd (Luffa cylindrica). Genomics 2024, 116, 110859. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Zhang, Z.; Tso, N.; Zhang, S.; Chen, Y.; Shu, Q.; Li, J.; Liang, Z.; Wang, R.; Wang, J.; et al. Complete mitochondrial genome of Hippophae tibetana: Insights into adaptation to high-altitude environments. Front. Plant Sci. 2024, 15, 1449606. [Google Scholar] [CrossRef]
- Miao, X.; Yang, W.; Li, D.; Wang, A.; Li, J.; Deng, X.; He, L.; Niu, J. Assembly and comparative analysis of the complete mitochondrial and chloroplast genome of Cyperus stoloniferus (Cyperaceae), a coastal plant possessing saline-alkali tolerance. BMC Plant Biol. 2024, 24, 628. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liang, L.; Ran, J.; Yang, F.; Ran, M.; Yong, X.; Kong, C.; Tang, Y.; Li, H. Re-Sequencing the Mitochondrial Genome Unveils a Novel Isomeric Form of NWB CMS Line in Radish and Functional Verification of Its Candidate Sterile Gene. Horticulturae 2024, 10, 395. [Google Scholar] [CrossRef]
- Hao, J.; Liang, Y.; Wang, T.; Su, Y. Correlations of gene expression, codon usage bias, and evolutionary rates of the mitochondrial genome show tissue differentiation in Ophioglossum vulgatum. BMC Plant Biol. 2025, 25, 134. [Google Scholar] [CrossRef]
- Ramadan, A.; Alnufaei, A.A.; Fiaz, S.; Khan, T.K.; Hassan, S.M. Effect of salinity on ccmfn gene RNA editing of mitochondria in wild barley and uncommon types of RNA editing. Funct. Integr. Genom. 2023, 23, 50. [Google Scholar] [CrossRef]
- Bi, C.; Qu, Y.; Hou, J.; Wu, K.; Ye, N.; Yin, T. Deciphering the Multi-Chromosomal Mitochondrial Genome of Populus simonii. Front. Plant Sci. 2022, 13, 914635. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, J.; He, W.; Kan, S.; Liao, X.; Jordan, D.R.; Mace, E.S.; Tao, Y.; Cruickshank, A.W.; Klein, R.; et al. Variation in mitogenome structural conformation in wild and cultivated lineages of sorghum corresponds with domestication history and plastome evolution. BMC Plant Biol. 2023, 23, 91. [Google Scholar] [CrossRef]
- Guo, G.Y. Famous tea of China: Xinyangmaojian. J. Xinyang Agric. Coll. 1999, 9, 49–52. [Google Scholar]
- Chen, J.-D.; Zheng, C.; Ma, J.Q.; Jiang, C.K.; Ercisli, S.; Yao, M.Z.; Chen, L. The chromosome-scale genome reveals the evolution and diversification after the recent tetraploidization event in tea plant. Hortic. Res. 2020, 7, 63. [Google Scholar] [CrossRef]
- Chen, S.; Wang, P.; Kong, W.; Chai, K.; Zhang, S.; Yu, J.; Wang, Y.; Jiang, M.; Lei, W.; Chen, X.; et al. Gene mining and genomics-assisted breeding empowered by the pangenome of tea plant Camellia sinensis. Nat. Plants 2023, 9, 1986–1999. [Google Scholar] [CrossRef]
- Xia, E.; Tong, W.; Hou, Y.; An, Y.; Chen, L.; Wu, Q.; Liu, Y.; Yu, J.; Li, F.; Li, R.; et al. The Reference Genome of Tea Plant and Resequencing of 81 Diverse Accessions Provide Insights into Its Genome Evolution and Adaptation. Mol. Plant 2020, 13, 1013–1026. [Google Scholar] [CrossRef]
- Wang, X.; Feng, H.; Chang, Y.; Ma, C.; Wang, L.; Hao, X.; Li, A.l.; Cheng, H.; Wang, L.; Cui, P.; et al. Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 2020, 11, 4447. [Google Scholar] [CrossRef]
- Kong, W.; Kong, X.; Xia, Z.; Li, X.; Wang, F.; Shan, R.; Chen, Z.; You, X.; Zhao, Y.; Hu, Y.; et al. Genomic analysis of 1,325 Camellia accessions sheds light on agronomic and metabolic traits for tea plant improvement. Nat. Genet. 2025, 57, 997–1007. [Google Scholar] [CrossRef]
- Li, L.; Li, X.; Liu, Y.; Li, J.; Zhen, X.; Huang, Y.; Ye, J.; Fan, L. Comparative analysis of the complete mitogenomes of Camellia sinensis var. sinensis and C. sinensis var. assamica provide insights into evolution and phylogeny relationship. Front Plant Sci. 2024, 15, 1396389. [Google Scholar]
- Zhang, F.; Li, W.; Gao, C.W.; Zhang, D.; Gao, L.Z. Deciphering tea tree chloroplast and mitochondrial genomes of Camellia sinensis var. assamica. Sci. Data 2019, 6, 209. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Yang, L.; Zhou, J.; Xiao, Z.; Lu, M.; Zeng, Y.; Tan, X. Mitochondrial genome study of Camellia oleifera revealed the tandem conserved gene cluster of nad5–nads in evolution. Front. Plant Sci. 2024, 15, 1396635. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Latif, A.; Osman, G. Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods 2017, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Bi, C.; Shen, F.; Han, F.; Qu, Y.; Hou, J.; Xu, K.; Xu, L.A.; He, W.; Wu, Z.; Yin, T. PMAT: An efficient plant mitogenome assembly toolkit using low-coverage HiFi sequencing data. Hortic. Res. 2024, 11, uhae023. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Brown, M.; Blaxter, M.; McCarthy, S.A.; Durbin, R. Oatk: A de novo assembly tool for complex plant organelle genomes. Genome Biol. 2025, 26, 235. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, w6–w11. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, H.; Ni, Y.; Wu, B.; Li, J.; Burzyński, A.; Liu, C. Plant mitochondrial genome map (PMGmap): A software tool for the comprehensive visualization of coding, noncoding and genome features of plant mitochondrial genomes. Mol. Ecol. Resour. 2024, 24, e13952. [Google Scholar] [CrossRef]
- Ankenbrand, M.J.; Hohlfeld, S.; Hackl, T.; Förster, F. AliTV—Interactive visualization of whole genome comparisons. PeerJ Comput. Sci. 2017, 3, e116. [Google Scholar] [CrossRef]
- Zhang, H.; Meltzer, P.; Davis, S. RCircos: An R package for Circos 2D track plots. BMC Bioinform. 2013, 14, 244. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ye, W.; Zhang, Y.; Xu, Y. High speed BLASTN: An accelerated MegaBLAST search tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef]
- Huang, L.; Yu, H.; Wang, Z.; Xu, W. CPStools: A package for analyzing chloroplast genome sequences. iMetaOmics 2024, 1, e25. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Proceedings of the Nucleic acids symposium series, Durham, NC, USA, 15–17 October 1999; pp. 95–98. [Google Scholar]
- Yan, M.H.; Liu, K.; Wang, M.; Lyu, Y.; Zhang, Q. Complete chloroplast genome of Camellia sinensis cv. Xinyang 10 and its phylogenetic evolution. Tea Sci. 2021, 41, 777–788. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Genovese, G.; Rockweiler, N.B.; Gorman, B.R.; Bigdeli, T.B.; Pato, M.T.; Pato, C.N.; Ichihara, K.; McCarroll, S.A. BCFtools/liftover: An accurate and comprehensive tool to convert genetic variants across genome assemblies. Bioinformatics 2024, 40, btae038. [Google Scholar] [CrossRef]
- Peden, J.F. Analysis of Codon Usage. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 2000. Available online: https://codonw.sourceforge.net/JohnPedenThesisPressOpt_water.pdf (accessed on 9 October 2025).
- Huang, X.; Jiao, Y.; Guo, J.; Wang, Y.; Chu, G.; Wang, M. Analysis of codon usage patterns in Haloxylon ammodendron based on genomic and transcriptomic data. Gene 2022, 845, 146842. [Google Scholar] [CrossRef]
- Wright, F. The ‘effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Xiao, W.; Wu, X.; Zhou, X.; Zhang, J.; Huang, J.; Dai, X.; Ren, H.; Xu, D. Assembly and comparative analysis of the first complete mitochondrial genome of zicaitai (Brassica rapa var. Purpuraria): Insights into its genetic architecture and evolutionary relationships. Front. Plant Sci. 2024, 15, 1475064. [Google Scholar] [CrossRef]
- Arimura, S.I. Fission and Fusion of Plant Mitochondria, and Genome Maintenance. Plant Physiol. 2018, 176, 152–161. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Mileshina, D.; Wallet, C.; Niazi, A.K.; Weber-Lotfi, F.; Dietrich, A. The plant mitochondrial genome: Dynamics and maintenance. Biochimie 2014, 100, 107–120. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Newton, K.J. Plant Mitochondrial Genomes: Dynamics and Mechanisms of Mutation. Annu. Rev. Plant Biol. 2017, 68, 225–252. [Google Scholar] [CrossRef]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019, 15, e1008373. [Google Scholar] [CrossRef]
- Zeng, W.J.; Zhu, Y.P.; Chen, J.X.; Li, H.Y.; Wang, S.H.; Gong, Y.H.; Chen, Z.Y. Analysis of Codon Usage Bias in Chloroplast and Mitochondrial Genomes of Camellia sinensis cv. ‘Zhuyeqi’. J. Tea Sci. 2025, 45, 201–218. [Google Scholar]
- Chen, Z.; Zhou, W.; Wang, Z.; Chen, Z.; You, X.; Gong, Y. Analysis of the Mitochondrial Genome of the Camellia sinensis cv. ‘Zhuyeqi’: Multichromosomal Structure, RNA Editing Sites, and Evolutionary Characterization. Front. Plant Sci. 2025, 16, 1644130. [Google Scholar] [CrossRef]
- Lu, C.; Gao, L.Z.; Zhang, Q.J. A high-quality genome assembly of the mitochondrial genome of the oil-tea tree Camellia gigantocarpa (Theaceae). Diversity 2022, 14, 850. [Google Scholar] [CrossRef]
- Wang, S.; Li, D.; Yao, X.; Song, Q.; Wang, Z.; Zhang, Q.; Zhong, C.; Liu, Y.; Huang, H. Evolution and Diversification of Kiwifruit Mitogenomes through Extensive Whole-Genome Rearrangement and Mosaic Loss of Intergenic Sequences in a Highly Variable Region. Genome Biol. Evol. 2019, 11, 1192–1206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, S.; Zhang, Y. The complete mitochondrial genome of a mangrove plant: Aegiceras corniculatum and its phylogenetic implications. Mitochondrial DNA Part B 2020, 5, 1502–1503. [Google Scholar] [CrossRef]
- Ammiraju, J.S.; Zuccolo, A.; Yu, Y.; Song, X.; Piegu, B.; Chevalier, F.; Walling, J.G.; Ma, J.; Talag, J.; Brar, D.S.; et al. Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza. Plant J. 2007, 52, 342–351. [Google Scholar] [CrossRef]
- Hu, J.; Gui, S.; Zhu, Z.; Wang, X.; Ke, W.; Ding, Y. Genome-Wide Identification of SSR and SNP Markers Based on Whole-Genome Re-Sequencing of a Thailand Wild Sacred Lotus (Nelumbo nucifera). PLoS ONE 2015, 10, e0143765. [Google Scholar] [CrossRef]
- Dong, S.; Zhao, C.; Chen, F.; Liu, Y.; Zhang, S.; Wu, H.; Zhang, L.; Liu, Y. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genom. 2018, 19, 614. [Google Scholar] [CrossRef]
- Yang, Z.; Ni, Y.; Lin, Z.; Yang, L.; Chen, G.; Nijiati, N.; Hu, Y.; Chen, X. De novo assembly of the complete mitochondrial genome of sweet potato (Ipomoea batatas [L.] Lam) revealed the existence of homologous conformations generated by the repeat-mediated recombination. BMC Plant Biol. 2022, 22, 285. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; Wang, Z.X.; Zhou, W.; Liu, S.J.; Xiao, Y.X.; Gong, Y.H. Complete sequencing of the mitochondrial genome of tea plant Camellia sinensis cv. ‘Baihaozao’: Multichromosomal structure, phylogenetic relationships, and adaptive evolutionary analysis. Front. Plant Sci. 2025, 16, 1604404. [Google Scholar] [CrossRef]
- Abdullah, M.; Sheraz, U.; Ain, A.T.; Nasir, B.; Hammad, S.; Shokat, S. Exploring the Strategies of Male Sterility for Hybrid Development in Hexaploid Wheat: Prevailing Methods and Potential Approaches. Rice 2025, 18, 53. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ni, Y.; Zhang, X.; Li, J.; Chen, H.; Liu, C. The mitochondrial genomes of Panax notoginseng reveal recombination mediated by repeats associated with DNA replication. Int. J. Biol. Macromol. 2023, 252, 126359. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kan, S.; Liao, X.; Zhou, J.; Tembrock, L.R.; Daniell, H.; Jin, S.; Wu, Z. Plant organellar genomes: Much done, much more to do. Trends Plant Sci. 2024, 29, 754–769. [Google Scholar] [CrossRef]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef]
- Zandueta-Criado, A.; Bock, R. Surprising features of plastid ndhD transcripts: Addition of non-encoded nucleotides and polysome association of mRNAs with an unedited start codon. Nucleic Acids Res. 2004, 32, 542–550. [Google Scholar] [CrossRef]
- Kubo, T.; Nishizawa, S.; Sugawara, A.; Itchoda, N.; Estiati, A.; Mikami, T. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA). Nucleic Acids Res. 2000, 28, 2571–2576. [Google Scholar] [CrossRef] [PubMed]
- Satoh, M.; Kubo, T.; Nishizawa, S.; Estiati, A.; Itchoda, N.; Mikami, T. The cytoplasmic male-sterile type and normal type mitochondrial genomes of sugar beet share the same complement of genes of known function but differ in the content of expressed ORFs. Mol. Genet. Genom. 2004, 272, 247–256. [Google Scholar] [CrossRef]
- Li, L.; Wang, B.; Liu, Y.; Qiu, Y.L. The complete mitochondrial genome sequence of the hornwort Megaceros aenigmaticus shows a mixed mode of conservative yet dynamic evolution in early land plant mitochondrial genomes. J. Mol. Evol. 2009, 68, 665–678. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, P.; Yi, B.; He, D. Current Advances in Plant Mitochondria: Application Revolution of Cytoplasmic Male Sterility. New Crops 2025, 3, 100081. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, N.; Ma, J.; Zhou, Y.; Wei, Q.; Tian, C.; Fang, Y.; Zhong, R.; Chen, G.; Zhang, S. Advances in cold-adapted enzymes derived from microorganisms. Front. Microbiol. 2023, 14, 1152847. [Google Scholar] [CrossRef]
- Hershberg, R.; Petrov, D.A. Selection on codon bias. Annu. Rev. Genet. 2008, 42, 287–299. [Google Scholar] [CrossRef]
- Tuller, T.; Waldman, Y.Y.; Kupiec, M.; Ruppin, E. Translation efficiency is determined by both codon bias and folding energy. Proc. Natl. Acad. Sci. USA 2010, 107, 3645–3650. [Google Scholar] [CrossRef]
- Mower, J.P. Variation in protein gene and intron content among land plant mitogenomes. Mitochondrion 2020, 53, 203–213. [Google Scholar] [CrossRef]
- Afzal Malik, W.; Afzal, M.; Chen, X.; Cui, R.; Lu, X.; Wang, S.; Wang, J.; Mahmood, I.; Ye, W. Systematic analysis and comparison of ABC proteins superfamily confer structural, functional and evolutionary insights into four cotton species. Ind. Crops Prod. 2022, 177, 114433. [Google Scholar] [CrossRef]
- He, D.; Li, Y.; Yuan, C.; Pei, X.; Damaris, R.N.; Yu, H.; Qian, B.; Liu, Y.; Yi, B.; Huang, C.; et al. Characterization of the CMS genetic regulation through comparative complete mitochondrial genome sequencing in Nicotiana tabacum. Plant Genome 2024, 17, e20409. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lu, S.S.; Bi, Y.; Jiang, Y.M.; Feng, L.D.; He, J. Assembly and Analysis of the Mitochondrial Genome of Hippophae rhamnoides subsp. sinensis, an Important Ecological and Economic Forest Tree Species in China. Plants 2025, 14, 2170. [Google Scholar] [CrossRef] [PubMed]
- Yisilam, G.; Liu, Z.; Turdi, R.; Chu, Z.; Luo, W.; Tian, X. Assembly and comparative analysis of the complete mitochondrial genome of Isopyrum anemonoides (Ranunculaceae). PLoS ONE 2023, 18, e0286628. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, R.; Yun, Q.; Xu, Y.; Zhao, G.; Liu, J.; Shi, S.; Chen, Z.; Jia, L. Comprehensive analysis of complete mitochondrial genome of Sapindus mukorossi Gaertn.: An important industrial oil tree species in China. Ind. Crops Prod. 2021, 174, 114210. [Google Scholar] [CrossRef]
- Han, F.; Bi, C.; Zhao, Y.; Gao, M.; Wang, Y.; Chen, Y. Unraveling the complex evolutionary features of the Cinnamomum camphora mitochondrial genome. Plant Cell Rep. 2024, 43, 183. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, Z.; Jiang, J.; Wu, W.; Yu, W.; Zhang, S.; Zeng, W. The assembly and comparative analysis of the first complete mitogenome of Lindera aggregata. Front. Plant Sci. 2024, 15, 1439245. [Google Scholar] [CrossRef]
- He, X.; Qian, Z.; Gichira, A.W.; Chen, J.; Li, Z. Assembly and comparative analysis of the first complete mitochondrial genome of the invasive water hyacinth, Eichhornia crassipes. Gene 2024, 914, 148416. [Google Scholar] [CrossRef]
- Yan, Y.; da Fonseca, R.R.; Rahbek, C.; Borregaard, M.K.; Davis, C.C. A new nuclear phylogeny of the tea family (Theaceae) unravels rapid radiations in genus Camellia. Mol. Phylogenet. Evol. 2024, 196, 108089. [Google Scholar] [CrossRef]
- Li, L.; Hu, Y.; He, M.; Zhang, B.; Wu, W.; Cai, P.; Huo, D.; Hong, Y. Comparative chloroplast genomes: Insights into the evolution of the chloroplast genome of Camellia sinensis and the phylogeny of Camellia. BMC Genom. 2021, 22, 138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Folk, R.A.; Mo, Z.Y.; Peng, H.; Zhao, J.L.; Yang, S.X.; Yu, X.Q. Phylotranscriptomic analyses reveal deep gene tree discordance in Camellia (Theaceae). Mol. Phylogenet. Evol. 2023, 188, 107912. [Google Scholar] [CrossRef]
- Pang, Z.; Wang, Y.L.; Mantri, N.; Wang, Y.; Hua, X.J.; Quan, Y.P.; Zhou, X.; Jiang, Z.D.; Qi, Z.C.; Lu, H.F. Molecular phylogenetic relationships and taxonomy position of 161 Camellia species in China. Taiwania 2022, 67, 560–570. [Google Scholar]
- Wu, Q.; Tong, W.; Zhao, H.; Ge, R.; Li, R.; Huang, J.; Li, F.; Wang, Y.; Mallano, A.I.; Deng, W.; et al. Comparative transcriptomic analysis unveils the deep phylogeny and secondary metabolite evolution of 116 Camellia plants. Plant J. 2022, 111, 406–421. [Google Scholar] [CrossRef]
- Zhu, Z.J.; Qin, X.M.; Li, P.W.; Lu, Y.B.; Mo, X.Y.; Fang, Y.; Zhang, Q. Characterize the Complete Mitogenome of Semiaquilegia guangxiensis and Assess the Efficiency of the Mitochondrial Genes in Ranunculales Phylogeny. Ecol. Evol. 2025, 15, e71165. [Google Scholar] [CrossRef]
- Liang, H.; Qi, H.; Wang, C.; Wang, Y.; Liu, M.; Chen, J.; Sun, X.; Xia, T.; Feng, S.; Chen, C.; et al. Analysis of the complete mitogenomes of three high economic value tea plants (Tea-oil Camellia) provide insights into evolution and phylogeny relationship. Front. Plant Sci. 2025, 16, 1549185. [Google Scholar] [CrossRef] [PubMed]
- Zan, T.; He, Y.T.; Zhang, M.; Yonezawa, T.; Ma, H.; Zhao, Q.M.; Kuo, W.Y.; Zhang, W.J.; Huang, C.H. Phylogenomic analyses of Camellia support reticulate evolution among major clades. Mol. Phylogenet. Evol. 2023, 182, 107744. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.J.; Zhang, X.J.; Cao, D.L.; Guo, X.X.; Mower, J.P.; Fan, S.J. Plastid and mitochondrial phylogenomics reveal correlated substitution rate variation in Koenigia (Polygonoideae, Polygonaceae) and a reduced plastome for Koenigia delicatula including loss of all ndh genes. Mol. Phylogenet. Evol. 2022, 174, 107544. [Google Scholar] [CrossRef] [PubMed]
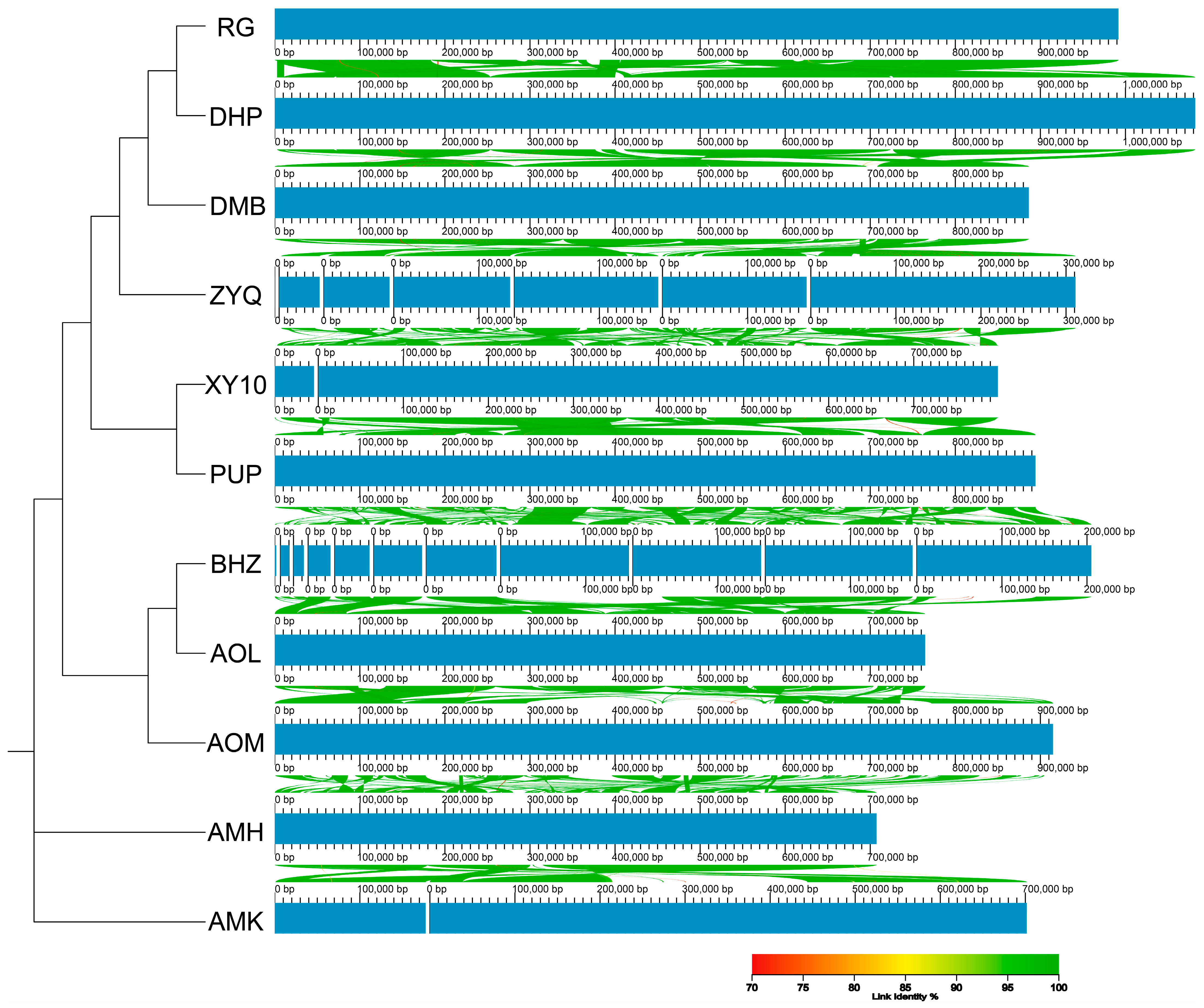
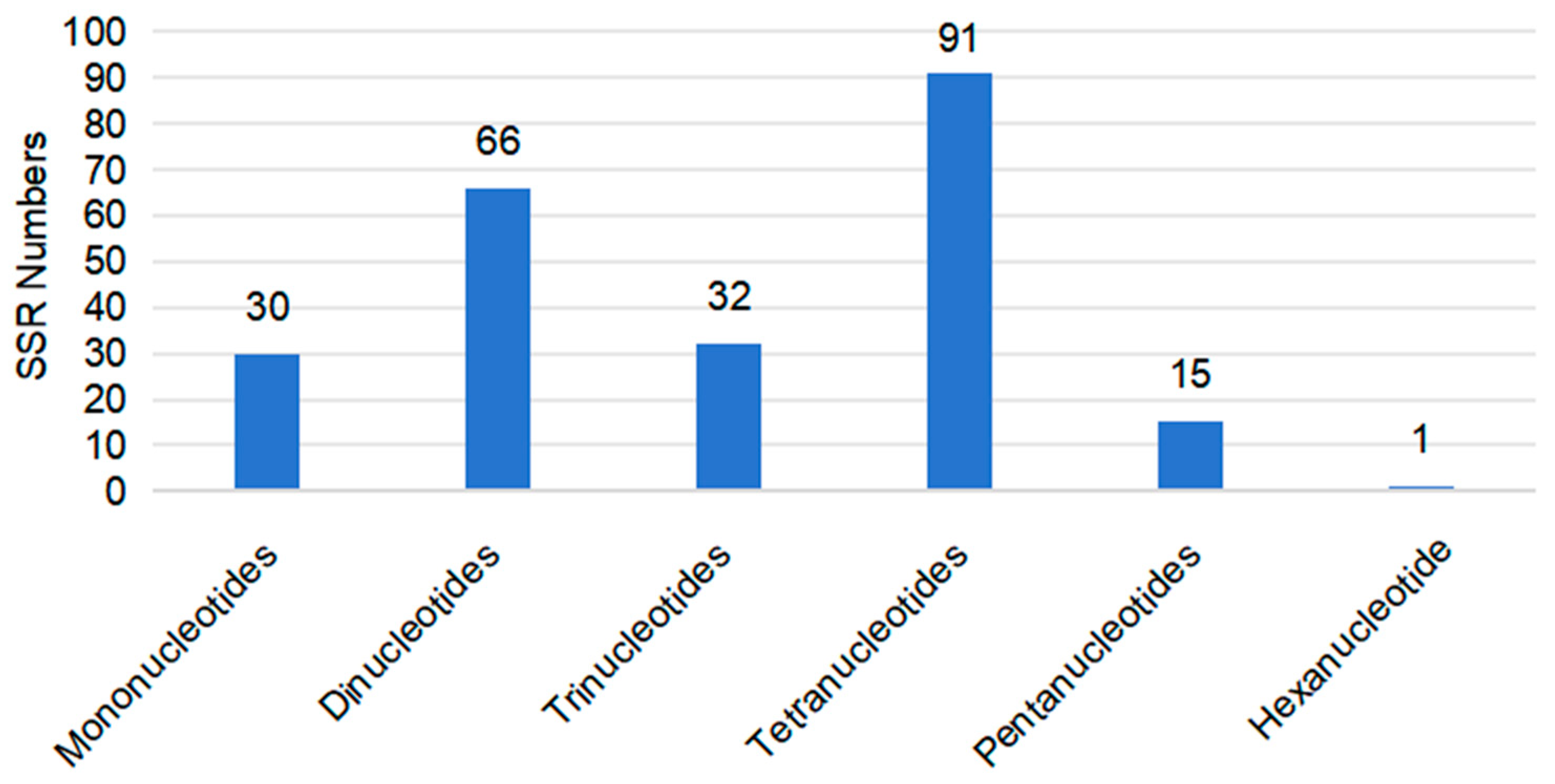
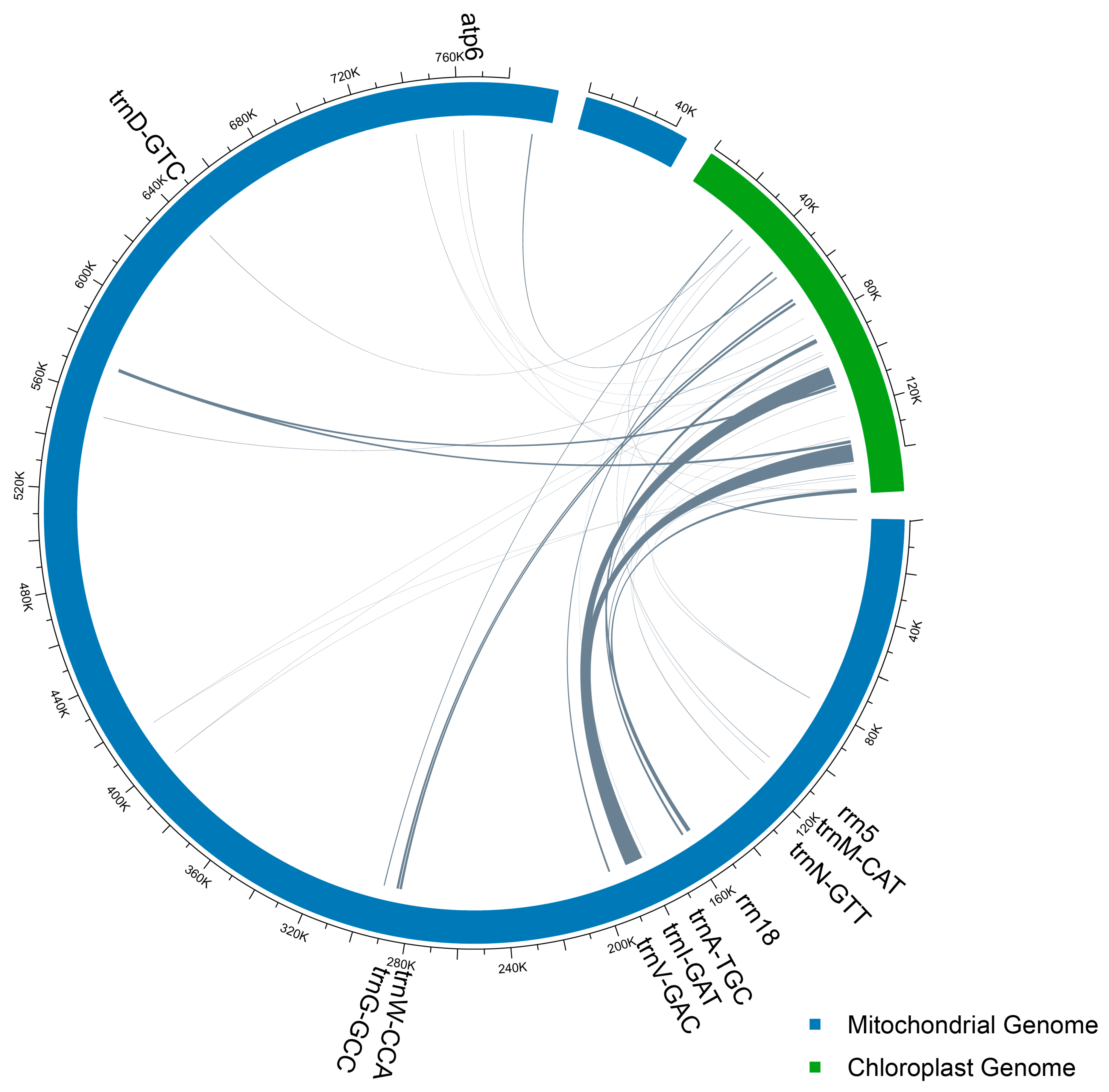
| Species | Species Code | Accession Number | Length (bp) |
|---|---|---|---|
| C. sinensis var. assamica | AMH | MH376284 | 707,441 |
| C. sinensis var. assamica cv. Yunkang10 | AMK | MK574876-MK574877 | 879,048 |
| C. sinensis var. assamica | AOM | OM809792 | 914,855 |
| C. sinensis var. assamica cv. Duntsch | AOL | OL989850 | 1,081,966 |
| C. sinensis sp. Baihaozao | BHZ | PV340641-PV340651 | 909,843 |
| C. sinensis sp. Zhuyeqi | ZYQ | PQ763484-PQ763490 | 911,255 |
| C. sinensis cv. Dahongpao | DHP | PP212895 | 1,082,025 |
| C. sinensis cv. Rougui | RG | PP212896 | 991,788 |
| C. sinensis cv. SRDMB | DMB | PP770440 | 886,354 |
| C. sinensis var. pubilimba | PUP | ON782577 | 894,250 |
| C. sinensis cv. Xinyang10 | XY10 | PX398764-PX398765 (current study) | 845,076 |
| Group of Genes | XY10 | |
|---|---|---|
| Core gene | ATP synthase | atp1, atp4, atp6, atp8, atp9 |
| NADH dehydrogenase | nad1 ****, nad2 ****, nad3, nad4 **, nad5 ****, nad6, nad7 ****, nad9, nad4L | |
| Cytochrome cbiogenesis | cob | |
| Ubiquinol cytochromre creductase | ccmC, ccmFN, ccmFC *, ccmB | |
| Cytochromre coxidase | cox1, cox2, cox3 | |
| Maturases | matR | |
| Variable gene | Transport membrane protein | mttB |
| Large subunit of ribosome | rpl2 *, rpl5, rpl10, rpl16 | |
| Small subun it of ribosome | rps1, rps3 *, rps4, rps7, rps12, rps13, rps14, rps19 | |
| Succinate dehydrogenasse | sdh3, sdh4 | |
| rRNA gene | Ribosome RNA | rrn5, rrn18, rrn26 |
| tRNA gene | Transfer RNA | trnM-CAT(4), trnK-TTT, trnC-GCA, trnN-GTT, trnY-GTA, trnS-GCT(2), trnF-GAA(2), trnP-TGG(2), trnS-TGA *(2), trnA-TGC *, trnV-GAC, trnI-GAT *(2), trnE-TTC, trnH-GTG, trnW-CCA, trnG-GCC(2), trnQ-TTG, trnF-AAA *, trnT-TGT *(2), trnN-ATT, trnD-GTC, trnT-GGT, trnS-GCT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, M.-H.; Du, Y.-R.; Tong, W.; Su, J.-M.; Pu, G.-Q.; Yan, L.-M.; Zhu, T.-T.; Wang, W.-W. The Multipartite Mitogenome of Camellia sinensis cv. Xinyang10 Reveals Frequent Reorganization and Hints at Phylogeographic History. Diversity 2025, 17, 705. https://doi.org/10.3390/d17100705
Yan M-H, Du Y-R, Tong W, Su J-M, Pu G-Q, Yan L-M, Zhu T-T, Wang W-W. The Multipartite Mitogenome of Camellia sinensis cv. Xinyang10 Reveals Frequent Reorganization and Hints at Phylogeographic History. Diversity. 2025; 17(10):705. https://doi.org/10.3390/d17100705
Chicago/Turabian StyleYan, Ming-Hui, Yan-Rong Du, Wei Tong, Jia-Meng Su, Guo-Qing Pu, Lu-Miao Yan, Tong-Tong Zhu, and Wen-Wen Wang. 2025. "The Multipartite Mitogenome of Camellia sinensis cv. Xinyang10 Reveals Frequent Reorganization and Hints at Phylogeographic History" Diversity 17, no. 10: 705. https://doi.org/10.3390/d17100705
APA StyleYan, M.-H., Du, Y.-R., Tong, W., Su, J.-M., Pu, G.-Q., Yan, L.-M., Zhu, T.-T., & Wang, W.-W. (2025). The Multipartite Mitogenome of Camellia sinensis cv. Xinyang10 Reveals Frequent Reorganization and Hints at Phylogeographic History. Diversity, 17(10), 705. https://doi.org/10.3390/d17100705






