Abstract
Alfalfa (Medicago sativa L.) is a vitally important perennial fodder legume worldwide. Given their particular traits, alfalfa crop wild relatives (CWRs) could be used to develop cultivars that can tolerate extreme environmental and climatic conditions. Until now, researchers have overlooked the composition and structure of bacterial communities in the root zone of alfalfa and its relevant CWRs and their influence on forage performance under actual field conditions. In this study, high-throughput sequencing of 16S rRNA analysis was performed to investigate the diversity and assemblies of bacterial communities in the bulk soil and in the root zone of individual field-grown Medicago plants arranged in a honeycomb selection design. The plants used in this study were M. sativa × M. arborea hybrids (Genotypes 6 and 8), the closely-related M. sativa nothosubsp. varia (Martyn) Arcang. (Genotype 13), and M. sativa ssp. sativa (Genotype 20). The bacterial communities in the root-zone samples and the assemblies in the bulk soil differed significantly. Genotype 13 was found to have distinct bacterial assemblies from the other genotypes while exhibiting the lowest forage productivity. These findings suggest that plant productivity may influence the composition of bacterial communities in the root zone. Biomarker analysis conducted using linear discriminant analysis (LDA) revealed that only members of the Rhizobiales order were enriched in the M. sativa nothosubsp. varia root zone whereas taxa belonging to Sphingomonas and various Bacteriodota were enriched in the other genotypes. Of the shared taxa identified in the root zone of the Medicago lines, the abundance of specific taxa, namely, Flavisolibacter, Stenotrophomonas, and Sphingomonas, were positively associated with forage yield. This pioneering study, in which the root zones of individual Medicago plants under actual field conditions were examined, offers evidence of differences in the bacterial composition of alfalfa genotypes with varying genetic backgrounds. Its findings indicate that particular bacterial taxa may favorably influence plant performance. This study covered the first six months of crop establishment and paves the way for further investigations to advance understanding of how shifts in bacterial assemblies in alfalfa roots affect plant performance over time.
1. Introduction
In agricultural ecosystems, plants co-exist and co-evolve dynamically with soil microbiomes, whose interactions with them may be beneficial, neutral, or pathogenic [1]. Key soil processes, such as nutrient cycling, soil formation, organic carbon (C) stability, transformation, and sequestration affect crop productivity and are governed by microbial communities (Fierer, 2017) [2]. Plant-associated microorganisms also promote plant growth through hormone production, enhanced nutrient and water uptake, and the alleviation of abiotic and biotic [3].
Previous studies on several plant species have shown that genotypic variation can have stronger or weaker effects on bacterial diversity [4,5,6,7]. For example, plant genetic variation in maize is associated with a decrease in bacterial diversity in the rhizosphere [8]. Similarly, wheat domestication is associated with a decrease in the abundance of specific bacteria taxa in the root zone compared with their abundance in the bulk soil, including those involved in nitrogen (N) cycling [9]. However, Leff et al. [10] found that sunflower domestication has no effect on root-zone bacterial communities. During the process of plant breeding, plants are generally selected on the basis of traits controlled by multiple genes that affect numerous pathways. These multigenic traits include increased yield, disease resistance, or tolerance to biotic stresses. They may be associated with several root morphological changes as well as alterations in the quantity and quality of root exudates, which in turn may influence bacterial assemblies in the soil [11,12]. Additionally, the soil bacterial composition is affected by the growth stage of plants, as their root architecture and exudates are continuously modified during their development. For example, a plant’s root exudates are more C-rich during its early growth than during its main growth, when N-rich compounds increase [13], leading to significant changes in bacterial communities [14].
Alfalfa (Medicago sativa L.) is one of the most important fodder legumes worldwide and is cultivated in over 80 countries. In 2019, the trade in alfalfa meal and pellets alone was estimated at USD 390 million, accounting for 12% of the global forage crop trade (http://oec.world/en/profile/hs92/forage-crops, accessed on 18 August 2023). Given its perennial nature and ability to access nutrients and water from the soil through an extended deep tap root system, alfalfa could be a key crop for climate change adaptation [15]. However, models of future climate scenarios have predicted prolonged drought periods in the Mediterranean region and in areas with a similar climate [16], posing significant risks for agricultural ecosystems. The development of alfalfa varieties with improved traits is therefore necessary to secure continuous feed production under extreme environmental conditions. For this purpose, crop wild relatives (CWRs) of alfalfa provide an important pool with species that are well adapted to such conditions [17,18].
A study on how alfalfa plants interact with bacterial communities in the soil is crucial for understanding what influences their persistence and eventual growth, especially in light of the economic significance of perennial alfalfa cultivation. Most of the previous studies on Medicago species have examined the bacterial composition of root compartments, such as the root endosphere and nodules [19,20,21,22]. Recent research on M. truncatula, considered a model plant for study, has shown that plant genotypes significantly influence the rhizosphere bacterial community composition [20]. Experiments developed for the above studies were designed and performed in well-controlled, artificial mesocosm or microcosm settings. Controlled experimental settings evidently work well for addressing specific scientific questions, especially given the complexity of plant processes and their interactions with microbial communities. However, they cannot address questions relating to field-established crops. Moreover, to the best of our knowledge, until now, no studies have provided a comprehensive phylogenetic characterization of the bacterial assemblages found in the root zones of M. sativa × M. arborea hybrids, and M. sativa subspecies.
Another important gap in the literature on root microbiomes concerns individual plants and their associated root systems grown in actual field conditions [23,24]. Specifically, root entanglement in conventional, dense crop stands hinders the study of individual root systems, whereas the ultra-wide distances employed in the honeycomb selection designs (HSDs) [25] are appropriate for such studies [26]. Therefore, microbiome studies in plants contrast with microbiome studies involving humans or animals, which can and do focus on the individual [27,28]. To overcome this challenge, alfalfa trials in this study were established applying one suitable honeycomb design, according to the specific alfalfa breeding guidelines detailed in Fasoula and Fasoula [29]. In the HSDs, ultra-wide spacings are maintained between growing plants to maximize their phenotypic expression and differentiation while eliminating the masking effects of interplant competition and soil/environmental heterogeneity [30,31].
Following the establishment of the field trial, this study’s specific objectives were pursued. The first was to examine the extent to which the genotypic profiles of four alfalfa lines belonging to M. sativa × M. arborea hybrids, and M. sativa species with differentiated pedigree histories [17] affect the individual root-zone bacterial microbiome. The second was to identify specific bacterial taxa for each genotype. Significant associations between the abundance of bacterial taxa and forage biomass were also examined. Lastly, the hypothesis that the bacterial composition of the root-zone region of M. sativa × M. arborea hybrids, and M. sativa species and the bulk soil differ during the first six months of crop establishment was tested.
2. Materials and Methods
2.1. Field Experimental Setup, Planting, Harvesting, and Yield Measurements
In April 2019, seedlings from four (4) alfalfa genetic lines, namely, Genotype 6, 8, 13, and 20, previously described by Humphries et al. (2020) [17], were transplanted in a field at the experimental station of the Agricultural Research Institute in Tochni, which is in Larnaca, Cyprus. Two lines (Genotypes 6 and 8) have introgressions from the woody shrub, M. arborea originating from the Greek islands. M. sativa nothosubpp. varia (Genotype 13) is a breeders’ line that was developed from a diverse range of wild accessions selected for their tolerance to drought and continuous grazing tolerance in Australia [17]. Lastly, SARDI 7 series 2 (Genotype 20) is a commercial Australian alfalfa cultivar (Table 1). A honeycomb selection design (Supplementary Figure S1) was applied to allow for unhindered growth of the individual root systems and avoid the negative effects of interplant competition and spatial heterogeneity [25]. In particular, the trial consisted of 20 rows and 35 plants per row (R0 design), whereas the plant-to-plant distance was 1 m to exclude interplant competition for growth resources (Supplementary Figure S1) [30]. A combination of motor cultivation with hand weeding was meticulously implemented at the early stages of weed development during all phases of the trial, so that the individual plants were not affected by interplant weed competition. Irrigation was initiated at 5 min/day during the first month, then 30 min/day for 4 days/week during the second month, and thereafter 45 min/day for 4 days/week. In October 2019, the plants were harvested and individually weighed. Plants did not receive any basal or side fertilization and no chemical control for pests and diseases was implemented.

Table 1.
Summary of alfalfa accessions evaluated at the Agricultural Research Institute, Cyprus in 2019.
2.2. Soil Sampling and Chemical Analysis
From each genotype, fourteen plants (n = 14) were chosen for sampling and root-zone soil samples were carefully collected in situ, a short distance from the base of the alfalfa plants (Figure 1). To ensure accuracy and prevent any contamination, the sampling from each plant was carried out using a tipless, sterile 1 mL syringe (Becton Dickinson, Vaud, Switzerland). Spots that were free of weeds and alfalfa within the experimental field were selected as collection sites for bulk soil samples (n = 11). All samples were transported to the laboratory where 3 g of each individual plant was stored at −20 °C prior to undergoing further processing. The remaining soil collected from each individual root-zone sample was combined in equal volumes to create a composite sample for chemical properties analysis (Supplementary Table S1). The pH [H+] and conductivity (mS/cm) of the soil samples were determined using calibrated pH and conductivity electrodes immersed in a stirred (for 30 min) and settled (for 10 min) suspension of 2 g of soil in 5 mL of deionized water. Total N content (2 g) was assessed using the Kjeldahl method [32], and organic matter (0.5 g) was determined using a modified version of the Dergtjareff method [33]. Available phosphorus extracted with sodium carbonate [34] was estimated (5 g), and exchangeable potassium (5 g) was determined using the ammonium acetate method [35]. Following potassium chloride extraction, colorimetry [36] and the cadmium reduction method [37] were, respectively, performed to determine ammonium and nitrate content. Atomic absorption spectroscopy (SOLAAR S series AA Spectrometer, Thermo Fisher Scientific Inc., Waltham, MA, USA) was used to determine elemental calcium and iron content from nitric/hydrochloric acid-digested soil samples.
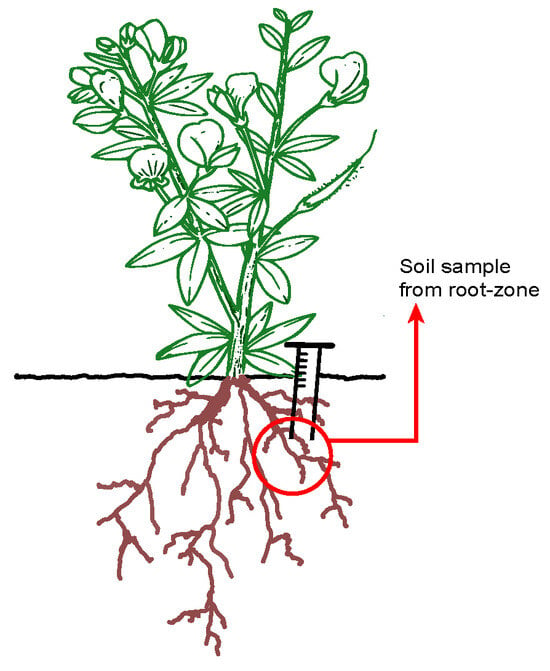
Figure 1.
The diagram shows an alfalfa plant and its root system. The red circle and arrow highlight a specific region of the root zone, which is the focus area for bacterial community analysis in our study. To ensure accuracy and prevent contamination, sampling from each plant was carried out using a tipless, sterile 1 mL syringe.
2.3. DNA Extraction and High-Throughput Sequencing
Soil DNA from all samples was extracted with a DNeasy PowerSoil kit using a TissueLyser II for conducting initial cell lysis (QIAGEN, Hilden, Germany) at 20 Hz for 12 min. Extracted DNA was eluted in 1 × Tris–EDTA (TE) buffer (pH 8), quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific Inc., Waltham, MA, USA), and stored at −20 °C. Bacterial communities in all the samples with the bacterial-specific 16S rRNA gene possessing variable regions 3 and 4 were amplified with the PCR primers 515/806 tagged with oligonucleotide indexing. A MiSeq (Illumina, San Diego, CA, USA) high-throughput amplicon sequencer (paired-end reads, 2 × 300 bp) was then run at the genome sequencing facility at the Environmental Microbiology and Biotechnology Center of the Agricultural Research Institute in Nicosia, Cyprus. A sequencing standard (mock community) was used to optimize data processing pipelines and detect possible PCR and sequencing biases that might hinder identification of bacterial taxa (Supplementary Table S2). The Quantitative Insights into Microbial Ecology (QIIME 2) microbiome bioinformatics platform [38] integrated into a Jupyter Notebook [39] web-based platform was used for the analyses. For bacterial taxonomic assignments, the SILVA database (version 138.99) [40] was used. Raw sequencing data and metadata files (Sequence Read Archive) and amplicon sequence variant (ASV) sequences were submitted to NCBI under BioProject PRJNA893879.
2.4. Statistical Analysis
All of the statistical analyses were performed using RStudio (version 2022.07.2). Data pre-processing effectively removed mock community samples and taxa with no sequence hits. The prevalence threshold was set at 0.05. Therefore, only ASVs occurring in more than 5% of the samples were retained. The ASVs were analyzed as a phyloseq object using the phyloseq package. The α-diversity indices (Fisher, Gini–Simpson, inverse Simpson, and Shannon) were calculated, and linear model regression analysis was performed. Bacterial ꞵ-diversity was calculated using Bray–Curtis distance metrics, and ordination was performed using principal coordinate analysis (PCoA). Pairwise comparisons of dissimilarity between the different samples (alfalfa genotypes and bulk soil) were performed with permutational multivariate analysis of variance (PERMANOVA) using the “adonis2” function of the vegan package [41]. The composition of bacterial communities between genotypes and bulk soil samples was determined and presented using ggplot2 and Venn packages in R. Linear discriminant analysis effect size (LDA-LefSe) was applied to identified bacterial biomarkers in each genotype and bulk soil sample [42]. In all cases, pairwise comparisons between groups were performed using the Kruskal–Wallis test corrected with the Benjamini–Hochberg procedure used for multiple comparisons (adjusting the false discovery rate). The Spearman rank correlation coefficient was utilized to identify associations between the abundance (log2 transformed) of ASVs present in all alfalfa genotypes and plant yield, aiming to discern the relationship between bacterial taxa and plant performance.
Co-occurrence bacterial patterns were also constructed to further reveal bacterial community differences using the network inference tool SPIEC-EASI (v0.1.4). Bacterial genera found in more that 25% of the samples with abundance higher than 0.01% were selected to decrease the impact of site specific genera on the network structure [43]. To infer inverse covariance matrices for the network, SPIEC-EASI was run with the “MB” neighborhood selection method [44]. Network visualization was performed using Gephi 0.10.0 [45] and single nodes were not included in the network. Network topological attributes and taxa roles were calculated using the functions included in the meconetcomp 0.4.1 package [46]. Relationships among the main phyla of bacterial communities were visualized with a chord diagram using the circlize package [47].
3. Results
3.1. Forage Productivity of Alfalfa Genotypes
Analysis of variance revealed significant effects of alfalfa genotype on yield (F = 58.05, p < 0.01), indicating substantial variability among the different lines. The yield of individual plants of Medicago sativa ssp. sativa (Genotype 20) ranged from 910 g to 1260 g per plant, significantly outperforming Genotypes 6 and 8, which showed no significant differences between each other, with yields ranging from 605 g to 1080 g per plant. Both were markedly more productive than Genotype 13 (Medicago sativa nothosubsp. varia), which exhibited the lowest performance with yields, ranging from 220 g to 405 g per plant (Figure 2).
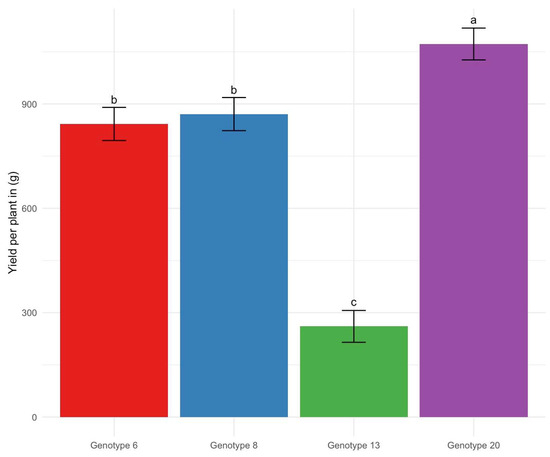
Figure 2.
Yield per plant (in grams) for Genotype 6, Genotype 8, Genotype 13, and Genotype 20. Bars represent the mean yield, with error bars indicating the standard error of the mean. Letters above the bars denote significant differences between genotypes (p < 0.05) using the Tukey HSD test.
3.2. Bacterial Assemblies Differ between Root-Zone and Bulk Soil Samples but Are Similar for the Different Genotypes
Overall, alfalfa genotypic variation was minimal in terms of α-diversity, and no differences were observed between genotypes for most of the indicators tested. Pairwise comparisons revealed that bulk soil samples exhibited lower α-diversity values for the Fisher and inverse Simpson indices compared with those observed for alfalfa Genotypes 6, 8, and 20. However, Genotype 13 exhibited similar values to those observed in bulk soil for these α-diversity indices (Figure 3A). The results of the PCoA of the Bray–Curtis distances conducted for each sample (Figure 3B) clearly showed that the bacterial communities of the bulk soil samples and that of Genotype 13 were distinct from those of the root-zone samples collected from Genotypes 6, 8, and 20. Moreover, the PERMANOVA analysis showed that the samples were substantially dissimilar (F = 8.32, R2 = 0.51, p < 0.01). Furthermore, pairwise comparisons showed that the bulk soil samples differed significantly from all the root-zone samples. Of the alfalfa genotypes, Genotype 13 exhibited a bacterial composition that differed from that of the other genotypes (Table 2).
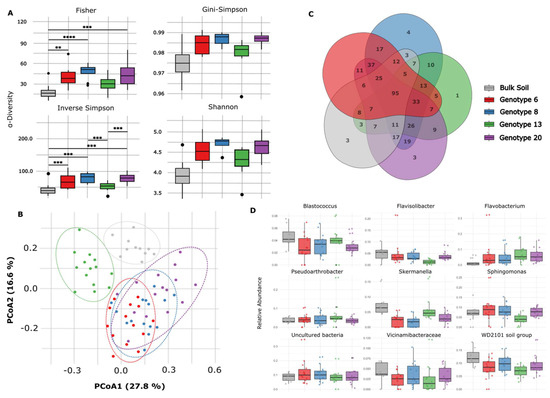
Figure 3.
(A) Alpha and beta diversity plots. The boxplots correspond to the following indices of α-diversity: Fisher, Gini–Simpson, inverse Simpson, and Shannon. A statistical analysis was performed with multiple comparisons using the Kruskal–Wallis test corrected for false discovery rates with the Benjamini–Hochberg procedure. Asterisks indicate corrected p-values: * p < 0.05, ** p < 0.05, *** p < 0.001, and **** p < 0.0001; (B) principal coordinate analysis corresponding to the Bray–Curtis dissimilarity index and permutational multivariate analysis of variance. (C) A Venn diagram depicting unique amplicon sequence variants (ASVs) shared among the four alfalfa genotypes and bulk soil samples; (D) a boxplot of the relative abundance of the main phyla representing the shared bacterial community present in alfalfa root-zone samples and bulk soil.

Table 2.
Pairwise comparisons of bacterial microbial communities found in different alfalfa root-zone and bulk soil samples based on permutational multivariate analysis of variance. p-values were adjusted using the Benjamini–Hochberg procedure to correct for false discovery rates.
The top five phyla present in the root zone of alfalfa genotypes were Pseudomonadota (~40%), Bacteroidota (~20%), Actinomycetota (~10%), Planctomycetota (~9%), and Acidobacteriota (~7%). The core bacterial communities of both the root-zone and bulk soil samples comprised a total of 403 key ASVs at a prevalence of 50% and a relative abundance of 0.01% (Genotype 6: 336 ASVs; Genotype 8: 351 ASVs; Genotype 13: 281 ASVs; Genotype 20: 346 ASVs; and bulk soil: 199 ASVs). In total, 113 ASVs (23.4%) were common to all alfalfa genotypes and bulk soil samples (Figure 3C), and this community mainly comprised genera belonging to the WD2101 soil group (6.9–12.3%), Sphingomonas (3.95–8.81%), Pseudarthrobacter (3.6–7.3%), Blastococcus (2.7–4.7%), Skermanella (1.9–7.2%), and Flavisolibacter (1.3–4.9%). It also included unassigned members of Bacteroidota (2.6–11.7%) and members of Vicinamibacteraceae (2.4–4.2%) (Figure 3D).
Network analysis further shed light on the intricate bacterial composition and the distinct communities associated with different alfalfa genotypes compared to bulk soil samples. Specifically, the analysis revealed a more complex bacterial network within the root-zone bacterial communities, as evidenced by a higher number of nodes and edges in alfalfa genotypes than in bulk soil samples (Figure 4a). This complexity is highlighted by an increased average degree in the alfalfa genotypes, pointing to a denser network of connections per node, indicative of a robust and highly interactive community. Furthermore, the alfalfa genotypes displayed a shorter average path length compared to bulk soil, suggesting a more tightly knit network that supports swift and efficient microbial interactions. Remarkably, Genotype 13 stands out with its unique network characteristics, compared to the rest of the alfalfa genotypes, including a high network diameter and modularity, hinting at a specialized microbial community niche or adaptation (Figure 4A).
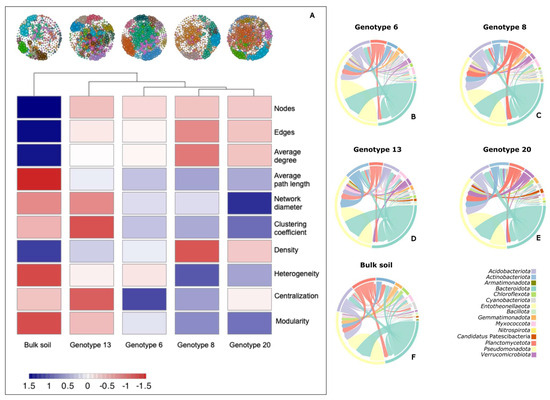
Figure 4.
(A) Co-occurrence networks of the bacterial community found in different alfalfa genotypes and bulk soil samples. Nodes represent the relative abundance of ASVs within the networks, with different colors indicating distinct modules. The heatmap shows various network attribute characteristics calculated for each network, along with their clustering patterns. (B–F) Chord diagrams represent the inter-phyla connections, highlighting the intricate relationships and community structure of bacteria within each genotype and the bulk soil environment.
Across the different alfalfa genotypes and bulk soil samples, our analysis revealed a bacterial network predominantly orchestrated by Bacteroidota and Pseudomonadota, suggesting a universal role in plant-associated bacterial interactions (Figure 4B–F). In detail, Bacteroidota demonstrated a high degree of connectivity, particularly with Pseudomonadota, ranging from 42% of the total links in soil to 53% in Genotype 6, indicating potential cross-phylum synergy. Also, in all samples, Bacteroidota showed a high inter-phylum connectivity ranking from 14 to 21%. The Pseudomonadota phylum was central in the constructed networks showing high inter- and intra-phylum connectivity in all samples. Interestingly, the connectivity of Planctomycetota with Actinobacteriota was substantially higher in bulk soil samples (23.4%) compared to the links found in the root-zone samples of alfalfa genotypes, ranging from 4.1% (Genotype 20) to 13.8% (Genotype 13).
3.3. Biomarker Taxa in the Root Zone of Alfalfa Genotypes and Bulk Soil
Linear discriminant analysis (LDA) was performed to identify bacterial taxa as biomarkers among the different alfalfa root-zone and bulk soil samples (Figure 5). In total, 53 ASVs were found to be significantly enriched (LDA > 2) in all samples, of which 10 bacterial taxa were enriched in the bulk soil samples. These ASVs were assigned to the following orders: Azospirillales, Burkholderiales, Solirubrobacterales, Micromonosporales, Tepidisphaerales, Chitinophagales, Candidatus Saccharimonadales, and Gemmatimonadales. Several of these ASVs were exclusively enriched in bulk soil samples. These bacteria included those belonging to the genera Acidibacter (ASV3652), Massila (ASV4082), and Actinoplanes (ASV2555). They also included uncultured members of Candidatus Saccharimonadales (ASV2074), Gemmatimonadaceae (ASV2344), and Solirubrobacterales bacterium 67-14 (ASV2683). The bacteria belonging to the phylum Planctomycetota in the WD2101 soil group were only enriched in Genotype 8 and in the bulk soil sample. Similarly, two Alphaproteobacteria ASVs assigned to the genus Skermanella were prolific in the bulk soil samples and in the root zone of Genotype 13 (Figure 5).
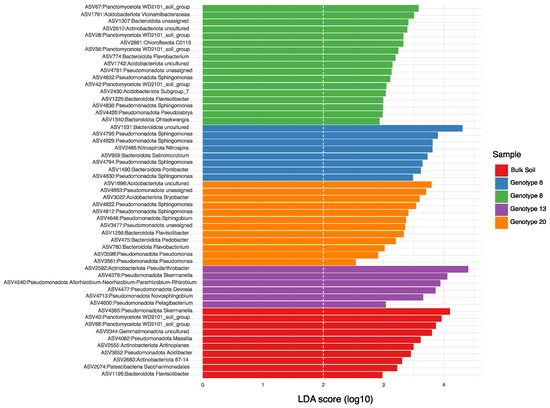
Figure 5.
A bar plot depicting linear discriminant analysis scores for differentially abundant bacterial taxa found in each alfalfa genotype root-zone sample and in bulk soil. ASV = amplicon sequence variant; LDA = linear discriminant analysis.
The 43 enriched ASVs found in alfalfa root-zone samples belonged to a variety of taxa: Acidobacteriota, Actinomycetota, Bacteroidota, Nitrospirota, Chloroflexota, Planctomycetota, and Pseudomonadota. The results of this study showed that genotypes had a substantial effect on the proliferation of specific bacterial taxa in the soil samples collected. Acidobacteriota species were mainly enriched in Genotypes 8 and 20. Two ASVs (ASV4540 and ASV4477), which are members of the Hyphomicrobiales order and belong to the genera Allorhizobium–Neorhizobium–Pararhizobium–Rhizobium and Devosia were exclusively enriched in the root zone of Genotype 13. ASVs belonging to the genus Sphingomonas were not enriched in Genotype 13 but were prolific in the root-zone samples of Genotypes 6, 8, and 20. A similar trend was observed for the different taxa belonging to the Bacteroidota phylum, which were mainly enriched in these alfalfa genotypes. Notably, taxa assigned to Pseudomonas (ASV561 and ASV598), Bryobacter (ASV3022), and an unassigned Enterobacteriaceae (ASV3477), all of which belonged to Gammaproteobacteria, were found only in the root-zone samples of Genotype 20.
Differential abundance analysis further shows significant variations between the alfalfa genotypes in bacterial taxa. For example, Genotype 6 had higher abundances of ASVs like Sphingomonas and Flavisolibacter. In contrast, Genotype 13, was enriched with specific ASVs such as Devosia and Rubellimicrobium. Members of WD2101 soil group, C0119 of Chloroflexota phylum and Bryobacter were characteristic for Genotype 8 and 20 (Figure 6).
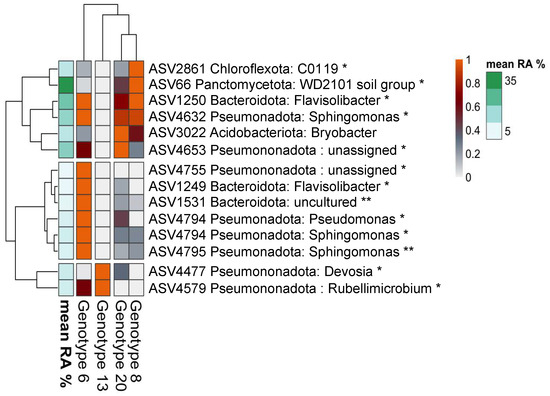
Figure 6.
Differential abundance heatmap showing amplicon sequence variants (ASVs) whose relative abundance exhibited a significant positive association with alfalfa genotypes (p < 0.05 * and p < 0.01 **). Each ASV is labeled with its taxonomic classification and the dendrogram on the left clusters ASVs based on their abundance profiles across the alfalfa genotypes.
3.4. Linking Bacterial Composition and Diversity Indicators with Alfalfa Yield
The Spearman correlation coefficient of alfalfa yield did not show any significant positive or negative association with any of the α-diversity indexes examined (Table 3). Contrasting with the α-diversity indexes, yield was significantly associated with ASVs characterized as core members of the bacterial communities found in the root-zone samples. ASVs assigned to Flavisolibacter, uncultured Vicinamibacteraceae, the Panctomycetota group of WD2101, Agromyces, Blastococcus, Rhizobacter, Microvirga, and Sphingomonas were all positively associated with alfalfa yield (Table 3).

Table 3.
Significant associations (p < 0.05) found between α-diversity indices (Shannon, Fisher, inverse Simpson, and Gini–Simpson) and the abundance of shared bacterial taxa in the alfalfa root zone and plant yields using the Spearman correlation coefficient (r).
4. Discussion
4.1. Bacterial Diversity Differs between Root-Zone and Bulk Soil Samples but Not among Different Alfalfa Genotypes
Network attribute and PERMANOVA analysis performed on the Bray–Curtis distances showed that the bacterial composition of the root-zone samples was substantially dissimilar (F = 12.42, R2 = 0.63, p < 0.05) from that of the bulk soil samples. Plant roots affect nutrient availability, oxygen levels, pH, and organic C quality and quantity in areas of the soil that are close to plant roots. They do so through a range of processes, which in turn affect the composition and functions of bacterial communities [48,49]. Several studies have shown that the root compartments of plants exhibit different bacterial assemblies and that plants develop a selection mechanism that enables them to recruit niche-compliant microbes. For example, the bacterial assembly associated with Glycine max is primarily determined by the root compartment [50]. Soil origin exhibits a stronger effect than root compartment on the structure of the rhizosphere bacterial community in M. truncatula [20]. However, the findings of the present study are in line with those of Xiong et al. (2021) [6], who reported that in Zea mays, Triticum aestivum, and Hordeum vulgare, the microbiome composition along the soil–root continuum is predominantly shaped by the compartment niche. The bacterial community that was common to the different alfalfa genotypes accounted for approximately 50% of the total ASVs identified and was phylogenetically assigned mainly to the following phyla: Pseudomonadota, Actinobacteriota, Bacteroidota, and Planctomycetota. Genotypes 6, 8, and 20 shared 9.1% of the core bacterial community, suggesting that these genotypes are similar. Previous studies have shown that plant genotypic variation determines the bacterial composition in the rhizosphere and that plants filter and select bacterial assemblies according to differences in root exudation [51,52]. The results of the current study suggest that the different origin of Genotype 13 (M. sativa nothosubsp. varia (Martyn) Arcang) could partially account for the dissimilarities among different alfalfa genotypes (Figure 3). Genotype 13 exhibited the lowest forage productivity among the alfalfa genotypes, suggesting that plant productivity may influence the composition of bacterial communities in the root zone. Graf et al. [53] showed that M. sativa altered the bacterial composition of denitrifiers in the root zone. They attributed this phenomenon to the production of secondary metabolites with antimicrobial activities [54]. In addition, the lack of differences between the β-diversity distance metrics of Genotypes 6 and 8 (M. sativa × M. arborea) and Genotype 20 (M. sativa ssp. sativa) implies that they could excrete similar root exudates. These exudates could then lead to a proliferation of specific taxa that shape the bacterial composition. In the future, more lines should be examined so that the effect of the alfalfa-genotypic variation on bacterial community composition can be assessed [20]. Assessment of root exudates could also shed light on genotypic differences accounting for the root-zone bacterial composition. In addition, the inclusion in the sampling process of more root compartment samples should increase the plant filtering effect and finetune any genotypic variation effects on bacterial assembly.
4.2. Keystone Bacterial Features in Alfalfa Genotypes
The bacterial taxa identified in the current study were similar to previously studied communities associated with model alfalfa lines of M. truncatula and M. sativa [20,55]. LDA performed on the rhizospheres and bulk soil (Figure 5) revealed the specific ASVs that were enriched in the bulk soil and those enriched or selected for each root-zone environment. In bulk soil samples, the enriched ASVs were assigned to taxa that were previously reported to grow and thrive under high temperatures and drought conditions. For example, members of Gemmatimonadaceae and Solirubrobacterales bacterium 67-14 that proliferated in the bulk soil samples used in the current study have been reported to be dominant in desert and arid soils (DeBruyn et al., 2011; Ullah et al., 2018) [56,57] and poor grassland soil [58]. Taxa of the ubiquitous but uncharacterized WD2101 soil group [59] belonging to the Planctomycetota phylum were only found in bulk soil samples and samples collected from the root zone of Genotype 8. Several members of the WD2101 soil group are known for their general metabolic function of degrading complex C-rich polymers, including cellulose. Thus, the potential role of these taxa on C-cycling in soils remains underexplored [60,61,62].
It is noteworthy that several genera thought to be characteristic of the rhizosphere and root nodules because of their N-fixing abilities, such as Ensifer species, were almost completely absent from the root zone of the alfalfa genotypes under study possibly due to soil effects and/or the early growth stage of the plants. Ensifer (taxonomically under Rhizobiaceae and Alphaproteobacteria) has been extensively studied in the root nodules of leguminous plants like alfalfa [19,21]. It appears to be a major part not only of the root nodule but also of the rhizosphere [20]. The most closely related sequence detected as a statistically important component of all root-zone bacterial communities in the current study was a species under the Allorhizobium–Neorhizobium–Pararhizobium–Rhizobium genus (ASV4540). In addition, many sequences were detected that seem to be preferentially associated with alfalfa genotypes but are not typically observed as major rhizosphere communities, including Sphingomonas, Pseudomonas, and uncultured members of the Vicinamibacterales order (Figure 5). In particular, many bacteria belonging to the Sphingomonas genus that promote plant growth have been identified as important members of bacterial assemblies in the rhizosphere of several crops, including alfalfa [63].
4.3. Linking Bacterial Composition with Alfalfa Forage Productivity
Given this study’s focus on the initial months of crop establishment, it was not possible to pinpoint a significant trend between the α-diversity indexes and plant performance. This issue was also encountered in previous studies, indicating that the effect of bacterial diversity in crop yield is less strong in arid regions [64]. However, other microcosm studies have shown that potato yields under low nutrient conditions are predicted by high initial microbial diversity [65], whereas in pea crops, an initially high level of bacterial diversity increases plant performance under drought conditions [66]. These findings indicate that factors such as plant species and water and nutrient availability can directly affect both bacterial composition and diversity as well as plant yield. In agreement with the above, each individual plant in this study inherently fosters unique bacterial assemblies, and despite the genotypic and phenotypic variations in alfalfa plants, a core bacterial community is consistently shared across all alfalfa genotypes (Figure 4 and Figure 5). This shared bacterial community is postulated to perform specific functions that can significantly influence (or not) plant performance [67]. The abundance of some of the shared bacterial taxa found in the alfalfa root zone was significantly and positively associated with the yield of individual plants (Table 3). Our research shed light on members of the common ASVs found in alfalfa root zone, appearing to correlate with plant yield, suggesting their potential role as key components of the “yield-related” microbiome. Such taxa could be included in synthetic communities for alfalfa since numerous studies have highlighted the potential of synthetic microbial communities in enhancing plant performance [68,69,70,71].
Previous studies have found that members of the identified taxa in our study are bacteria that promote plant growth. For example, in a pot trial, and in line with the results of the current study, it has been shown that Flavisolibacter is significantly associated with alfalfa productivity [72]. These findings suggest that members of this genus that are associated with alfalfa genotypes could have the effect of promoting plant growth. Similarly, Agromyces has been reported to produce indole-3-acetic acid and siderophores. These molecules are known to promote plant growth [73]. The isolation of several strains and ecotypes of Stenotrophomonas has been found to be positively associated with the promotion of plant growth in several crops, especially under abiotic and biotic stress conditions [74,75,76,77,78]. In the root zone of the different alfalfa genotypes under study, the abundance of ASVs belonging to Microvirga was also associated with increased plant yield (Table 3). Recently, the inoculation of rice with a Microvirga isolate was also found to improve shoot and root growth substantially in a genotype-dependent manner [79]. Members of the Microvirga genus are known to nodulate different legumes and were shown to be positively associated with increased yield in a recent study on alfalfa [80].
5. Conclusions
This pioneering study examined the root zones of individual Medicago plants grown under actual field conditions in an HSD, using ultra-wide spacings to eliminate the masking effect of interplant competition. The findings of the current study have shown that the alfalfa genotypic profile can have a significant impact on the composition of a bacterial community in the root zone. Specifically, the bacterial assembly of M. sativa nothosubsp. varia (Martyn) Arcang (Genotype 13) was observed to be highly dissimilar to that found in M. sativa × M. arborea hybrids and in M. sativa ssp. sativa. Besides differences found among the alfalfa genotypes, the bacterial communities in the root-zone and bulk soil samples were also dissimilar, demonstrating the role of the presence of plants in a bacterial assembly. Keystone taxa enriched in each alfalfa genotype were identified. Moreover, specific bacterial taxa were confirmed to be positively associated with fodder yield. These results advance understanding of how alfalfa CWRs regulate bacterial assemblies in the root zone even during the early stages of crop establishment. Furthermore, they highlight the potential influence of specific core bacterial taxa on plant performance and vice versa, which is a critical factor in sustainable production. Future studies should focus on elucidating how shifts in the bacterial assemblies associated with various alfalfa genetic backgrounds affect plant performance over time.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d16070410/s1, Figure S1: The honeycomb field design demonstrates the different moving-ring sizes that a breeder can apply to sample soil heterogeneity more effectively and maximize genetic gain and response to selection; Table S1: Chemical properties of bulk and root-zone soil samples.; Table S2: Composition of the mock community used in the current MiSeq run.
Author Contributions
Conceptualization, M.O. and D.A.F.; methodology, U.M., M.O. and D.F; formal analysis, M.O. and U.M.; resources, A.H., B.K. and D.A.F.; writing—original draft preparation, U.M. and M.O.; writing—review and editing, all authors; supervision, D.A.F. and M.O.; funding acquisition, M.O., D.A.F. and B.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study is part of the MAGNET (INFRASTRUCTURES/1216/0032) project funded by the Cyprus Research and Innovation Foundation (RIF). The RIF is supported by the European Regional Development Fund and the Republic of Cyprus. This work was undertaken as part of the Research Project AGRICYGEN funded by the Republic of Cyprus and the initiative “Adapting Agriculture to Climate Change: Collecting, Protecting and Preparing Crop Wild Relatives” and as part of the initiative “Biodiversity for Opportunities, Livelihoods and Development (BOLD)”, both supported by the Government of Norway. Grant numbers from the Norwegian Government: *QZA-14/0005* and *QZA-20/0154*. The funders had no involvement in the study design; collection, analysis, and interpretation of the data; or the writing of the manuscript.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Supplementary data related to this article can be found in the online version of this manuscript. Raw sequencing data (FASTQ files) have been deposited at the National Center for Biotechnology Information under BioProject PRJNA893879.
Acknowledgments
The authors would like to thank Vasilia Fasoula for providing the figure of the honeycomb design as well as helpful feedback. They also thank Louiza Constantinou for assistance with DNA extraction and Feidias Nicolaou and Zacharias Sofokleous for assistance with field trials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hassani, M.; Durán, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Kavadia, A.; Omirou, M.; Fasoula, D.; Ioannides, I.M. The importance of microbial inoculants in a climate-changing agriculture in Eastern Mediterranean Region. Atmosphere 2020, 11, 1136. [Google Scholar] [CrossRef]
- Liu, F.; Hewezi, T.; Lebeis, S.L.; Pantalone, V.; Grewal, P.S.; Staton, M.E. Soil indigenous microbiome and plant genotypes cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiol. 2019, 19, 201. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Jaramillo, J.E.; Mendes, R.; Raaijmakers, J.M. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 2016, 90, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.; Zhu, Y.G.; Wang, J.T.; Singh, B.; Han, L.L.; Shen, J.P.; Li, P.P.; Wang, G.B.; Wu, C.F.; Ge, A.H. Host selection shapes crop microbiome assembly and network complexity. New Phytol. 2021, 229, 1091–1104. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Yue, W.; Jiao, S.; Kim, H.; Lee, Y.-H.; Wei, G.; Song, W.; Shu, D. Plant domestication shapes rhizosphere microbiome assembly and metabolic functions. Microbiome 2023, 11, 70. [Google Scholar] [CrossRef] [PubMed]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef] [PubMed]
- Spor, A.; Roucou, A.; Mounier, A.; Bru, D.; Breuil, M.-C.; Fort, F.; Vile, D.; Roumet, P.; Philippot, L.; Violle, C. Domestication-driven changes in plant traits associated with changes in the assembly of the rhizosphere microbiota in tetraploid wheat. Sci. Rep. 2020, 10, 12234. [Google Scholar] [CrossRef]
- Leff, J.W.; Lynch, R.C.; Kane, N.C.; Fierer, N. Plant domestication and the assembly of bacterial and fungal communities associated with strains of the common sunflower, Helianthus annuus. New Phytol. 2017, 214, 412–423. [Google Scholar] [CrossRef]
- Huang, X.-F.; Chaparro, J.M.; Reardon, K.F.; Zhang, R.; Shen, Q.; Vivanco, J.M. Rhizosphere interactions: Root exudates, microbes, and microbial communities. Botany 2014, 92, 267–275. [Google Scholar] [CrossRef]
- Mus, F.; Crook, M.B.; Garcia, K.; Garcia Costas, A.; Geddes, B.A.; Kouri, E.D.; Paramasivan, P.; Ryu, M.-H.; Oldroyd, G.E.; Poole, P.S.; et al. Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl. Environ. Microbiol. 2016, 82, 3698–3710. [Google Scholar] [CrossRef] [PubMed]
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 2013, 8, e55731. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; McConkey, B.; Wang, H.; Janzen, H. Root distribution by depth for temperate agricultural crops. Field Crops Res. 2016, 189, 68–74. [Google Scholar] [CrossRef]
- Tramblay, Y.; Koutroulis, A.; Samaniego, L.; Vicente-Serrano, S.M.; Volaire, F.; Boone, A.; Le Page, M.; Llasat, M.C.; Albergel, C.; Burak, S.; et al. Challenges for drought assessment in the Mediterranean region under future climate scenarios. Earth-Sci. Rev. 2020, 210, 103348. [Google Scholar] [CrossRef]
- Humphries, A.W.; Ovalle, C.; Hughes, S.; del Pozo, A.; Inostroza, L.; Barahona, V.; Yu, L.; Yerzhanova, S.; Rowe, T.; Hill, J.; et al. Characterization and pre-breeding of diverse alfalfa crop wild relatives originating from drought-stressed environments. Crop Sci. 2020, 61, 69–88. [Google Scholar] [CrossRef]
- Innes, L.A.; Denton, M.D.; Dundas, I.S.; Peck, D.M.; Humphries, A.W. The effect of ploidy number on vigor, productivity, and potential adaptation to climate change in annual Medicago species. Crop Sci. 2020, 61, 89–103. [Google Scholar] [CrossRef]
- Bromfield, E.S.; Tambong, J.T.; Cloutier, S.; Prévost, D.; Laguerre, G.; Van Berkum, P.; Thi, T.T.; Assabgui, R.; Barran, L. Ensifer, Phyllobacterium and Rhizobium species occupy nodules of Medicago sativa (alfalfa) and Melilotus alba (sweet clover) grown at a Canadian site without a history of cultivation. Microbiology 2010, 156, 505–520. [Google Scholar] [CrossRef]
- Brown, S.P.; Grillo, M.A.; Podowski, J.C.; Heath, K.D. Soil origin and plant genotype structure distinct microbiome compartments in the model legume Medicago truncatula. Microbiome 2020, 8, 139. [Google Scholar] [CrossRef]
- Martínez-Hidalgo, P.; Humm, E.A.; Still, D.W.; Shi, B.; Pellegrini, M.; de la Roca, G.; Veliz, E.; Maymon, M.; Bru, P.; Huntemann, M. Medicago root nodule microbiomes: Insights into a complex ecosystem with potential candidates for plant growth promotion. Plant Soil 2022, 471, 507–526. [Google Scholar] [CrossRef]
- Offre, P.; Pivato, B.; Siblot, S.; Gamalero, E.; Corberand, T.; Lemanceau, P.; Mougel, C. Identification of bacterial groups preferentially associated with mycorrhizal roots of Medicago truncatula. Appl. Environ. Microbiol. 2007, 73, 913–921. [Google Scholar] [CrossRef] [PubMed]
- Fasoula, D.A.; Ioannides, I.M.; Omirou, M. Phenotyping and plant breeding: Overcoming the barriers. Front. Plant Sci. 2020, 10, 1713. [Google Scholar] [CrossRef] [PubMed]
- Omirou, M.; Fasoula, D.A. Creating a Paradigm Shift in Plant Breeding and Plant Phenotyping. Research OUTREACH 119. 2020. Available online: https://researchoutreach.org/articles/creating-a-paradigm-shift-in-plant-breeding-and-plant-phenotyping/ (accessed on 18 August 2023).
- Fasoulas, A.C.; Fasoula, V.A. Honeycomb selection designs. In Plant Breeding Reviews; Janick, J., Ed.; John Wiley and Sons, Inc.: New York, NY, USA, 1995; Volume 13, pp. 87–139. [Google Scholar] [CrossRef]
- Omirou, M.; Ioannides, I.M.; Fasoula, D.A. Optimizing resource allocation in a cowpea (Vigna unguiculata L. Walp.) landrace through whole-plant field phenotyping and non-stop selection to sustain increased genetic gain across a decade. Front. Plant Sci. 2019, 10, 949. [Google Scholar] [CrossRef]
- Chen, L.; Wang, D.; Garmaeva, S.; Kurilshikov, A.; Vich Vila, A.; Gacesa, R.; Sinha, T.; Segal, E.; Weersma, R.K.; Wijmenga, C.; et al. The long-term genetic stability and individual specificity of the human gut microbiome. Cell 2021, 184, 2302–2315. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Solvi, C.; Zhang, F.; Qi, Z.; Chittka, L.; Zhao, W. Gut microbiome drives individual memory variation in bumblebees. Nat. Commun. 2021, 12, 6588. [Google Scholar] [CrossRef] [PubMed]
- Fasoula, V.A.; Fasoula, D.A. Honeycomb breeding: Principles and applications. In Plant Breeding Reviews; Janick, J., Ed.; John Wiley and Sons, Inc.: New York, NY, USA, 2000; Volume 18, pp. 177–250. [Google Scholar] [CrossRef]
- Fasoula, D.A.; Fasoula, V.A. Competitive ability and plant breeding. In Plant Breeding Reviews; Janick, J., Ed.; John Wiley and Sons, Inc.: New York, NY, USA, 1997; Volume 14, pp. 89–138. [Google Scholar] [CrossRef]
- Fasoula, V.A. Prognostic breeding: A new paradigm for crop improvement. In Plant Breeding Reviews; Wiley-Blackwell: New York, NY, USA, 2013; Volume 37, pp. 297–347. [Google Scholar] [CrossRef]
- Bremner, J. Determination of nitrogen in soil by the Kjeldahl method. J. Agric. Sci. 1960, 55, 11–33. [Google Scholar] [CrossRef]
- Walkley, A.; Black, I.A. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Olsen, S.R.; Cole, C.V.; Watanabe, F.S. Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate; US Department of Agriculture. Circular No. 939, US Government Printing Office: Washington, DC, USA, 1954.
- Thomas, G.W. Exchangeable cations. In Methods of Soil Analysis: Part 2 Chemical and Microbiological Properties; American Society of Agronomy, Inc., Soil Science Society of America, Inc.: Madison, WI, USA, 1983; pp. 159–165. [Google Scholar] [CrossRef]
- Mulvaney, R.L. Nitrogen—Inorganic forms. In Methods of Soil Analysis: Part 3 Chemical Methods; Sparks, D.L., Page, A.L., Helmke, P.A., Loeppert, R.H., Soltanpour, P.N., Tabatabai, M.A., Johnston, C.T., Sumner, M.E., Eds.; Soil Science Society of America, Inc. and American Society of Agronomy, Inc.: Madison, WI, USA, 1996; pp. 1123–1184. [Google Scholar] [CrossRef]
- Dorich, R.; Nelson, D. Evaluation of manual cadmium reduction methods for determination of nitrate in potassium chloride extracts of soils. Soil Sci. Soc. Am. J. 1984, 48, 72–75. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Kluyver, T.; Ragan-Kelley, B.; Pérez, F.; Granger, B.E.; Bussonnier, M.; Frederic, J.; Kelley, K.; Hamrick, J.B.; Grout, J.; Corlay, S. Jupyter notebooks—A publishing format for reproducible computational workflows. In Positioning and Power in Academic Publishing: Players, Agents and Agendas; Loizides, F., Schmidt, B., Eds.; IOS Press: Amsterdam, The Netherlands, 2016; pp. 87–90. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, P.F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O‘Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R package Version 2.4-4. 2017. Available online: https://CRAN.R-project.org/package=vegan (accessed on 18 August 2023).
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Abdullaeva, Y.; Ratering, S.; Rosado-Porto, D.; Ambika Manirajan, B.; Glatt, A.; Schnell, S.; Cardinale, M. Domestication caused taxonomical and functional shifts in the wheat rhizosphere microbiota, and weakened the natural bacterial biocontrol against fungal pathogens. Microbiol. Res. 2024, 281, 127601. [Google Scholar] [CrossRef] [PubMed]
- Meinshausen, N.; Bühlmann, P. Stability Selection. J. R. Stat. Soc. Ser. B (Stat. Methodol. ) 2010, 72, 417–473. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. In Proceedings of the International AAAI Conference on Web and Social Media, San Jose, CA, USA, 17–20 May 2009; Volume 3, pp. 361–362. [Google Scholar]
- Liu, C.; Li, C.; Jiang, Y.; Zeng, R.J.; Yao, M.; Li, X. A guide for comparing microbial co-occurrence networks. iMeta 2023, 2, e71. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. “Circlize” Implements and Enhances Circular Visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Van Overbeek, L.; Van Elsas, J.D. Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.). FEMS Microbiol. Ecol. 2008, 64, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Wang, J.; Li, Y.; Wang, C.; Yang, S.; Jiao, S.; Wei, G.; Chen, W. Local domestication of soybean leads to strong root selection and diverse filtration of root-associated bacterial communities. Plant Soil 2022, 480, 439–455. [Google Scholar] [CrossRef]
- Lareen, A.; Burton, F.; Schäfer, P. Plant root–microbe communication in shaping root microbiomes. Plant Mol. Biol. 2016, 90, 575–587. [Google Scholar] [CrossRef]
- Rolfe, S.A.; Griffiths, J.; Ton, J. Crying out for help with root exudates: Adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr. Opin. Microbiol. 2019, 49, 73–82. [Google Scholar] [CrossRef]
- Graf, D.R.; Saghaï, A.; Zhao, M.; Carlsson, G.; Jones, C.M.; Hallin, S. Lucerne (Medicago sativa) alters N2O-reducing communities associated with cocksfoot (Dactylis glomerata) roots and promotes N2O production in intercropping in a greenhouse experiment. Soil Bio. Biochem. 2019, 137, 107547. [Google Scholar] [CrossRef]
- Gholami, A.; De Geyter, N.; Pollier, J.; Goormachtig, S.; Goossens, A. Natural product biosynthesis in Medicago species. Nat. Prod. Rep. 2014, 31, 356–380. [Google Scholar] [CrossRef] [PubMed]
- Pini, F.; Frascella, A.; Santopolo, L.; Bazzicalupo, M.; Biondi, E.G.; Scotti, C.; Mengoni, A. Exploring the plant-associated bacterial communities in Medicago sativa L. BMC Microbiol. 2012, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- DeBruyn, J.M.; Nixon, L.T.; Fawaz, M.; Johnson, A.M.; Radosevich, M. Global biogeography and quantitative seasonal dynamics of Gemmatimonadetes in soil. Appl. Environ. Microbiol. 2011, 77, 6295–6300. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Akbar, A.; Luo, Q.; Khan, A.H.; Manghwar, H.; Shaban, M.; Yang, X. Microbiome diversity in cotton rhizosphere under normal and drought conditions. Microb. Ecol. 2018, 77, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Whitton, M.M.; Ren, X.; Yu, S.J.; Irving, A.D.; Trotter, T.; Bajagai, Y.S.; Stanley, D. Humate application alters microbiota–mineral interactions and assists in pasture dieback recovery. Heliyon 2023, 9, e13327. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N.; Beletsky, A.V.; Ivanova, A.A.; Kulichevskaya, I.S.; Suzina, N.E.; Philippov, D.A.; Rakitin, A.L.; Mardanov, A.V.; Ravin, N.V. Wide distribution of Phycisphaera-like planctomycetes from WD2101 soil group in peatlands and genome analysis of the first cultivated representative. Environ. Microbiol. 2021, 23, 1510–1526. [Google Scholar] [CrossRef] [PubMed]
- Probst, M.; Ascher-Jenull, J.; Insam, H.; Gómez-Brandón, M. The molecular information about deadwood bacteriomes partly depends on the targeted environmental DNA. Front. Microbiol. 2021, 12, 640386. [Google Scholar] [CrossRef]
- Probst, M.; Gómez-Brandón, M.; Bardelli, T.; Egli, M.; Insam, H.; Ascher-Jenull, J. Bacterial communities of decaying Norway spruce follow distinct slope exposure and time-dependent trajectories. Environ. Microbiol. 2018, 20, 3657–3670. [Google Scholar] [CrossRef]
- Tláskal, V.; Zrůstová, P.; Vrška, T.; Baldrian, P. Bacteria associated with decomposing dead wood in a natural temperate forest. FEMS Microbiol. Ecol. 2017, 93, fix157. [Google Scholar] [CrossRef] [PubMed]
- Kisiel, A.; Kępczyńska, E. Medicago truncatula Gaertn. as a model for understanding the mechanism of growth promotion by bacteria from rhizosphere and nodules of alfalfa. Planta 2016, 243, 1169–1189. [Google Scholar] [CrossRef] [PubMed]
- Stefan, L.; Hartmann, M.; Engbersen, N.; Six, J.; Schöb, C. Positive effects of crop diversity on productivity driven by changes in soil microbial composition. Front. Microbiol. 2021, 12, 660749. [Google Scholar] [CrossRef]
- Lankau, R.A.; George, I.; Miao, M. Crop performance is predicted by soil microbial diversity across phylogenetic scales. Ecosphere 2022, 13, e4029. [Google Scholar] [CrossRef]
- Prudent, M.; Dequiedt, S.; Sorin, C.; Girodet, S.; Nowak, V.; Duc, G.; Salon, C.; Maron, P.A. The diversity of soil microbial communities matters when legumes face drought. Plant Cell Environ. 2020, 43, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Van Der Heijden, M.G.; Hartmann, M. Networking in the plant microbiome. PLoS Biol. 2016, 14, e1002378. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.; Tyagi, R.; Sharma, S. Combating biotic stresses in plants by synthetic microbial communities: Principles, applications and challenges. J. Appl. Microbiol. 2022, 133, 2742–2759. [Google Scholar] [CrossRef] [PubMed]
- Schlatter, D.; Yin, C.; Hulbert, S.H.; Paulitz, T.C. Core rhizosphere microbiomes of dryland wheat are influenced by location and land use history. Appl. Environ. Microbiol. 2020, 86, e02135-19. [Google Scholar] [CrossRef] [PubMed]
- Shayanthan, A.; Ordoñez, P.A.C.; Oresnik, I.J. The role of synthetic microbial communities (syncom) in sustainable agriculture. Front. Agron. 2022, 4, 896307. [Google Scholar] [CrossRef]
- Vorholt, J.A.; Vogel, C.; Carlström, C.I.; Müller, D.B. Establishing causality: Opportunities of synthetic communities for plant microbiome research. Cell Host Microbe 2017, 22, 142–155. [Google Scholar] [CrossRef]
- Xiao, X.; Fan, M.; Wang, E.; Chen, W.; Wei, G. Interactions of plant growth-promoting rhizobacteria and soil factors in two leguminous plants. Appl. Microbiol. Biotechnol. 2017, 101, 8485–8497. [Google Scholar] [CrossRef]
- Wu, C.H.; Bernard, S.M.; Andersen, G.L.; Chen, W. Developing microbe–plant interactions for applications in plant-growth promotion and disease control, production of useful compounds, remediation and carbon sequestration. Microb. Biotechnol. 2009, 2, 428–440. [Google Scholar] [CrossRef]
- Alexander, A.; Singh, V.K.; Mishra, A.; Jha, B. Plant growth promoting rhizobacterium Stenotrophomonas maltophilia BJ01 augments endurance against N2 starvation by modulating physiology and biochemical activities of Arachis hypogea. PLoS ONE 2019, 14, e0222405. [Google Scholar] [CrossRef]
- Alijani, Z.; Amini, J.; Ashengroph, M.; Bahramnejad, B. Volatile compounds mediated effects of Stenotrophomonas maltophilia strain UN1512 in plant growth promotion and its potential for the biocontrol of Colletotrichum nymphaeae. Physiol. Mol. Plant Pathol. 2020, 112, 101555. [Google Scholar] [CrossRef]
- Manh Tuong, H.; Garcia Mendez, S.; Vandecasteele, M.; Willems, A.; Luo, D.; Beirinckx, S.; Goormachtig, S. Stenotrophomonas sp. SRS1 promotes growth of Arabidopsis and tomato plants under salt stress conditions. Plant Soil 2022, 473, 547–571. [Google Scholar] [CrossRef]
- Ulrich, K.; Kube, M.; Becker, R.; Schneck, V.; Ulrich, A. Genomic analysis of the endophytic Stenotrophomonas strain 169 reveals features related to plant-growth promotion and stress tolerance. Front. Microbiol. 2021, 12, 687463. [Google Scholar] [CrossRef]
- Zhuang, X.; Liu, Y.; Fang, N.; Bai, Z.; Gao, J. Quorum sensing improves the plant growth-promoting ability of Stenotrophomonas rhizophila under saline-alkaline stress by enhancing its environmental adaptability. Front. Microbiol. 2023, 14, 1155081. [Google Scholar] [CrossRef]
- Balasjin, N.M.; Maki, J.S.; Schläppi, M.R.; Marshall, C.W. Plant growth-promoting activity of bacteria isolated from Asian rice (Oryza sativa L.) depends on rice genotype. Microbiol. Spectr. 2022, 10, e0278721. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, Y.; Zhang, F. Community assembly correlates with alfalfa production by mediating rhizosphere soil microbial community composition in different planting years and regimes. Plant Soil 2022, 479, 355–370. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).