The Bear Truth: Analyzing Genetic Variability and Population Structure in Sloth Bear across the Vidarbha Landscape Using Microsatellite Markers
Abstract
1. Introduction
2. Materials and Methods
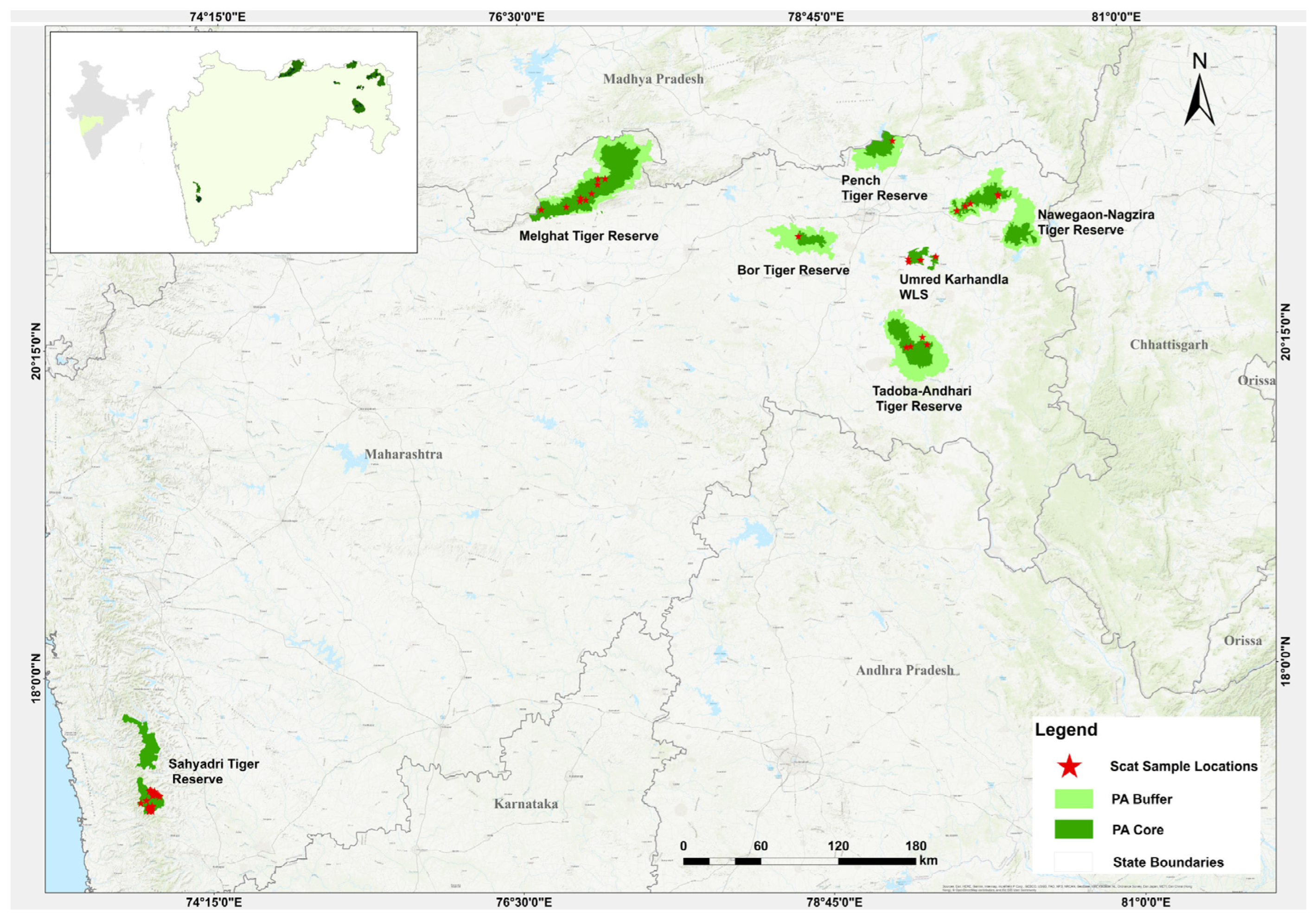
2.1. Sampling Area
2.2. Field Sampling
2.3. Primer Selection
2.4. DNA Extraction
2.5. PCR Standardization and Data Validation
2.6. Data Analyses
3. Results
3.1. Microsatellite Genotyping
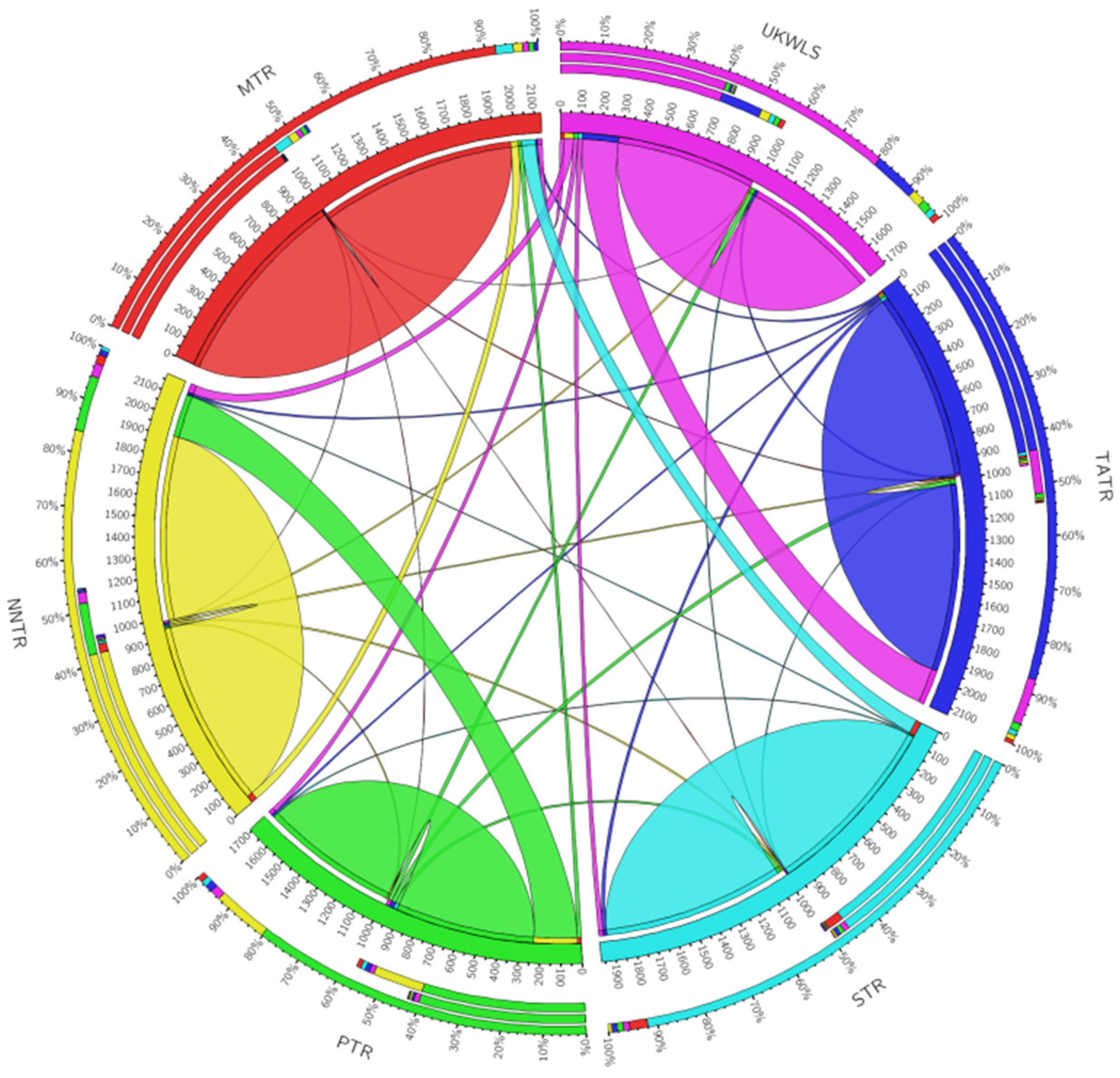
3.2. Population Structuring and Variability
4. Discussion
4.1. Primer Standardization and Data Analysis
4.2. Population Structuring and Variability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ouborg, N. Isolation, Population Size and Extinction: The Classical and Metapopulation Approaches Applied to Vascular Plants along the Dutch Rhine-System. Oikos 1993, 66, 298–308. [Google Scholar] [CrossRef]
- Pacifici, M.; Rondinini, C.; Rhodes, J.; Vurbidge, A.; Cristiano, A.; Watson, J.; Woinarski, J.C.Z.; Di Marco, M. Global correlates of range contractions and expansions in terestrial mammals. Nat. Commun. 2020, 11, 2840. [Google Scholar] [CrossRef] [PubMed]
- Wolf, C.; Ripple, W. Range Contractions of the world’s large carnivores. R. Soc. Open Sci. 2017, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ripple, W.; Newsome, T.; Wolf, C.; Dirzo, R.; Everatt, K.; Galetti, M.; Hayward, M.W.; Kerley, G.I.H.; Levi, T.; Lindsey, P.A.; et al. Collapse of World’s largest herbivores. Sci. Adv. 2015, 1, e1400103. [Google Scholar] [CrossRef]
- Fisher, D.; Owens, I. The comparative methods in conservation biology. Trends Ecol. Evol. 2004, 6, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Crooks, K.; Burdett, C.; Theobald, D.; Rondinini, C.; Boitani, L. Global patterns of fragmentation and connectivity of mammalian carnivore habitat. Philos. Trans. R. Soc. B: Biol. Sci. 2011, 366, 2642–2651. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, H.; Durant, S.; Woodroffe, R. What wild dogs want: Habitat selection differs across life stages and orders of selection in a wide-ranging carnivore. BMC Zool. 2020, 5, 2–11. [Google Scholar] [CrossRef]
- Sacco, T.; Valkenburgh, B. V Ecomorphological indicators of feeding behaviour in the bears (Carnivora: Ursidae). J. Zool. 2006, 263, 41–54. [Google Scholar] [CrossRef]
- Islam, M.; Uddin, M.; Aziz, M.; Muzaffar, S.; Chakma, S.; Chowdhury, U.; Chowdhury, G.W.; Rashid, M.A.; Samiul, M.; Jahan, I.; et al. Status of bears in Bangladesh: Going, going, gone? Ursus 2013, 24, 83–90. [Google Scholar] [CrossRef]
- Dhraiya, N.; Baragli, H.; Sharp, T. Melursus ursinus (Amended Version of 2016 Assessment). 2020. Available online: https://www.iucnredlist.org/species/13143/166519315#geographic-range (accessed on 25 November 2023).
- Yoganand, K.; Rice, C.; Seidensticker, J.; Johnsingh, A. Is the sloth bear in India secure? A preliminary report on distribution, threats and conservation requirements. J. Bombay Nat. Hist. Soc. 2006, 103, 172–181. [Google Scholar]
- Shankar, K.; Murthy, R.S. Assessment of Bear-Man Conflict in North Bilaspur Forest Division, Bilaspur, Madhya Pradesh; Wildlife Institute of India: Dehradun, India, 1995.
- Frankham, R. Genetics and Extinction. Biol. Conserv. 2005, 126, 131–140. [Google Scholar] [CrossRef]
- Frankham, R Genetic rescue of small inbred populations: Meta-analysis reveals large and consistent benefits of gene flow. Mol. Ecol. 2015, 24, 2610–2618. [CrossRef] [PubMed]
- Ralls, K.; Ballou, J.; Dudash, M.; Elridge, M.; Fenster, C.; Lacy, R.; Sunucks, R.; Frankham, R. Call for a paradigm shift in the genetic management of fragmented populations: Genetic management. Conserv. Lett. 2018, 11, 1–6. [Google Scholar] [CrossRef]
- Tallmon, D.A.; Bellemain, E.; Swenson, J.E.; Taberlet, P. Genetic monitoring of Scandinavian brown bear effective population size and immigration. J. Wildl. Manag. 2009, 68, 960–965. [Google Scholar] [CrossRef]
- Kruckenhauser, L.; Rauer, G.; Daubl, B.; Haring, E. Genetic monitoring of a founder population of brown bears (Ursus arctos) in central Austria. Conserv. Genet. 2008, 10, 1223–1233. [Google Scholar] [CrossRef]
- Carroll, E.; Bruford, M.; DeWoody, J.; Leroy, G.; Strand, A.; Waits, L.; Wang, J. Genetic and genomic monitoring with minimally invasive sampling methods. Evol. Appl. 2018, 11, 1094–1119. [Google Scholar] [CrossRef] [PubMed]
- Procter, M.; Kasworm, W.; Teisberg, J.; Servheen, C.; Radandt, T.; Lamb, C.T.; Kendall, K.C.; Mace, R.D.; Paetkau, D.; Boyce, M. American black bear population fragmentation detected with pedigrees in the transborder Canada–United States region. Ursus 2020, 31, 1–15. [Google Scholar] [CrossRef]
- Dutta, T.; Sharma, S.; Maldonado, J.; Seidensticker, J. Genetic Variation, Structure, and Gene Flow in a Sloth Bear (Melursus ursinus) Meta-Population in the Satpura-Maikal Landscape of Central India. PLoS ONE 2020, 10, e0123384. [Google Scholar] [CrossRef]
- Sharma, S.; Dutta, T.; Maldonando, J.E.; Wood, T.C. Selection of microsatellite loci for genetic monitoring of sloth bears. Ursus 2013, 24, 164–169. [Google Scholar] [CrossRef]
- Thatte, P.; Chandramouli, A.; Tyagi, A.; Patel, K.; Baro, P.; Chhattani, H.; Ramakrishnan, U. Human footprint differentially impacts genetic connectivity of four wide-ranging mammals in a fragmented landscape. Divers. Distrib. 2020, 26, 299–314. [Google Scholar] [CrossRef]
- Wang, H.; Yang, B.; Wang, H.; Xiao, H. Impact of different numbers of microsatellite markers on population genetic results using SLAF-seq data for Rhododendron species. Sci. Rep. 2021, 11, 8597. [Google Scholar] [CrossRef] [PubMed]
- Landguth, E.; Fedy, B.; Oyler-McCance, S.J.; Garey, A.L.; Emel, S.L.; Mumma, M.; Wagner, H.H.; Fortin, M.J. Effects of sample size, number of markers, and allelic richness on the detection of spatial genetic pattern. Mol. Ecol. Resour. 2011, 12, 276–284. [Google Scholar] [CrossRef]
- Hale, M.; Burg, T.; Steeves, T. Sampling for Microsatellite-Based Population Genetic Studies: 25 to 30 Individuals per Population Is Enough to Accurately Estimate Allele Frequencies. PLoS ONE 2012, 7, e45170. [Google Scholar] [CrossRef] [PubMed]
- Sathyakumar, S.; Kaul, R.; Ashraf, N.; Mookherjee, A.; Menon, V. National Bear Conservation and Welfare Action Plan; Wildlife Institute of India and Wildlife Trust of India: Dehradun, India, 2012.
- Forest Survey of India. Indian State Forest Report. 2021. Available online: https://fsi.nic.in/forest-report-2021-details (accessed on 3 January 2023).
- Laurie, A.; Seidensticker, J. Behavioural ecology of the Sloth Bear (Melursus ursinus). J. Zool. 1977, 182, 187–204. [Google Scholar] [CrossRef]
- Paetkau, D.; Strobeck, C.S.; Calvert, W. Microsatellite analysis of population-structure in Canadian Polar Bears. Mol. Ecol. 1995, 4, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Camarra, J.; Grifin, S.; Uhres, E.; Hanotte, O.; Waits, L.; Dubois-Paganon, D.; Burke, T.; Bouvet, J. Noninvasive genetic tracking of the endangered Pyrenean brown bear population. Mol. Ecol. 1997, 6, 869–876. [Google Scholar] [CrossRef]
- Kitahara, E.; Isagi, Y.I.; Saitoh, T. Polymorphic microsatellite DNA markers in the asiatic black bear Ursus thibetanus. Mol. Ecol. 2000, 9, 1661–1662. [Google Scholar] [CrossRef] [PubMed]
- Shih, C.; Huang, C.; Li, S.; Hwang, M. Ten novel tetranucleotide microsatellite DNA markers from Asiatic black bear, Ursus thibetanus. Conserv. Genet. 2009, 10, 1845–1847. [Google Scholar] [CrossRef]
- Kleven, O.; Hallstrom, B.; Hailer, F.; Janke, A.; Hagen, S.; Kopatz, A.; Eiken, H. Identification and evaluation of novel di- and tetranucleotide microsatellite markers from the brown bear (Ursus arctos). Conserv. Genet. Resour. 2012, 4, 737–741. [Google Scholar] [CrossRef]
- Meredith, E.; Rodzen, J.; Banks, J.D.; Jones, K. Characterization of 29 tetranucleotide microsatellite loci in black bear (Ursus americanus) for use in forensic and population applications. Conserv. Genet. 2008, 10, 693–696. [Google Scholar] [CrossRef]
- Mondol, S.; Karanth, K.; Samba Kumar, N.; Gopalswamy, A.; Andheria, A.; Ramakrishnan, U. Evaluation of non-invasive genetic sampling methods for estimating tiger population size. Biol. Conserv. 2009, 142, 2350–2360. [Google Scholar] [CrossRef]
- Miquel, C.; Bellemain, E.; Poillot, C.; Bessière, J.; Durand, A.; Taberlet, P. Quality indexes to assess the reliability of genotypes in studies using noninvasive sampling and multiple-tube approach. Mol. Ecol. Notes 2006, 6, 985–988. [Google Scholar] [CrossRef]
- Broquet, T. Petit, Quantifying genotyping errors in noninvasive population genetics. Mol. Ecol. 2004, 13, 3601–3608. [Google Scholar] [CrossRef]
- Oosterhout, C.V.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Kalinowski, S.; Taper, M.; Marshall, T. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Valiere, N. Gimlet: A computer program for analysing genetic individual identification data. Mol. Ecol. Resour. 2002, 2, 377–379. [Google Scholar] [CrossRef]
- Wright, S. The genetical structure of populations. Ann. Eugen. 1951, 15, 323–354. [Google Scholar] [CrossRef]
- Nei, M. F-statistics and analysis of gene diversity in subdivided populations. Ann. Hum. Genet. 1977, 41, 225–233. [Google Scholar] [CrossRef]
- Rousset, F. Genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Caye, K.; Deist, T.M.; Martins, H.; Michel, O.; François, O. TESS3: Fast inference of spatial population structure and genome scans for selection. Mol. Ecol. Resour. 2016, 16, 540–548. [Google Scholar] [CrossRef]
- Walsh, P.S.; Fildes, N.J.; Reynolds, R. Sequence analysis and characterization of stutter products at the tetranucleotide repeat locus. Nucleic Acids Res. 1996, 24, 2807–2812. [Google Scholar] [CrossRef]
- Rannala. BayesAss Edition 3.0 User’s Manual; University of California: Davis, CA, USA, 2007. [Google Scholar]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize: Implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]
- Waits, L.; Luikart, G.; Taberlet, P. Estimating the probability of identity among genotypes in natural populations: Cautions and guidelines. Mol. Ecol. 2001, 10, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, L.; Holloway, J.; Patterson, B.; White, B. An improved field method to obtain DNA for individual identification from wold scat. J. Wildl. Manag. 2009, 73, 1430–1435. [Google Scholar] [CrossRef]
- Mondal, I.; Habib, B.; Talukdar, G.; Nigam, P. Triage of means: Options for conserving tiger corridors beyond designated protected lands in India. Front. Ecol. Evol. 2016, 4, 2–7. [Google Scholar] [CrossRef]
- Reddy, P.A.; Gour, D.S.; Bhavanishankar, M.; Jaggi, K.; Hussain, S.M.; Harika, K.; Shivaju, S. Spatial genetic analysis reveals high connectivity of tiger (Panthera tigris) populations in the Satpura–Maikal landscape of Central India. PLoS ONE 2012, 3, 48–60. [Google Scholar]
- Sharma, S.; Dutta, T.; Maldonado, J.E.; Wood, T.C.; Panwar, H.S.; Seidenstick, J. Genetic Evidence of Tiger Population Structure and Migration within an Isolated and Fragmented Landscape in Northwest India. PLoS ONE 2013, 3, 48–60. [Google Scholar]
- Yumnam, B.; Jhala, Y.; Qureshi, Q.; Maldonado, J.; Gopal, R.; Saini, S.; Srinivas, Y.; Fleischer, R. Prioritizing tiger conservation through landscape genetics and habitat linkages. PLoS ONE 2014, 9, e111207. [Google Scholar] [CrossRef] [PubMed]
- Dutta, T.; Sharma, S.; DeFries, R. Targeting restoration sites to improve connectivity in a tiger conservation landscape in India. PeerJ 2018, 6, 587. [Google Scholar] [CrossRef]
- Dutta, T.; Sharma, S.; Maldonado, J.; Wood, T.; Panwar, H.; Seidensticker, J. Fine-scale population geneic structure in a wide-ranging carnivore, the leopard (Panthera pardus fusca) in central India. Divers. Distrib. 2012, 19, 760–771. [Google Scholar] [CrossRef]
- Ratnayeke, S.; Van Manen, F.T.; Padmalal, U.K.G.K. Home ranges and habitat use of sloth bears Melursus ursinus inornatus in Wasgomuwa National Park. Sri Lanka. Wildl. Biol. 2007, 13, 272–284. [Google Scholar] [CrossRef]
- Joshi, A.R.; Garshelis, D.L.; Smith, J.L.D. Home Ranges of Sloth bears in Nepal: Implications for Conservation. J. Wildl. Manag. 1995, 59, 204–214. [Google Scholar] [CrossRef]
- Diniz, M.F.; Coehlo, M.T.P.; Sousa, F.G.; Hasui, E.; Loyola, R. The underestimated role of small fragments for carnivore dispersal in the Atlantic Forest. Perspect. Ecol. Conserv. 2021, 19, 81–89. [Google Scholar] [CrossRef]
- Murphy, S.; Augustine, B.; Ulrey, W.; Guthrie, J.; McCown, J.; Cox, J. Consequences of severe habitat fragmentation on density, genetics, and spatial capture-recapture analysis of a small bear population. PLoS ONE 2017, 12, e0181849. [Google Scholar] [CrossRef]
- Maduna, S.; Aars, J.; Fløystad, I.; Klütsch, C.; Zeyl Fiskebeck, E.; Wiig, O.; Ehrich, D.; Anderson, M.; Bachmann, L.; Derocher, A.E.; et al. Sea ice reduction drives genetic differentiation among Barents Sea polar bears. Proc. R. Soc. B Biol. Sci. 2021, 288, 1–9. [Google Scholar] [CrossRef]
- Crooks, K.R.; Burdett, C.L.; Theobald, D.M.; King, S.R.; Di Marco, M.; Rondinini, C.; Boitani, L. Quantification of habitat fragmentation reveals extinction risk in terrestrial mammals. Proc. Natl. Acad. Sci. USA 2017, 114, 7635–7640. [Google Scholar] [CrossRef]

| Locus | Primer Sequence | Multiplex Panel Group | Repeat Type |
|---|---|---|---|
| UarT838 | 5′-3′ TCTCTACATCCTTGCCAGC CGCAAATCAAAACCACAATG | 1 | Tetra |
| UT1 | 5′-3′ ACAACTCTTCTCAGATGTTCACAAA CCCAGGTCAGCACTTGGCATAC | 1 | Tetra |
| UT38 | 5′-3′ ATTATTGATGAGCAGGGACAG CTAAAGCAACAACATGTGAATG | 2 | Tetra |
| UarT259 | 5′-3′ CTCTGGACTTCTGGCTCAGG TGAAGCCATCAACATTGCTC | 2 | Tetra |
| Umar2 | 5′-3′ TCACGGGTTTGTAGTAAACA CACAAAGTGGATGCTAAGAA | 3 | Di |
| UT4 | 5′-3′ GAGTTATTGGCACTAAAATCTAATG CTGCAAATCCCTGCTCAACTTTC | 3 | Tetra |
| UamD112 | 5′-3′ GAATCCTCTCCAAGACCTATG GTTTTCCTTATCCCTGAACTG | 3 | Di |
| G1D | 5′-3′ GATCTGTGGGTTTATAGGTTACA CTACTCTTCCTACTCTTTAAGAG | 4 | Di |
| G10L | 5′-3′ GTACTGATTTTATTCACATTTCCC GAAGATACAGAAACCTACCCATGC | 4 | Di |
| G10B | 5′-3′ AAGCCTTTTAATGTTCTGTTG AGGACAAATCACAGAAACCT | 5 | Di |
| UT29 | 5′-3′ GACATTGCCTTTTACAGAGCAG GGGCAGATCTCAACCACCATAAGC | 6 | Di |
| CXX203 | 5′-3′ TTGATCTGAATAGTCCTCTGCG AGCAACCCCTCCCATTTACT | 6 | Tetra |
| Mu23 | 5′-3′ GCCTGTGTGCTATTTTATCC TTGCTTGCCTAGACCACC | 5 | Di |
| Locus | Ta | Allelic Size | Number of Alleles | Ho | HE | PID a | PID(sibs) b | PID(cum) c | PID (Sibs-cum) d | ADO | FA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UarT838 | 57 °C | 97–145 | 4 | 0.66 | 0.80 | 1.777 × 10−1 | 4.649 × 10−1 | 1.777 × 10−1 | 4.649 × 10−1 | 0.183 | 0.024 |
| UT1 | 57 °C | 176–192 | 5 | 0.63 | 0.36 | 1.786 × 10−1 | 4.825 × 10−1 | 3.172 × 10−2 | 2.243 × 10−1 | 0 | 0.028 |
| UT38 | 53 °C | 196–232 | 12 | 0.68 | 0.24 | 1.118 × 10−1 | 4.396 × 10−1 | 3.548 × 10−3 | 9.860 × 10−2 | 0 | 0.038 |
| UarT259 | 53 °C | 153–177 | 10 | 0.46 | 0.18 | 2.923 × 10−1 | 5.975 × 10−1 | 1.037 × 10−3 | 5.891 × 10−2 | 0 | 0.038 |
| Umar2 | 53 °C | 185–227 | 10 | 0.65 | 0.31 | 1.472 × 10−1 | 4.632 × 10−1 | 1.527 × 10−4 | 2.729 × 10−2 | 0 | 0.056 |
| UT4 | 55 °C | 157–182 | 8 | 0.75 | 0.55 | 9.237 × 10−2 | 3.984 × 10−1 | 1.410 × 10−5 | 1.087 × 10−2 | 0.225 | 0.054 |
| UamD112 | 55 °C | 142–210 | 12 | 0.84 | 0.29 | 3.785 × 10−2 | 3.411 × 10−1 | 5.339 × 10−7 | 3.708 × 10−3 | 0 | 0.042 |
| G1D | 59 °C | 176 | 5 | 0.60 | 0.47 | 2.031 × 10−1 | 5.048 × 10−1 | 1.084 × 10−7 | 1.872 × 10−3 | 0.072 | 0.035 |
| G10L | 59 °C | 165 | 7 | 0.73 | 0.84 | 1.063 × 10−1 | 4.149 × 10−1 | 1.153 × 10−8 | 7.766 × 10−4 | 0.026 | 0.012 |
| G10B | 56 °C | 133–143 | 14 | 0.89 | 0.45 | 1.910 × 10−2 | 3.123 × 10−1 | 2.20 × 10−10 | 2.42 × 10−4 | 0.063 | 0.051 |
| UT29 | 58 °C | 168–192 | 10 | 0.85 | 0.53 | 3.598 × 10−2 | 3.353 × 10−1 | 7.927 × 10−12 | 8.131 × 10−5 | 0.105 | 0.025 |
| CXX203 | 58 °C | 122–146 | 7 | 0.69 | 0.46 | 1.366 × 10−1 | 4.404 × 10−1 | 1.08 × 10−12 | 3.581 × 10−5 | 0.288 | 0.024 |
| Mu23 | 56 °C | 164–180 | 11 | 0.73 | 0.25 | 9.926 × 10−2 | 4.093 × 10−1 | 1.075 × 10−13 | 1.46 × 10−5 | 0.015 | 0.024 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomes, L.; Modi, S.; Nigam, P.; Habib, B. The Bear Truth: Analyzing Genetic Variability and Population Structure in Sloth Bear across the Vidarbha Landscape Using Microsatellite Markers. Diversity 2024, 16, 74. https://doi.org/10.3390/d16020074
Gomes L, Modi S, Nigam P, Habib B. The Bear Truth: Analyzing Genetic Variability and Population Structure in Sloth Bear across the Vidarbha Landscape Using Microsatellite Markers. Diversity. 2024; 16(2):74. https://doi.org/10.3390/d16020074
Chicago/Turabian StyleGomes, Lynette, Shrushti Modi, Parag Nigam, and Bilal Habib. 2024. "The Bear Truth: Analyzing Genetic Variability and Population Structure in Sloth Bear across the Vidarbha Landscape Using Microsatellite Markers" Diversity 16, no. 2: 74. https://doi.org/10.3390/d16020074
APA StyleGomes, L., Modi, S., Nigam, P., & Habib, B. (2024). The Bear Truth: Analyzing Genetic Variability and Population Structure in Sloth Bear across the Vidarbha Landscape Using Microsatellite Markers. Diversity, 16(2), 74. https://doi.org/10.3390/d16020074






