Abstract
The complete mitogenome of Doleschallia bisaltide was sequenced with a size of 16,389 bp. Gene orientation and arrangement in the newly sequenced mitogenome are the same as other mitogenomes in Lepidoptera. Except for trnS1(AGN), which lacks the dihydrouridine (DHC) arm, the other 21 tRNA genes all contain a typical cloverleaf structure. Ka/Ks ratio analysis of 13 protein-coding genes (PCGs) from 23 Nymphalinae species indicates that the evolutionary rate of COX1 was slowest, while that of ATP8, ND5, and ND6 was substantially high. Phylogenetic analysis revealed that Nymphalinae and Kallimini were nonmonophyletic. Trees constructed only from the nuclear DNA (nDNA) dataset had lower support than mitochondrial or combined datasets. The addition of RNA genes did not improve the phylogenetic signal, and nodal support decreased. These data provide important information for future studies into the phylogeny of Nymphalinae.
1. Introduction
The subfamily Nymphalinae (Lepidoptera, Nymphalidae) is distributed worldwide and consists of 496 species in 56 genera [1]. Nymphalinae contributed to studies of speciation, plant-insect interactions, biogeography, community ecology, and climate change and includes many model organisms and pest species [2,3,4,5]. The composition and tribal classification of Nymphalinae has been controversial, with authors recognizing from two to six tribes. Ackery [6] divided Nymphalinae into two tribes based on host plants: the Nymphalini and Coloburini. Based on morphological characters, the Nymphalinae in China were divided into three tribes: Nymphalini, Hypolimni, and Melitaeini [7,8]. Based on the arrangement of filiform setae of larvae, the Nymphalinae was divided into three tribes: Nymphalini, Melitaeini, and Kallimini [9]. This classification has been recognized by other taxonomists, with Coeini later added [10,11,12,13,14]. Subsequently, Wahlberg et al. [1] divided Nymphalinae into six tribes: Melitaeini, Nymphalini, Junoniini, Victorinini, Kallimini, and Coeini based on DNA sequence data (COX1, EF-1α and wingless) although some clades were not stable. Zhang et al. [15] divided Nymphalinae into 11 tribes: Nymphalini, Junoniini, Victorinini, Kallimini, Melitaeini, Coeini, Pycinini, Rhinopalpini, Kallimoidini, Vanessulini, and Doleschalliaini based on genomic data, with four genera Pycina, Rhinopalpa, Kallimoides, and Vanessula unplaced to a tribe. Zhang et al. [15] placed Doleschallia in a new tribe known as Doleschalliaini.
Previous phylogenetic studies sampled enough representatives of Nymphalidae to test the monophyly and circumscription of the Nymphalinae [4,12,14,16,17,18]. However, in these studies, there were only a few representatives of Nymphalinae, and some tribes had unstable taxonomic status. Neither DNA sequences [11,12,19] nor morphological characters [14] recovered Kallimini as a monophyletic group. Based on a more taxon-rich phylogenetic analysis of Nymphalinae using DNA sequences, Wahlberg et al. [1] proposed that the tribe Coeini, as currently constituted, is untenable. Based on analysis of the genomic data, Zhang et al. [15] proposed that Kallimoides rumia and Vanessula milca were placed in the clade composed of Melitaeini, Junoniini, Victorinini, and Doleschallia, with their exact phylogenetic position weakly supported.
Mitochondrial genomes and nuclear genes have been widely used in phylogenetic studies of Lepidoptera [16,20,21,22]. In this study, we conducted a phylogenetic analysis of Nymphalinae using ML and BI analyses of several different datasets assembled from complete mitochondrial genome sequences and four nuclear genes. The impact of inclusion vs. exclusion of RNA genes and nuclear genes on the phylogenetic resolution was tested. Our aim was to test the monophyly of tribes and relationships among the major lineages of this subfamily.
2. Materials and Methods
2.1. Taxon Sampling, Identification and DNA Extraction
We sampled 23 taxa representing 21 genera and 6 tribes within the Nymphalinae. Three genera, Ariadne, Cyrestis, and Dichorragia, from the related subfamilies (Biblidinae, Cyrestinae, and Pseudergolinae, respectively) were used as outgroups. The species sampled are summarized in Table S1. Among these species, five species (Doleschallia bisaltide, Kallima inachus, Junonia lemonias, Junonia iphita, and Stibochiona nicea) were collected in China (collection details are shown in Table S2) and identified based on morphological descriptions and illustrations [7,8,23]. Voucher specimens were deposited in the Entomological Museum of Northwest A&F University in Yangling, Shaanxi Province, China. Genomic DNA was extracted from the frozen legs of every single individual using EasyPure R Genomic DNA Kit (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. The DNA was stored at −20 °C until further use.
2.2. Nuclear DNA Amplification and Sequencing
Elongation factor 1 alpha (EF-1α), wingless (wgl), glyceraldehydes-3-phosphate dehydrogenase (GAPDH), and Ribosomal Protein S5 (RpS5) of five collected species were amplified via polymerase chain reaction (PCR) employing universal primers [16,24,25]. The cycling profile was: an initial denaturation for 7 min at 95 °C, followed by 40 cycles of 95 °C for 30 s, 50 °C for 30 s, 72 °C for 15 s, and a final extension of 72 °C for 10 min. Amplification products were detected by 1% (w/v) agarose gel electrophoresis and Sanger sequencing (Tsingke Biotechnology Co., Ltd., Xi’an, China). Primers and PCR recipes are listed in Tables S3 and S4. Sequencing chromatograms were checked using SeqMan 7.1.0 from DNAStar (Lasergene, GATC Biotech, Konstanz, Germany). Sequences were corrected with Geneious 8.1.3 (Biomatters, Auckland, New Zealand) or SeqMan 7.1.0. New sequences were submitted to GenBank (accession numbers: Table S1). Sequences of five nuclear genes for other taxa were downloaded from GenBank, and gene sequences of EF-1α, GAPDH, and RpS5 were missing in some species (Table S1).
2.3. Mitogenome Sequencing, Annotation and Analyses
DNA samples were randomly sheared with an ultrasonic crusher (Covaris, Woburn, MA, USA). Libraries were prepared by terminal repair, ligating adapters onto the 3′ end of the sheared fragments, purification, and PCR amplification. Libraries were sequenced on the Illumina HiseqTM Xten platform (Novogene Technologies, Beijing, China) for 150 bp paired-end reads. FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc, accessed on 5 September 2022) was used for read cleaning. Clean paired reads were used to reconstruct mitogenomes in Geneious 8.1.3 [26] software with default parameters. The mitogenome of Kallima inachus (GenBank accession number: NC_016196.1) was used as reference.
The mitogenome was annotated with the Geneious 8.1.3 software with default parameters. Protein-coding genes (PCGs) were annotated by alignment to homologous sequences of Kallima inachus. The MITOS server (http://mitos.bioinf.uni-leipzig.de/index.py, accessed on 10 November 2022) [27] was used to identify tRNA genes. tRNA secondary structures were drawn using MITOS predictions in Adobe Illustrator 2021. CGView server (https://cgview.ca/, accessed on 12 November 2022) [28] was used to visualize a circular mitogenomic map.
PhyloSuite 1.2.2 [29] was used to calculate base composition, the codon usage of PCGs, and relative RSCU (synonymous codon usage) values. Nucleotide compositional bias was calculated as AT-skew [(A − T)/(A + T)] and GC-skew [(G − C)/(G + C)] [30]. Sliding window analysis (window = 200 bp, step size = 20 bp) was used to determine nucleotide diversity (Pi value) of PCGs in DnaSP 6 [31]. The non-synonymous (Ka) and synonymous substitution rates (Ks) for each PCG were determined in DnaSP 6. Genetic distances for each PCG were calculated in MEGA X with the Kimura 2-parameter substitution model [32]. GraphPad Prism 8.0.1 (San Diego, CA, USA) was used to plot genetic distances and Ka/Ks ratios (ω). The newly sequenced mitogenome has been submitted to GenBank (accession number: OP748221).
2.4. Dataset Partitioning and Model Selection
Using codon alignment mode and the G-INS-I strategy, MAFFT 7.313 integrated into PhyloSuite 1.2.2 was used to align each PCG and nuclear DNA (nDNA). The MAFFT 7 online service (https://mafft.cbrc.jp/alignment/server/, accessed on 24 November 2022) [33] was used to align all RNAs using Q-INS-I. Poorly aligned regions were removed by Gblocks 0.91b [34] with default parameters. Xia’s [35] index of substitution saturation (Iss) in DAMBE 7 [36,37] was used to analyze substitution saturation.
To assess the effect of data partitioning and incorporation of RNAs and nDNA on phylogeny, seven datasets were used for phylogenetic inferences: the nuclear DNA dataset (nDNA), PCG123 dataset (13 PCGs), the PCG123R dataset (13 PCGs and two rRNAs), the PCG123RT dataset (13 PCGs, two rRNAs, and 22 tRNAs), the PCG123N dataset (13 PCGs and nDNA), the PCG123RN dataset (13 PCGs, two rRNAs, and nDNA), and the PCG123RTN dataset (13 PCGs, two rRNAs, 22 tRNAs, and nDNA). Datasets were partitioned in PartitionFinder 2.1.1 [38] and integrated into PhyloSuite 1.2.2, using the “greedy” search algorithm and Bayesian Information Criterion (BIC). Partitions and best-fit models for each dataset are listed in Table S5.
2.5. Phylogenetic Inference
Under an edge-linked partition model, Maximum Likelihood (ML) analysis was performed in IQ-TREE 2.2.0 [39]. A total of 5000 ultrafast bootstrap (UFB) replicates were used to assess bootstrap support (BS) [40]. MrBayes 3.2.6 [41] implemented in the CIPRES Science Gateway (www.phylo.org, accessed on 1 December 2022) was used for Bayesian Inference (BI). BI analysis of each dataset was carried out with four chains and run for 5–10 million generations. Trees were sampled every 1000 generations, with the first 25% discarded as burn-in. The convergence of the independent runs was indicated by a standard deviation of split frequencies below 0.01 and an estimated sample size (ESS) greater than 200. Convergence between runs was examined with Tracer 1.7 software [42].
3. Results and Discussion
3.1. Basic Characteristics of the Mitochondrial Genome
The complete mitogenome of the Doleschallia bisaltide is 16,389 bp in length, containing 37 genes and a control region (CR) (Figure 1). A total of 23 genes (9 PCGs and 14 tRNAs) are located on the majority strand (J-strand), while the minority strand (N-strand) encodes another 14 genes (4 PCGs, 2 rRNAs, and 8 tRNAs) (Table S6). As is typical for Lepidoptera [43], a significant AT bias is present in the mitogenome of Doleschallia bisaltide with a nucleotide composition of 79.2% A + T. The AT- and GC-skew are 0 and −0.22, respectively. Ten gene overlaps, ranging in length from 1 bp to 8 bp, and twelve intergenic spacers, ranging from 1 bp to 48 bp, were identified in the mitogenome (Table S6).
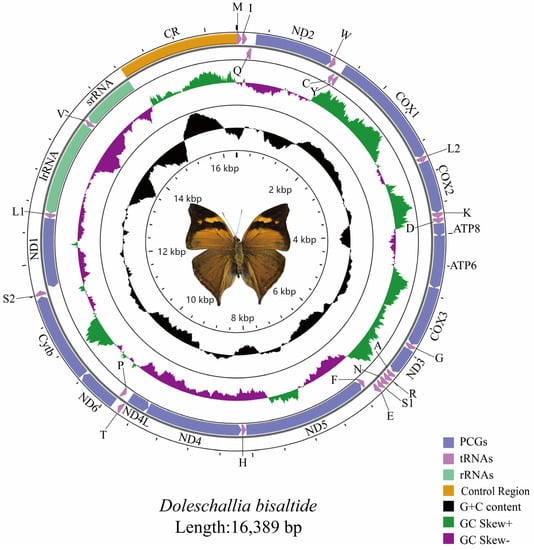
Figure 1.
The mitogenome of Doleschallia bisaltide.
3.2. Protein-coding Genes, tRNAs, rRNAs and Control Region
The length of the 13 PCGs was 11,203 bp. AT content of the PCGs was 78.7%. Except for COX1 (with CGA as start codon), all other PCGs initiate strictly with ATT or ATG (Table S6). It is common for Lepidoptera COX1 genes to use CGA as the start codon [44,45,46,47,48]. The stop codon of most PCGs was TAA; four PCGs (COX1, COX2, ND4, and ND5) terminated with an incomplete T residue. Incomplete termination codons of PCGs are converted into TAA by post-transcriptional polyadenylation [49]. RSCU of the PCGs (Figure S1) showed that the four most frequently utilized amino acids were Ile (AUU), Leu (UUA), Met (AUA), and Phe (UUU). Nucleotide diversity (Figure S2) showed that COX1 and ND1 were the most conserved (Pi = 0.103), while ATP8 was the most variable (Pi = 0.163). Congruent results were obtained from pairwise genetic distances (Figure S3). Ka/Ks (ω) ratios (Figure S3) were low (0 < ω < 1) for all 13 PCGs, suggesting that these genes experienced purifying selection. COX1 showed an extremely low evolutionary rate (ω = 0.049). By contrast, the ATP8 (ω = 0.499) shows a relatively fast evolutionary rate, as dose ND6 (ω = 0.237) and ND5 (ω = 0.233). Overall, the evolutionary rates of NADH dehydrogenase genes are faster than that of cytochrome oxidase genes.
Except for trnS1, which lacks the dihydrouridine (DHU) arm, all other tRNA genes had canonical cloverleaf structures (Figure S4). The absence of the DHU arm in trnS1 is common across metazoan mitogenomes [50,51]. The anticodon and amino acid acceptor arms are highly conserved, while the pseudouridine (TΨC) and DHU arms are variable. lrRNA (16S rRNA) is 1326 bp in length and is located between trnL1 and trnV. srRNA (12S rRNA) is 775 bp in length and is located between trnV and the control region. AT content of the rRNAs is 84.4%.
The control region (1542 bp) is located between srRNA and trnM. As in other lepidopteran mitogenomes [48,52], the control region of the Doleschallia bisaltide mitogenome contains the motif ATAGA between the 5′-end of the 12S rRNA and poly-T stretch, which is proposed to be the replication origin for the minority strand.
3.3. Phylogenetic Analysis
Based on seven datasets, phylogenetic trees were generated by BI and ML (Figure 2, Figure 3 and Figure 4). Nymphalinae was not a monophyletic group in any dataset. Baeotus (Coeini) clusters within the outgroup taxa (Biblidinae, Cyrestinae, and Pseudergolinae). These results are in accordance with previous studies [19,53]. In all analyses, Nymphalini, Melitaeini, and Junoniini form stable monophyletic groups with strong support. Our results show that Kallimini is not monophyletic; Doleschallia bisaltide clusters with species of the Melitaeini, consistent with Zhang et al. [15] and Su et al. [19], but contra the results of Wahlberg et al. [1].
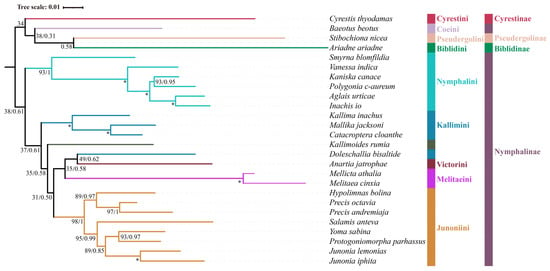
Figure 2.
Phylogenetic analysis using the ML and BI methods based on the nDNA dataset. Bootstrap support (BS) values and posterior probability (BPP) are indicated at nodes (from left to right). Maximum support values produced are indicated by star symbols.

Figure 3.
Phylogenetic analysis using the ML method based on the datasets of PCG123, PCG123R, PCG123RT, PCG123N, PCG123RN, and PCG123RTN. Bootstrap support (BS) values at nodes (from left to right) represent support for PCG123, PCG123R, PCG123RT, PCG123N, PCG123RN, and PCG123RTN datasets, respectively. The “-” indicates the clades are different. Maximum support values produced are indicated by star symbols.

Figure 4.
Phylogenetic analysis using the BI method based on the datasets of PCG123, PCG123R, PCG123RT, PCG123N, PCG123RN, and PCG123RTN. Posterior probability (BPP) values at nodes (from left to right) represent support for PCG123, PCG123R, PCG123RT, PCG123N, PCG123RN, and PCG123RTN datasets, respectively. The “-” indicates the clades are different. Maximum support values produced are indicated by star symbols.
The monophyly of Nymphalini was supported in six datasets (PCG123, PCG123R, PCG123RT, PCG123N, PCG123RN, PCG123RTN) with strong support (BS = 100, PP = 1.0), but only moderate support in nDNA datasets (BS = 93). Nymphalini was sister to the remaining Nymphalinae (except Coeini) in all phylogenetic analyses, with low support (BS = 38, PP = 0.61) using nDNA dataset and high support (BS = 100, PP = 1.0) using mitochondrial or combined datasets. These results are consistent with previous publications [1,19,53,54,55]. Smyrna blomfildia was a sister to the rest of Nymphalini with moderate to strong support (BS = 93, PP = 1.0; BS = 100, PP = 1.0).
Victorinini contains four South American genera Siproeta, Anartia, Metamorpha, and Napeocles. Only Anartia was included in this study. The nDNA dataset found Anartia and Doleschallia to be sisters but with low branch support (BS = 49, PP = 0.62). The PCG123, PCG123R, PCG123N, and PCG123RN datasets without tRNA genes, found Anartia to be sister to the clade (Kallimini + (Melitaeini + Junoniini) across ML analyses. However, Anartia was a sister to Kallimoides in BI analyses with lower support (PP = 0.56, 0.66, 0.59, 0.64) in these same datasets.
Due to superficial similarity, Doleschallia was previously placed in Kallimini. Currently, there are four genera (Doleschallia, Kallima, Catacroptera, and Mallika) in Kallimini. Both ML and BI analyses of six datasets (PCG123, PCG123R, PCG123RT, PCG123N, PCG123RN, and PCG123RTN) found Doleschallia to be separated from the Kallimini and instead clusters with the Melitaeini. Genomic data found Doleschallia to be sister to Melitaeini, which is consistent with our results; thus, Doleschallia was assigned the status of a tribe [15]. Our analyses based on the nDNA dataset found that the Melitaeini was sister to Doleschallia + Anartia. Analyses of all datasets reveal that the remaining members of the Kallimini (Catacroptera, Kallima, and Mallika) always form a monophyletic group with strong support (BS = 100, PP = 1.0).
The monophyly of Melitaeini was confirmed. This tribe has five major clades, the Gnathotriche-group, the Melitaea-group (Melitaeina), the Chlosyne-group, the Euphydryas-group (Euphydryina), and the Phyciodes-group (Phyciodina) [1]. Mellicta has been synonymized with Melitaea [56]; however, Lang [13] recognized both genera, Melitaea and Mellicta, the former with a tegumen as a narrow transverse band and the latter with a small canopy tegumen. In this study, only two Melitaeini genera (Melitaea and Mellicta) were included and were monophyletic in all trees with strong support (BS = 100, PP = 1.0).
The Junoniini currently comprises six genera (Yoma, Hypolimnas, Junonia, Precis, Salamis, and Protogoniomorpha) [1]. In this study, all six genera were monophyletic with high support (BS = 98–100, PP = 1.0). Junonia and Precis, two genera that have been used interchangeably for several hundred years, are not each a sister group; rather, Precis is sister to Hypolimnas. In this study, both ML and BI analyses also confirm that Precis is a sister to Hypolimnas based on all datasets. Strong branch support (BS = 100, PP = 1.0) was obtained from PCG123, PCG123R, PCG123RT, PCG123N, PCG123RN, and PCG123RTN, while branch support in the nDNA dataset was moderate (BS = 89, PP = 0.97), in line with previous research [1].
Based on a more taxon-rich analysis of Nymphalinae using three genes (COXI, EF-1α, and wingless), Su et al. [19] proposed Kallimoides to belong to the Victorinini. However, among the analyses in this study, the position of Kallimoides was not stable and showed weak support. These results are consistent with previous publications [1,54]. However, Zhang et al. [15] proposed Kallimoides to represent a monotypic tribe due to only moderate support for its association with Victorinini.
4. Conclusions
In this study, we sequenced the mitochondrial genome of Doleschallia bisaltide. It has the same gene orientation and arrangement as other mitogenomes from Lepidoptera. ATP8, ND6, and ND5 could be potential DNA markers for species delimitation due to their higher variance. By comparing trees constructed from different datasets, we found that trees constructed from the nDNA dataset had lower nodal support than other datasets, suggesting that the mitochondrial genome provides more phylogenetic resolution compared to a single nuclear gene or multiple genes [57,58]. Phylogenetic analysis of the PCG123N dataset (13 PCGs and nDNA) had the highest support. Our results indicate that the addition of RNA genes did not improve the phylogenetic signal, as nodal support decreased. In a future study, increasing the taxa sampling is necessary to test the monophyly of tribal groups in the Nymphalinae.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/d15040558/s1. Figure S1: Relative synonymous codon usage (RSCU) of the mitogenome of Doleschallia bisaltide; Figure S2: Nucleotide diversity on 13 PCGs in Nymphalinae; Figure S3: Genetic distances and non-synonymous (Ka) to synonymous (Ks) substitution rates of 13 protein-coding among Nymphalinae; Figure S4: Predicted secondary cloverleaf structure for the tRNAs of Doleschallia bisaltide; Table S1: List of taxa used for the phylogenetic analyses in this study; Table S2: Collection information of five species; Table S3: The primer information; Table S4: The PCR reaction system; Table S5: Best partitioning scheme and nucleotide substitution models for different datasets selected by PartitionFinder; Table S6: Mitogenomic organization of Doleschallia bisaltide.
Author Contributions
Conceptualization, N.L., L.F. and Y.Z.; specimen collection and identification, N.L., H.W., L.F. and Y.Z.; methodology and experiments, N.L., H.W. and Y.Z.; Data analysis, N.L.; writing—original draft preparation, N.L.; writing—review and editing, N.L., L.F. and Y.Z.; funding acquisition, L.F. and Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32170469, 31750002), the National Key Research and Development Program of the Ministry of Science and Technology, China (2022YFE0115200), Biodiversity Survey and Assessment, Project of the Ministry of Ecology and Environment, China (2019HJ2096001006), National Animal Collection Resource Center, China, and the Natural Science Foundation of Shaanxi Province (2019JQ-195).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We are grateful to John Richard Schrock (Emporia State University, USA) for revising the manuscript. We sincerely thank Long Liu and Deliang Xu for the revisions of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wahlberg, N.; Brower, A.V.Z.; Nylin, S. Phylogenetic relationships and historical biogeography of tribes and genera in the subfamily Nymphalinae (Lepidoptera: Nymphalidae). Biol. J. Linn. Soc. 2005, 86, 227–251. [Google Scholar] [CrossRef]
- Boggs, C.L.; Watt, W.B.; Ehrlich, P.R. Butterflies—Ecology and evolution—Taking flight. Science 2004, 303, 174. [Google Scholar]
- Roe, A.D.; Weller, S.J.; Baixeras, J.; Brown, J.; Cummings, M.P.; Davis, D.R.; Kawahara, A.Y.; Parr, C.S.; Regier, J.C.; Rubinoff, D.; et al. Evolutionary framework for Lepidoptera model systems. In Molecular Biology and Genetics of the Lepidoptera, 1st ed.; Goldsmith, M.R., Marec, F., Eds.; CRC Press: Boca Raton, FL, USA, 2009; pp. 1–24. [Google Scholar]
- Espeland, M.; Breinholt, J.; Willmott, K.R.; Warren, A.D.; Vila, R.; Toussaint, E.F.A.; Maunsell, S.C.; Aduse-Poku, K.; Talavera, G.; Eastwood, R.; et al. A comprehensive and dated phylogenomic analysis of butterflies. Curr. Biol. 2018, 28, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.T.; Teng, D.Q.; Li, X.Y.; Yang, P.W.; Da, W.; Zhang, Y.M.; Zhang, Y.B.; Liu, G.C.; Zhang, X.S.; Wan, W.T.; et al. The evolution and diversification of oakleaf butterflies. Cell 2022, 185, 3138–3152. [Google Scholar] [CrossRef] [PubMed]
- Ackery, P.R. Hostplants and classification: A review of nymphalid butterflies. Biol. J. Linn. Soc. 1988, 33, 95–203. [Google Scholar] [CrossRef]
- Chou, I. Classification and Identification of Chinese Butterflies; Henan Scientific and Technological Publishing House: Zhengzhou, China, 1998. [Google Scholar]
- Chou, I. Monograph of Chinese Butterflies, Second Volume; Henan Scientific and Technological Publishing House: Zhengzhou, China, 1994. [Google Scholar]
- Harvey, D.J. Higher classification of the Nymphalidae, Appendix B. In The Development and Evolution of Butterfly Wing Patterns; Nijhout, H.F., Ed.; Smithsonian Institution Press: Washington, DC, USA, 1991; pp. 255–273. [Google Scholar]
- Ackery, P.R.; de Jong, R.; Vane-Wright, R.I. The Butterflies: Hedyloidea, Hesperioidea and Papilionoidea. In Lepidoptera, Moths and Butterflies; Volume Evolution, Systematics and Biogeography; Kristensen, N.P., Ed.; De Gruyter: Berlin, Germany, 1999; pp. 263–300. [Google Scholar]
- Brower, A.V.Z. Phylogenetic relationships among the Nymphalidae (Lepidoptera) inferred from partial sequences of the wingless gene. Proc. Roy. Soc. B Biol. Sci. 2000, 267, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Wahlberg, N.; Weingartner, E.; Nylin, S. Towards a better understanding of the higher systematics of Nymphalidae (Lepidoptera: Papilionoidea). Mol. Phylogenet. Evol. 2003, 28, 473–484. [Google Scholar] [CrossRef]
- Lang, S.Y. The Nymphalidae of China (Lepidoptera, Rhopalocera) Part I; Tshikolovets Publications: Pardubice, Czech Republic, 2012. [Google Scholar]
- Freitas, A.V.L.; Brown, K.S.J. Phylogeny of the Nymphalidae (Lepidoptera). Syst. Biol. 2004, 53, 363–383. [Google Scholar] [CrossRef]
- Zhang, J.; Cong, Q.; Shen, J.; Opler, P.A.; Grishin, N.V. Genomics-guided refinement of butterfly taxonomy. Taxon. Rep. Int. Lepid. Surv. 2021, 9, 1–112. [Google Scholar]
- Wahlberg, N.; Wheat, C.W. Genomic outposts serve the phylogenomic pioneers: Designing novel nuclear markers for genomic DNA extractions of Lepidoptera. Syst. Biol. 2008, 57, 231–242. [Google Scholar] [CrossRef]
- Wahlberg, N.; Leneveu, J.; Kodandaramaiah, U.; Pena, C.; Nylin, S.; Freitas, A.V.L.; Brower, A.V.Z. Nymphalid butterflies diversify following near demise at the Cretaceous/Tertiary boundary. P. Roy. Soc. B Biol. Sci. 2009, 276, 4295–4302. [Google Scholar] [CrossRef]
- Wu, L.W.; Lin, L.H.; Lees, D.C.; Hsu, Y.F. Mitogenomic sequences effectively recover relationships within brush-footed butterflies (Lepidoptera: Nymphalidae). BMC Genomics 2014, 15, 468. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Shi, Q.; Sun, X.; Ma, J.; Li, C.; Hao, J.; Yang, Q. Dated phylogeny and dispersal history of the butterfly subfamily Nymphalinae (Lepidoptera: Nymphalidae). Sci. Rep. 2017, 7, 8799. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, I.; Tanikawa-Dodo, Y.; Saigusa, T.; Nishiyama, T.; Kitani, M.; Hasebe, M.; Mohri, H. Phylogeny, biogeography, and host-plant association in the subfamily Apaturinae (Insecta: Lepidoptera: Nymphalidae) inferred from eight nuclear and seven mitochondrial genes. Mol. Phylogenet. Evol. 2010, 57, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Peña, C.; Wahlberg, N.; Weingartner, E.; Kodandaramaiah, U.; Nylin, S.; Freitas, A.V.; Brower, A.V. Higher level phylogeny of Satyrinae butterflies (Lepidoptera: Nymphalidae) based on DNA sequence data. Mol. Phylogenet. Evol. 2006, 40, 29–49. [Google Scholar] [CrossRef]
- Wu, L.-W.; Chiba, H.; Lees, D.C.; Ohshima, Y.; Jeng, M.-L. Unravelling relationships among the shared stripes of sailors: Mitogenomic phylogeny of Limenitidini butterflies (Lepidoptera, Nymphalidae, Limenitidinae), focusing on the genera Athyma and Limenitis. Mol. Phylogenet. Evol. 2019, 130, 60–66. [Google Scholar] [CrossRef]
- Wu, C.S.; Hsu, Y.F. Butterflies of China; The Straits Publishing House: Fuzhou, China, 2017. [Google Scholar]
- Brower, A.V.Z.; DeSalle, R. Patterns of mitochondrial versus nuclear DNA sequence divergence among nymphalid butterflies: The utility of wingless as a source of characters for phylogenetic inference. Insect Mol. Biol. 1998, 7, 73–82. [Google Scholar] [CrossRef]
- Cho, S.W.; Mitchell, A.; Regier, J.C.; Mitter, C.; Poole, R.W.; Friedlander, T.P.; Zhao, S.W. A Highly Conserved Nuclear Gene for Low-Level Phylogenetics—Elongation Factor-1-Alpha Recovers Morphology-Based Tree for Heliothine Moths. Mol. Biol. Evol. 1995, 12, 650–656. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Juhling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Putz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.L.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of Nucleotide Composition at Fourfold Degenerate Sites of Animal Mitochondrial Genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Xia, X.H.; Xie, Z.; Salemi, M.; Chen, L.; Wang, Y. An index of substitution saturation and its application. Mol. Phylogenet. Evol. 2003, 26, 1–7. [Google Scholar] [CrossRef]
- Xia, X.H. DAMBE5: A comprehensive software package for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2013, 30, 1720–1728. [Google Scholar] [CrossRef]
- Xia, X.H. DAMBE6: New tools for microbial genomics, phylogenetics and molecular evolution. J. Hered. 2017, 108, 431–437. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Z.M.; Gu, X.S.; Wang, M.; Huang, G.H.; Zwick, A. Phylogenetic relationships among Bombycidae s.l. (Lepidoptera) based on analyses of complete mitochondrial genomes. Syst. Entomol. 2019, 44, 490–498. [Google Scholar] [CrossRef]
- Liu, N.; Li, N.; Yang, P.; Sun, C.; Fang, J.; Wang, S. The complete mitochondrial genome of Damora sagana and phylogenetic analyses of the family Nymphalidae. Genes Genom. 2018, 40, 109–122. [Google Scholar] [CrossRef]
- Tian, L.; Sun, X.-Y.; Chen, M.; Gai, Y.-H.; Hao, J.-S.; Yang, Q. Complete mitochondrial genome of the Five-dot Sergeant Parathyma sulpitia (Nymphalidae: Limenitidinae) and its phylogenetic implications. Zool. Res. 2012, 33, 133–143. [Google Scholar] [CrossRef]
- Wang, X.C.; Sun, X.Y.; Sun, Q.Q.; Zhang, D.X.; Hu, J.; Yang, Q.; Hao, J.S. Complete mitochondrial genome of the laced fritillary Argyreus hyperbius (Lepidoptera: Nymphalidae). Zool. Res. 2011, 32, 465–475. [Google Scholar]
- Kim, M.I.; Baek, J.Y.; Kim, M.J.; Jeong, H.C.; Kim, K.G.; Bae, C.H.; Han, Y.S.; Jin, B.R.; Kim, I. Complete nucleotide sequence and organization of the mitogenome of the red-spotted apollo butterfly, Parnassius bremeri (Lepidoptera: Papilionidae) and comparison with other lepidopteran insects. Mol. Cells 2009, 28, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.S.; Song, L.; Zhou, L.; Shi, Y.X.; Song, N.; Zhang, Y.L. Mitochondrial genomes of four satyrine butterflies and phylogenetic relationships of the family Nymphalidae (Lepidoptera: Papilionoidea). Int. J. Biol. Macromol. 2020, 145, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Garey, J.R.; Wolstenholme, D.R. Platyhelminth mitochondrial DNA: Evidence for early evolutionary origin of a tRNA (serAGN) that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J. Mol. Evol. 1989, 28, 374–387. [Google Scholar] [CrossRef]
- Chen, L.; Wahlberg, N.; Liao, C.Q.; Wang, C.B.; Ma, F.Z.; Huang, G.H. Fourteen complete mitochondrial genomes of butterflies from the genus Lethe (Lepidoptera, Nymphalidae, Satyrinae) with mitogenome-based phylogenetic analysis. Genomics 2020, 112, 4435–4441. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.H. Molecular phylogeny and historical biogeography of the main groups of nymphalid butterflies (Lepidoptera: Papilionoidea: Nymphalidae). Ph.D. Thesis, Anhui Normal University, Wuhu, China, 2015. [Google Scholar]
- Wahlberg, N. That awkward age for butterflies: Insights from the age of the butterfly subfamily Nymphalinae (Lepidoptera: Nymphalidae). Syst. Biol. 2006, 55, 703–714. [Google Scholar] [CrossRef]
- Nylin, S.; Wahlberg, N. Does plasticity drive speciation? Host-plant shifts and diversification in nymphaline butterflies (Lepidoptera: Nymphalidae) during the tertiary. Biol. J. Linn. Soc. 2008, 94, 115–130. [Google Scholar] [CrossRef]
- Leneveu, J.; Chichvarkhin, A.; Wahlberg, N. Varying rates of diversification in the genus Melitaea (Lepidoptera: Nymphalidae) during the past 20 million years. Biol. J. Linn. Soc. 2009, 97, 346–361. [Google Scholar] [CrossRef]
- Qin, J.; Zhang, Y.Z.; Zhou, X.; Kong, X.B.; Wei, S.J.; Ward, R.D.; Zhang, A.B. Mitochondrial phylogenomics and genetic relationships of closely related pine moth (Lasiocampidae: Dendrolimus) species in China, using whole mitochondrial genomes. BMC Genomics 2015, 16, 428. [Google Scholar] [CrossRef]
- Cheng, Y.C.; Chen, M.Y.; Wang, J.F.; Liang, A.P.; Lin, C.P. Some mitochondrial genes perform better for damselfly phylogenetics: Species- and population-level analyses of four complete mitogenomes of Euphaea sibling species. Syst. Entomol. 2018, 43, 702–715. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).