Stenomitos nagquensis sp. nov. (Leptolyngbyaceae, Cyanobacteria) from a Meadow Wetland in the Tibet Plateau: A Novel Species Studied Based on a Polyphasic Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Site
2.2. Culturing and Morphological Description
2.3. DNA Extraction, PCR Amplification, and Gene Sequencing
2.4. Phylogenetic Analyses
2.5. Secondary Structure Analyses of the 16S–23S Internal Transcribed Spacer (ITS) Region
2.6. Calculation of p-Distance for the 16S rRNA Gene and 16S–23S ITS Region; Nomenclature
3. Results
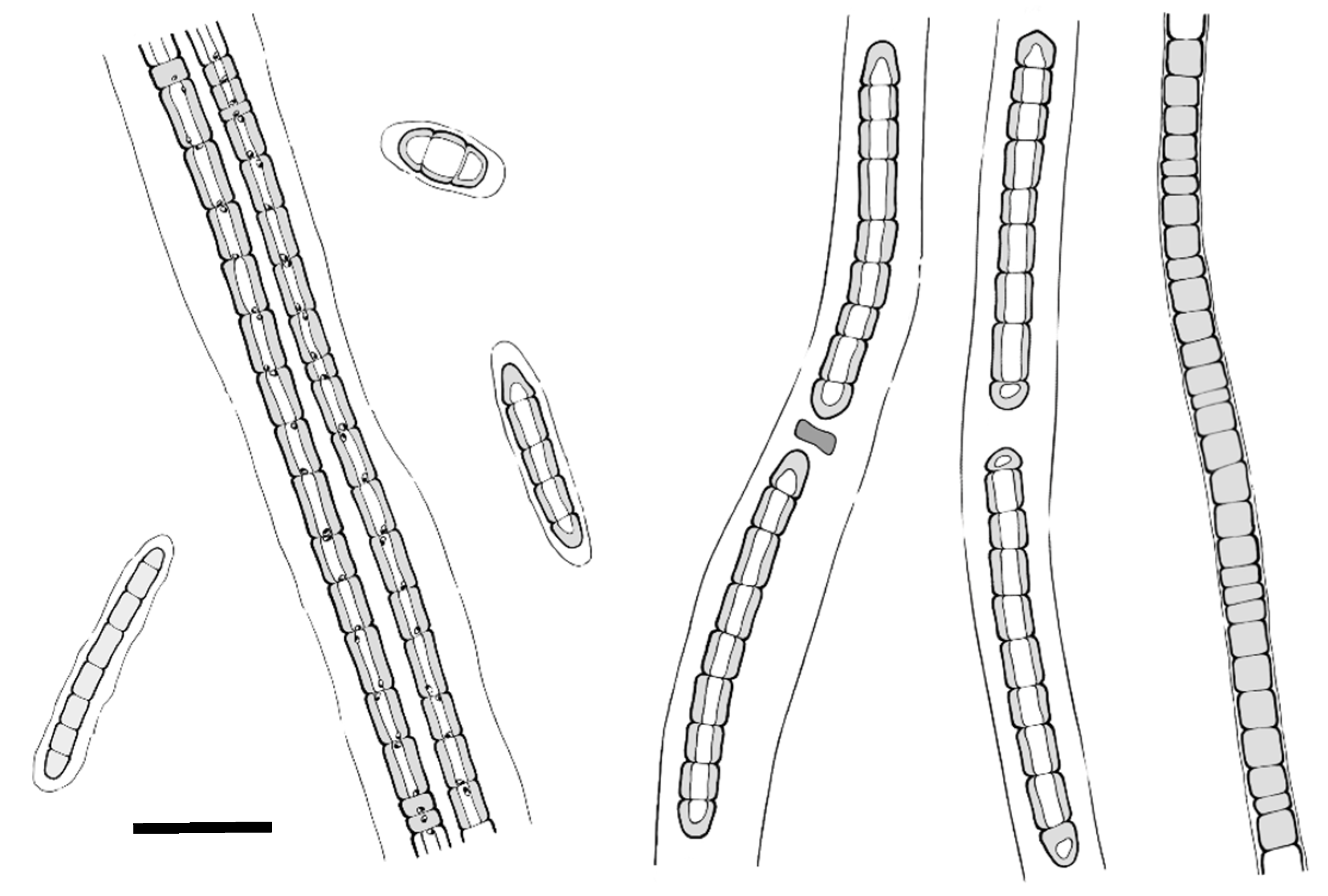
3.1. Taxonomic Treatment
3.2. 16S rRNA Gene Sequence Similarity and Phylogenetic Analyses
3.3. ITS p-Distance Dissimilarity and Secondary Structures of 16S–23S ITS
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Komárek, J. Cyanobacterial taxonomy current problems and prospects for the integration of traditional and molecular approaches. Algae 2006, 2, 249–375. [Google Scholar] [CrossRef]
- Taton, A.; Grubisic, S.; Ertz, D.; Hodgson, D.A.; Piccardi, R.; Biondi, N.; Tredici, M.R.; Mainini, M.; Losi, D.; Marinelli, F.; et al. Polyphasic study of Antarctic Cyanobacterial Strains. J. Phycol. 2006, 42, 1257–1270. [Google Scholar] [CrossRef]
- Johansen, J.R.; Casamatta, D.A. Recognizing cyanobacterial diversity through adoption of a new species paradigm. Algol. Stud. 2005, 117, 71–93. [Google Scholar] [CrossRef]
- Komárek, J. Recent changes (2008) in cyanobacteria taxonomy based on a combination of molecular background with phenotype and ecological consequences (genus and species concept). Hydrobiologia 2010, 1, 245–259. [Google Scholar] [CrossRef]
- Johansen, J.R.; Bohunická, M.; Lukešová, A.; Hrčková, K.; Vaccarino, M.A.; Chesarino, N.M. Morphological and molecular characterization within 26 strains of the genus Cylindrospermum (Nostocaceae, Cyanobacteria), with descriptions of three new species. J. Phycol. 2014, 50, 187–202. [Google Scholar] [CrossRef]
- Davydov, D.; Vilnet, A. Review of the Cyanobacterial Genus Phormidesmis (Leptolyngbyaceae) with the Description of Apatinema gen. nov. Diversity 2022, 14, 731. [Google Scholar] [CrossRef]
- Wilmotte, A.; Herdman, M. Phylogenetic relationships among cyanobacteria based on 16S rRNA sequences. In Bergey’s Manual of Systematic Bacteriology; Boone, D.R., Castenholz, R.W., Eds.; Springer: New York, NY, USA, 2001; pp. 487–493. [Google Scholar]
- Robertson, B.R.; Tezuka, N.; Watanabe, M. Phylogenetic analyses of Synechococcus strains (cyanobacteria) using sequences of 16S rDNA and part of the phycocyanin operon reveal multiple evolutionary lines and reflect phycobilin content. Int. J. Syst. Evol. Microbiol. 2001, 51, 861–871. [Google Scholar] [CrossRef]
- Komárek, J.; Kastovsky, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Sciuto, K.; Moro, I. Detection of the new cosmopolitan genus Thermoleptolyngbya (Cyanobacteria, Leptolyngbyaceae) using the 16S rRNA gene and 16S–23S ITS region. Mol. Phylogenetics Evol. 2016, 105, 15–35. [Google Scholar] [CrossRef]
- Mai, T.; Johansen, J.R.; Pietrasiak, N.B.M.; Martin, M.P. Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and Trichocoleaceae fam. nov. and six new genera containing 14 species. Phytotaxa 2018, 365, 1–59. [Google Scholar] [CrossRef]
- Caires, T.A.; Lyra, G.D.; Hentschke, G.S.; da Silva, A.M.S.; de Araujo, V.L.; Sant’Anna, C.L.; Nunes, J.M.D. Polyphasic delimitation of a filamentous marine genus, Capillus gen. nov. (Cyanobacteria, Oscillatoriaceae) with the description of two Brazilian species. Algae 2018, 33, 291–304. [Google Scholar] [CrossRef]
- Maltseva, S.; Kezlya, E.; Krivova, Z.; Gusev, E.; Kulikovskiy, M.; Maltsev, Y. Phylogeny and fatty acid profiles of Aliinostoc vietnamicum sp. nov. (Cyanobacteria) from soils of Vietnam. J. Phycol. 2022, 58, 789–803. [Google Scholar] [CrossRef] [PubMed]
- Strunecký, O.; Ivanovaš, A.P.; Mareš, J. An updated classification of cyanobacterial orders and families based on phylogenomic and polyphasic analysis. J. Phycol. 2022, 59, 12–51. [Google Scholar] [CrossRef] [PubMed]
- Casamatta, D.A.; Johansen, J.R.; Vis, M.L.; Broadwater, S.T. Molecular and morphological characterization of ten polar and near-polar strains within the Oscillatoriales (Cyanobacteria). J. Phycol. 2005, 41, 421–438. [Google Scholar] [CrossRef]
- Komárek, J.; Anagnostidis, K. Cyanoprokaryota II. In Süsswasserflora von Mittleuropa 19/2; Büdel, B., Krienitz, L., Gärtner, G., Schagerl, M., Eds.; Elsevier/Spektrum: München, Germany, 2005; p. 759. [Google Scholar]
- Johansen, J.R.; Olsen, C.E.; Lowe, R.L.; Fučíková, K.; Casamatta, D.A. Leptolyngbya species from selected seep walls in the Great Smoky Mountains National Park. Algol. Stud. 2008, 126, 21–36. [Google Scholar] [CrossRef]
- Perkerson, R.B.; Johansen, J.R.; Kovacik, L.; Brand, J.; Kastovsky, J.; Casamatta, D.A. A unique pseudanabaenalean (Cyanobacteria) genus Nodosilinea gen. nov. based on morphological and molecular data. J. Phycol. 2011, 47, 1397–1412. [Google Scholar] [CrossRef]
- Dadheech, P.W.; Mahmoud, H.; Kotut, K.; Krienitz, L. Haloleptolyngbya alcalis gen.et sp. nov., a new filamentous cyanobacterium from the soda lake Nakuru, Kenya. Hydrobiologia 2011, 637, 269–283. [Google Scholar]
- Vaz, M.G.M.V.; Genuário, D.B.; Andreote, A.P.D.; Malone, C.F.S.; Sant´Anna, C.L.; Barbiero, L.; Fiore, M.F. Pantanalinema gen. nov. and Alkalinema gen. nov.: Novel pseudanabaenacean genera (Cyanobacteria) isolated from saline-alkaline lakes. Int. J. Syst. Evol. Microbiol. 2015, 65, 298–308. [Google Scholar] [CrossRef]
- Miscoe, L.H.; Johansen, J.R.; Vaccarino, M.A.; Pietrasiak, N.; Sherwood, A.R. The diatom flora and cyobacteria from caves on Kauai, Hawaii. II. Novel cyanobacteria from caves on Kauai, Hawaii. Bibl. Phycol. 2016, 123, 75–152. [Google Scholar]
- Dvořák, P.; Hašler, P.; Pitelková, P.; Tabáková, P.; Casamatta, D.A.; Poulíčková, A. A new cyanobacterium from the Everglades, Florida—Chamaethrix gen. nov. Fottea 2017, 17, 269–276. [Google Scholar] [CrossRef]
- Becerra-Absalón, I.; Johansen, J.R.; Muñoz-Martín, M.A.; Gustavo, M. Chroakolemma gen. nov. (Leptolyngbyaceae, Cyanobacteria) from soil biocrusts in the semi-desert Central Region of Mexico. Phytotaxa 2018, 367, 201–218. [Google Scholar] [CrossRef]
- Zammit, G. Systematics and biogeography of sciophilous cyanobacteria; an ecological and molecular description of Albertania skiophila (Leptolyngbyaceae) gen. & sp. nov. Phycologia 2018, 57, 481–491. [Google Scholar]
- Chakraborty, S.; Maruthanayagam, V.; Achari, A.; Mahansaria, R.; Pramanik, A.; Jaisankar, P.; Mukherjee, J. Euryhalinema mangrovii gen. nov., sp. nov., and Leptoelongatus litoralis gen. nov., sp. nov. (Leptolyngbyaceae) isolated from an Indian mangrove forest. Phytotaxa 2019, 422, 58–74. [Google Scholar] [CrossRef]
- Radzi, R.; Muangmai, N.; Broady, P.; Wan, M.; Merican, F. Nodosilinea signiensis sp. nov. (Leptolyngbyaceae, Synechococcales), a new terrestrial cyanobacterium isolated from mats collected on signy island, south orkney islands, antarctica. PLoS ONE 2019, 14, e0224395. [Google Scholar] [CrossRef]
- Song, G.; Jiang, Y.; Li, R. Scytolyngbya timoleontis gen et sp. nov. (Leptolyngbyaceae, Cyanobacteria): A novel false branching Cyanobacteria from China. Phytotaxa 2015, 224, 72–84. [Google Scholar] [CrossRef]
- Geng, R.; Wang, Y.; Cai, F.; Zhang, Y.; Yang, P.; Dai, G.; Li, R.; Yu, G. Neochroococcus gongqingensis gen. et sp. nov., a new member of coccoid cyanobacteria from a watercourse, Eastern China. Fottea 2021, 21, 44–52. [Google Scholar] [CrossRef]
- Geng, R.; Li, W.; Chao, A.; Guo, X.; Li, H.; Yu, G.; Li, R. Establishment of a New Filamentous Cyanobacterial Genus, Microcoleusiopsis gen. nov. (Microcoleaceae, Cyanobacteria), from Benthic Mats in Open Channel, Jiangxi Province, China. Diversity 2021, 13, 548. [Google Scholar] [CrossRef]
- Geng, R.Z.; Wen, Q.Z.; Wang, Y.L.; Yang, P.; Dai, G.F.; Li, R.H.; Yu, G.L. A novel filamentous cyanobacterium Microseira minor sp. nov. (Oscillatoriaceae, Cyanobacteria) from the Ganfu Channel, Jiangxi, China. Phytotaxa 2021, 524, 178–190. [Google Scholar] [CrossRef]
- Wu, Q.L.; Xing, P.; Liu, W.-T. East Tibetan lakes harbour novel clusters of picocyanobacteria as inferred from the 16S–23S rRNA internal transcribed spacer sequences. Microb. Ecol. 2010, 59, 614–622. [Google Scholar] [CrossRef]
- Cai, F.; Wang, Y.; Yu, G.; Wang, J.; Pen, X.; Li, R. Proposal of Purpurea gen. nov. (Nostocales, Cyanobacteria), a novel cyanobacterial genus from wet soil samples in Tibet, China. Fottea 2020, 20, 86–97. [Google Scholar] [CrossRef]
- Cai, F.; Li, R. Purpureonostoc, a new name for a recently described genus of Nostoc-like cyanobacteria. Fottea 2020, 20, 111. [Google Scholar] [CrossRef]
- Pecundo, M.H.; Cai, F.; Chang, A.C.G.; Ren, H.; Li, N.; Li, R.; Chen, T. Polyphasic approach identifies two new species of Desmonostoc (Nostocales, Cyanobacteria) in the coralloid roots of Cycas fairylakea (Cycadales). Phycologia 2021, 60, 653–668. [Google Scholar] [CrossRef]
- Maltseva, S.; Bachura, Y.; Erst, T.; Kulikovskiy, M.; Maltsev, Y. Description of Desmonostoc caucasicum sp. nov. (Cyanobacteria) using an integrative taxonomic approach. Phycologia 2022, 61, 514–527. [Google Scholar] [CrossRef]
- Iteman, I.; Rippka, R.; Tandeau De Marsac, N.; Herdman, M. Comparison of conserved structural and regulatory domains within divergent 16S rRNA–23S rRNA spacer sequences of cyanobacteria. Microbiology 2000, 146, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Fatma, A.; Mishra, A.K. Molecular phylogeny and evogenomics of heterocystous cyanobacteria using rbcl gene sequence data. Ann. Microbiol. 2015, 65, 799–807. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQTREE: A fast-online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, 232–235. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES science gateway: A community resource for phylogenetic analyses. In Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery, Salt Lake City, UT, USA, 18-21 July 2011; pp. 1–8. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Mathews Lab. RNAstructure, 1996–2019, Version 6.2. Available online: http://rna.urmc.rochester.edu/RNAstructure.html (accessed on 10 June 2021).
- Shalygin, S.; Shalygina, R.R.; Redkina, V.V.; Gargas, C.B.; Johansen, J.R. Description of Stenomitos kolaensis and S. hiloensis sp. nov. (Leptolyngbyaceae, Cyanobacteria) with an emendation of the genus. Phytotaxa 2020, 440, 108–128. [Google Scholar] [CrossRef]
- Panou, M.; Gkelis, S. Unravelling unknown cyanobacteria diversity linked with HCN production. Mol. Phylogenetics Evol. 2022, 166, 107322. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, F.E. Freshwater algae. In National Antarctic Expedition 1901–1904 Natural History Zoology and Botany [Part 3]; Anon, Ed.; British Museum: London, UK, 1912; Volume VI, pp. 1–66. [Google Scholar]
- Stackebrandt, E.; Ebers, J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 2006, 33, 152–155. [Google Scholar]
- Staley, J.T. The bacterial species dilemma and the genomic–phylogenetic species concept. Philos. Trans. R. Soc. 2006, 361, 1899–1909. [Google Scholar] [CrossRef]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.D.; Kandler, O.; Krichevsky, M.I.; Starr, M.P. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Evol. Microbiol. 1987, 37, 463–464. [Google Scholar] [CrossRef]
- Pietrasiak, N.; Mühlsteinová, R.; Siegesmund, M.A.; Johansen, J.R. Phylogenetic placement of Symplocastrum (Phormidiaceae, Cyanophyceae) with a new combination S. californicum and two new species: S. fletchnerae and S. torsivum. Phycologia 2014, 53, 529–541. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Shalygin, S.; Huang, I.; Allen, E.H.; Burkholder, J.M.; Zimba, P.V. Odorella benthonica gen. & sp. nov. (Pleurocapsales, Cyanobacteria): An odor and prolific toxin producer isolated from a California aqueduct. J. Phycol. 2019, 55, 509–520. [Google Scholar]
- Jung, P.; Mikhailyuk, T.; Emrich, D.; Baumann, K.; Dultz, S.; Büdel, B. Shifting boundaries: Ecological and geographical range extension based on three new species in the cyanobacterial genera Cyanocohniella, Oculatella, and Aliterella. J. Phycol. 2020, 56, 1216–1231. [Google Scholar] [CrossRef]
- Rasouli-Dogaheh, S.; Komárek, J.; Chatchawan, T.; Hauer, T. Thainema gen. nov. (Leptolyngbyaceae, Synechococcales): A new genus of simple trichal cyanobacteria isolated from a solar saltern environment in Thailand. PLoS ONE 2022, 17, e0261682. [Google Scholar] [CrossRef] [PubMed]
- Cai, F.; Li, S.; Zhang, H.; Yu, G.; Li, R. Nodosilinea hunanesis sp. nov. (Prochlorotrichaceae, Synechococcales) from a Freshwater Pond in China Based on a Polyphasic Approach. Diversity 2022, 14, 364. [Google Scholar] [CrossRef]
- Johansen, J.R.; Kovacik, L.; Casamatta, D.A.; Fučiková, K.; Kaštovský, J. Utility of 16S-23S ITS sequence and secondary structure for recognition of intrageneric and intergeneric limits within cyanobacterial taxa: Leptolyngbya corticola sp. nov. (Pseudanabanaceae, Cyanobacteria). Nova Hedwig. 2011, 92, 283–302. [Google Scholar] [CrossRef]
- Osorio-Santos, K.; Pietrasiak, N.; Bohunická, M.; Miscoe, L.H.; Kovaácik, L.; Martin, M.P.; Johansen, J.R. Seven new species of Oculatella (Pseudanabaenales, Cyanobacteria). Eur. J. Phycol. 2014, 49, 450–470. [Google Scholar] [CrossRef]
- Pietrasiak, N.; Osorio-Santos, K.; Shalygin, S.; Martin, M.P.; Johansen, J.R. When is a lineage a species? A case study in Myxacorys gen. nov. (Synechococcales: Cyanobacteria) with the description of two new species from the Americas. J. Phycol. 2019, 55, 976–996. [Google Scholar] [CrossRef]


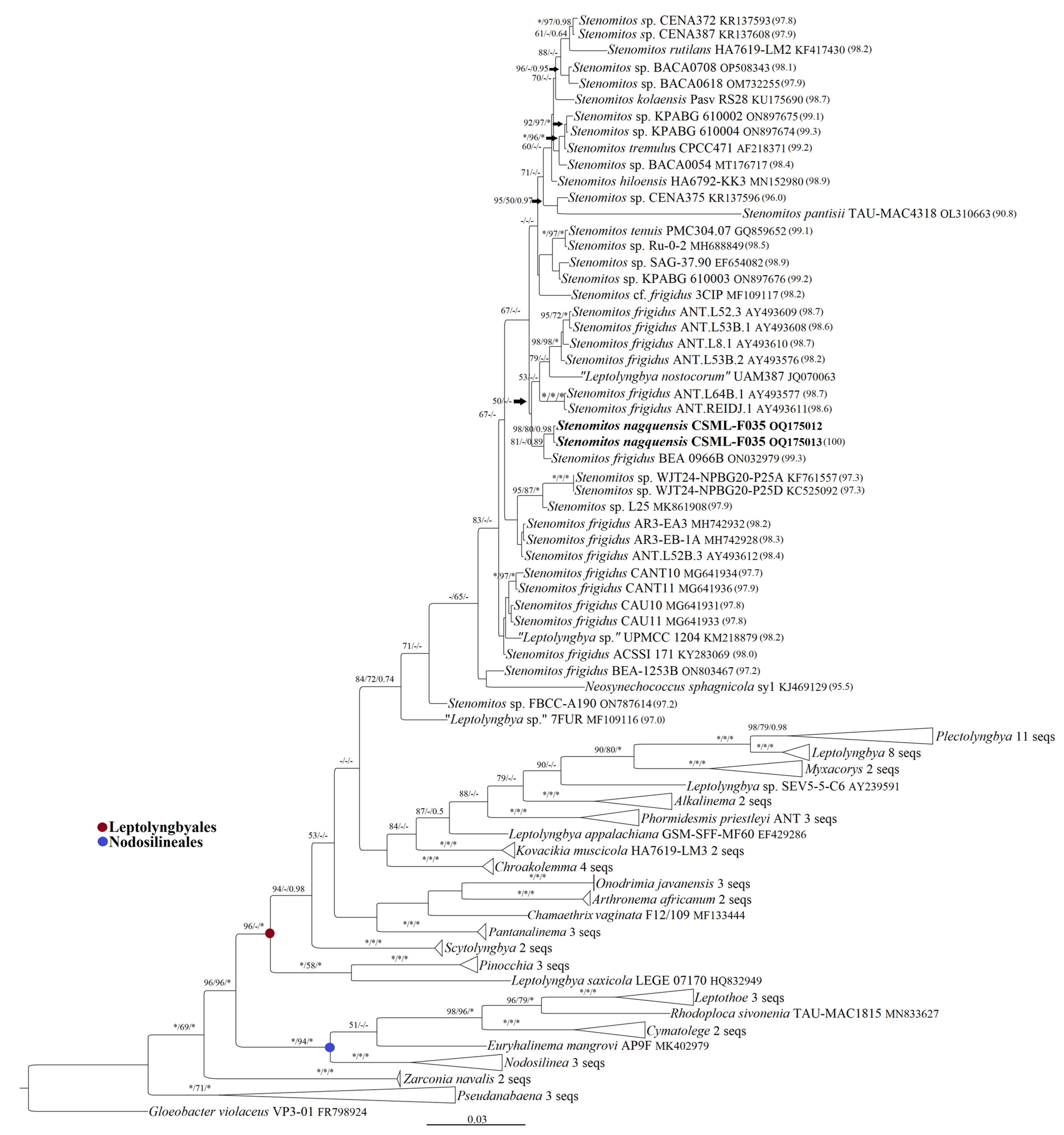



| Strain | Stenomitos nagquensis CSML-F035 | Stenomitos rutilans HA7619-LM2 | Stenomitos hiloensis HA6792-KK3 | Stenomitos kolaensis Pasv RS28 | Stenomitos frigidus ANT.L52B.3 | Stenomitos frigidus ANT.L8.1 | Stenomitos frigidus ANT.L53B.1 | Stenomitos sp. BACA0708 | Stenomitos sp. BACA0054 | Stenomitos sp. BACA0618 | Stenomitos sp. WJT24NPBG20 | Stenomitos sp. SAG 37.90 | Stenomitos sp. KPABG 610003 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stenomitos nagquensis CSML-F035 | |||||||||||||
| Stenomitos rutilans HA7619-LM2 | 15.1 | ||||||||||||
| Stenomitos hiloensis HA6792-KK3 | 14.1 | 12.3 | |||||||||||
| Stenomitos kolaensis Pasv RS28 | 11.2 | 11.6 | 8.6 | ||||||||||
| Stenomitos frigidus ANT.L52B.3 | 22.8 | 20.9 | 23.3 | 20.3 | |||||||||
| Stenomitos frigidus ANT.L8.1 | 14.1 | 15.6 | 13.8 | 12.6 | 22.8 | ||||||||
| Stenomitos frigidus ANT.L53B.1 | 14.1 | 15.3 | 13.6 | 12.6 | 22.8 | 0 | |||||||
| Stenomitos sp. BACA0708 | 12.6 | 10.9 | 3.9 | 7.7 | 22.2 | 14.2 | 14.0 | ||||||
| Stenomitos sp. BACA0054 | 16.3 | 12.3 | 11.7 | 11.6 | 23.9 | 16.5 | 16.2 | 11.4 | |||||
| Stenomitos sp. BACA0618 | 17.2 | 12.0 | 10.7 | 10.8 | 23.5 | 18.1 | 17.8 | 10.1 | 10.8 | ||||
| Stenomitos sp. WJT24NPBG20 | 20.1 | 14.2 | 17.5 | 15.2 | 20.7 | 23.8 | 23.8 | 17.2 | 17.2 | 19.1 | |||
| Stenomitos sp. SAG 37.90 | 15.7 | 10.6 | 12.7 | 9.3 | 23.3 | 16.6 | 16.3 | 12.2 | 13.0 | 11.8 | 18.7 | ||
| Stenomitos sp. KPABG 610003 | 15.7 | 12.9 | 14.3 | 9.9 | 20.7 | 15.0 | 14.7 | 13.9 | 16.3 | 16.3 | 21.3 | 15.8 | |
| Stenomitos sp. KPABG 610004 | 15.0 | 15.5 | 13.5 | 12.2 | 24.0 | 11.7 | 11.7 | 13.2 | 15.4 | 16.3 | 21.0 | 15.6 | 14.2 |
| Strain | D1-D1′ Helix | Spacer + D2 | D3 + Spacer | tRNA–Ile | V2 Helix | tRNA–Ala | Spacer | Box–B Helix | Spacer + Box–A | D4 + Spacer | V3 Helix | D5 region |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stenomitos nagquensis CSML-F035 | 65 | 38 | 10 | 79 | 65 | 73 | 39 | 39 | 26 | 13 | 94 | 28 |
| Stenomitos rutilans HA7619-LM2 | 65 | 36 | 10 | 79 | – | 73 | 48 | 40 | 26 | 14 | 92 | 28 |
| Stenomitos hiloensis HA6792-KK3 | 65 | 36 | 10 | 79 | 69 | 73 | 40 | 39 | 26 | 13 | 93 | 28 |
| Stenomitos kolaensis Pasv_RS28 | 65 | 37 | 10 | 79 | – | 73 | 42 | 40 | 26 | 13 | 93 | 28 |
| Stenomitos frigidus ANT.L52B.3 | 65 | 36 | 10 | 79 | 77 | 73 | 41 | 42 | 26 | 14 | 94 | 17 |
| Stenomitos frigidus ANT.L8.1/ANT L53B.1 | 65 | 36 | 10 | 79 | 69 | 73 | 40 | 40 | 26 | 13 | 96 | 17 |
| Stenomitos sp. BACA0708 | 65 | 36 | 10 | 79 | 69 | 73 | 37 | 39 | 26 | 13 | 93 | 28 |
| Stenomitos sp. BACA0054 | 65 | 36 | 10 | 79 | 75 | 73 | 41 | 39 | 26 | 14 | 93 | 28 |
| Stenomitos sp. WJT24NPBG20 | 65 | 36 | 10 | 79 | 74 | 73 | 40 | 43 | 26 | 14 | 93 | 27 |
| Stemonitos sp. SAG 37.90 | 65 | 37 | 10 | 79 | 74 | 73 | 41 | 40 | 26 | 14 | 94 | 26 |
| Stenomitos sp. KPABG 610003 | 65 | 37 | 10 | 79 | 73 | 73 | 42 | 39 | 26 | 14 | 94 | 28 |
| Stenomitos sp. KPABG 61004 | 64 | 37 | 10 | 79 | 68 | 73 | 40 | 40 | 26 | 13 | 96 | 25 |
| Stenomitos sp. BACA0618 | 65 | 37 | 10 | 79 | 74 | 73 | 43 | 40 | 26 | 14 | 93 | 28 |
| Strain | Color and Appearance of Trichomes | Size of Vegetative Cells Length (µm) | Shapes of Cells | Apical Cells | Appearance of Mucilaginous Sheath | Presence of Necridia | Isolation Source/Habitat Locality | Reference |
|---|---|---|---|---|---|---|---|---|
| Stenomitos nagquensis CSML-F035 | Bright-to-pale blue–green/short, unbranched, constricted at the cross-walls | 0.4–2.2 | Isodiametric, rarely elongated | Round to slightly conical | Colorless, firm, hyaline | Present | Freshwater, meadow wetland, Tibet, China | This study |
| Stenomitos pantisii TAU-MAC 4318 | Blue–green/constricted at the cross-walls | 1.5–2.5 | Isodiametric-to-elongated | Round to slightly conical, elongate | Soft, colorless, hyaline-to-firm | Present | Rocks, grava cave in Corfu Island, Greece | Panou and Gkelis [47] |
| Stenomitos hiloensis HA6792-KK3 | Blue–green/isopolar, unbranched, untapered, constricted near the cell walls | 0.5–1.2 | Isodiametric | Round | Firm, thin, colorless | Present, frequent | Seep wall, Hilo, Hawaii | Shalygin et al. [46] |
| Stenomitos kolaensis Pasv_RS28 | Bright-to-pale blue–green/constricted at the cross-walls | 1.5–2.5 | Isodiametric-to-elongated | Round to slightly conical, elongated | Soft, colorless, hyaline-to-firm | Present | Soil, Kola, Russia | Shalygin et al. [46] |
| Stenomitos rutilans HA7619-LM2 | Red–brown/short, tapered, unconstricted at the cross-wall | 2.8–4.8 | Thin, longer than broad | Thin, longer than broad | Absent or present as thin and colorless | Absent | Cave wall, Waikapala’e Cave, Oahu, Hawaii | Miscoe et al. [21] |
| Stenomitos tremulus UTCC471 | Blue–green/short, straight, bent | 3.0–7.0 | Longer than broad | Bluntly round | Colorless, open | Absent | Pond, Bylot Island, Northwest Canada | Casamatta et al. [15] |
| Stenomitos frigidus | Blue–green/constricted | 1.0–2.4 | – | Round | Hyaline | Absent | Streams, Antarctica | Miscoe et al.; Fritsch [21,48] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pecundo, M.H.; Chen, T.; Wang, Y.; Wen, X.; Hu, Z.; Chen, H.; Li, N. Stenomitos nagquensis sp. nov. (Leptolyngbyaceae, Cyanobacteria) from a Meadow Wetland in the Tibet Plateau: A Novel Species Studied Based on a Polyphasic Approach. Diversity 2023, 15, 536. https://doi.org/10.3390/d15040536
Pecundo MH, Chen T, Wang Y, Wen X, Hu Z, Chen H, Li N. Stenomitos nagquensis sp. nov. (Leptolyngbyaceae, Cyanobacteria) from a Meadow Wetland in the Tibet Plateau: A Novel Species Studied Based on a Polyphasic Approach. Diversity. 2023; 15(4):536. https://doi.org/10.3390/d15040536
Chicago/Turabian StylePecundo, Melissa H., Tao Chen, Yunhua Wang, Xuemei Wen, Zhangli Hu, Huirong Chen, and Nan Li. 2023. "Stenomitos nagquensis sp. nov. (Leptolyngbyaceae, Cyanobacteria) from a Meadow Wetland in the Tibet Plateau: A Novel Species Studied Based on a Polyphasic Approach" Diversity 15, no. 4: 536. https://doi.org/10.3390/d15040536
APA StylePecundo, M. H., Chen, T., Wang, Y., Wen, X., Hu, Z., Chen, H., & Li, N. (2023). Stenomitos nagquensis sp. nov. (Leptolyngbyaceae, Cyanobacteria) from a Meadow Wetland in the Tibet Plateau: A Novel Species Studied Based on a Polyphasic Approach. Diversity, 15(4), 536. https://doi.org/10.3390/d15040536








