Semicryptic Diversity around Chaetoceros elegans (Bacillariophyta, Mediophyceae), and the Description of Two New Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain Isolates and Maintenance
2.2. Morphological Observations
2.3. Phylogenetic Analyses Inferred from Nucleic Genes
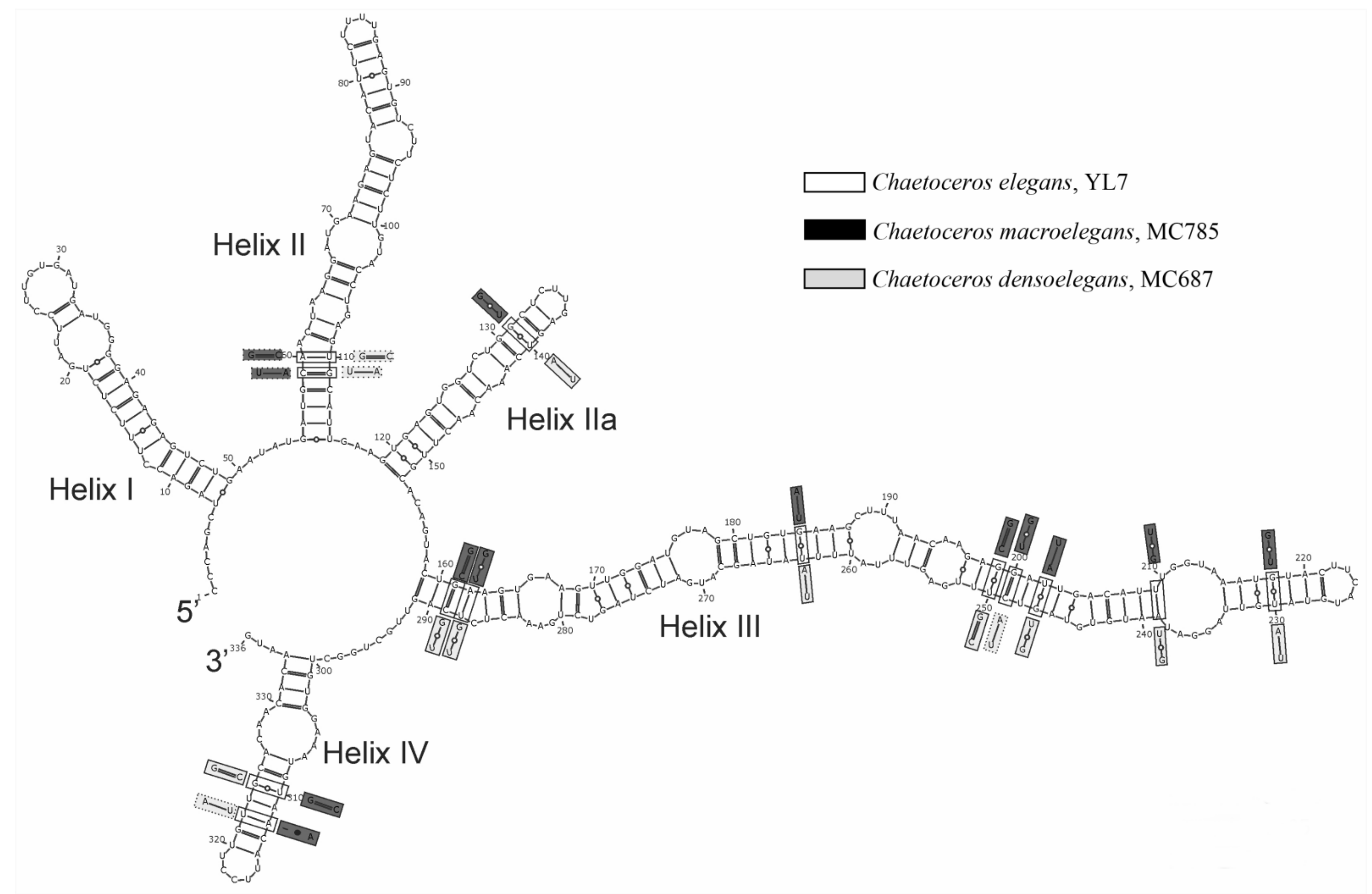
2.4. ITS2 Secondary Structure Comparison
3. Results
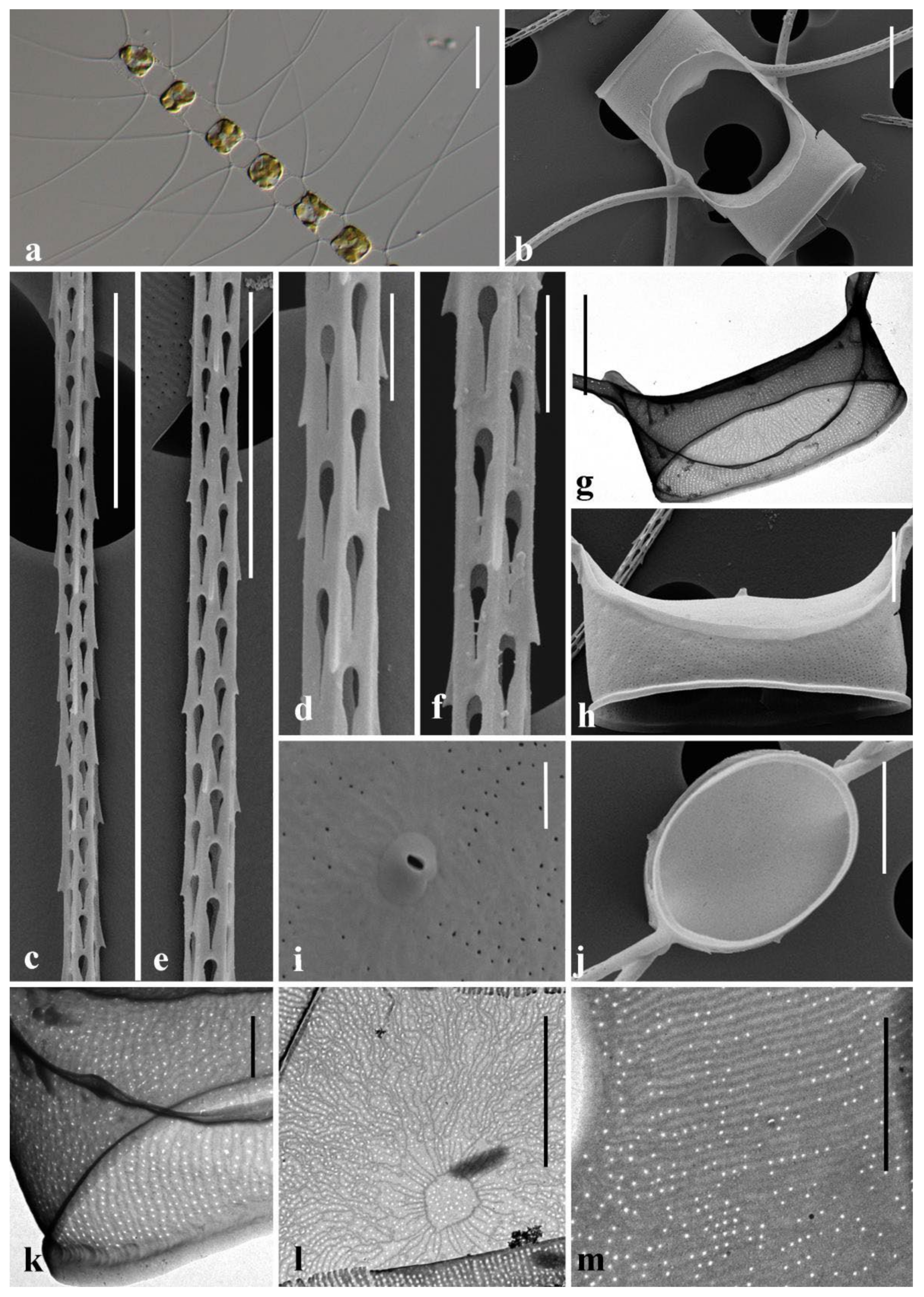
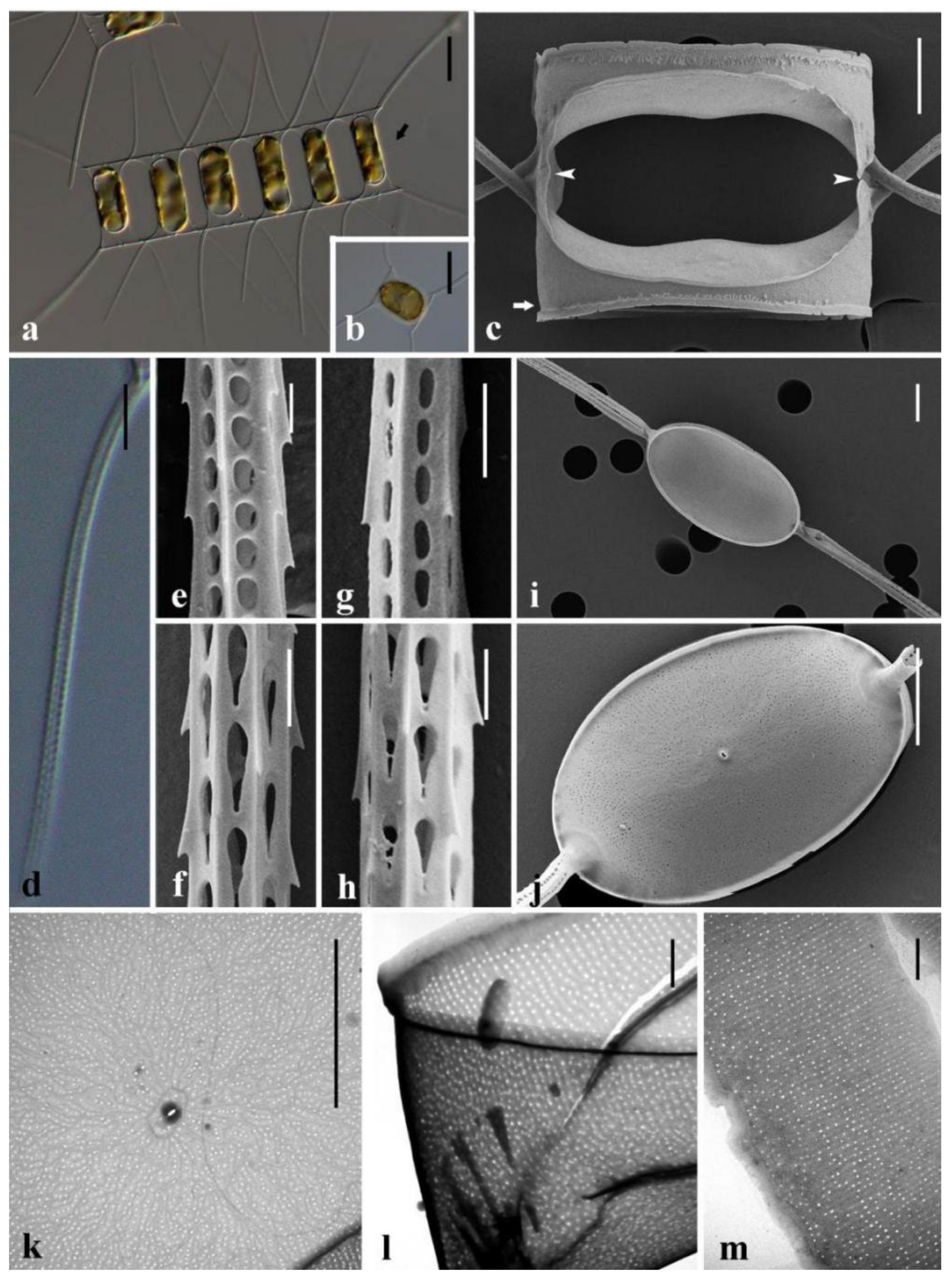
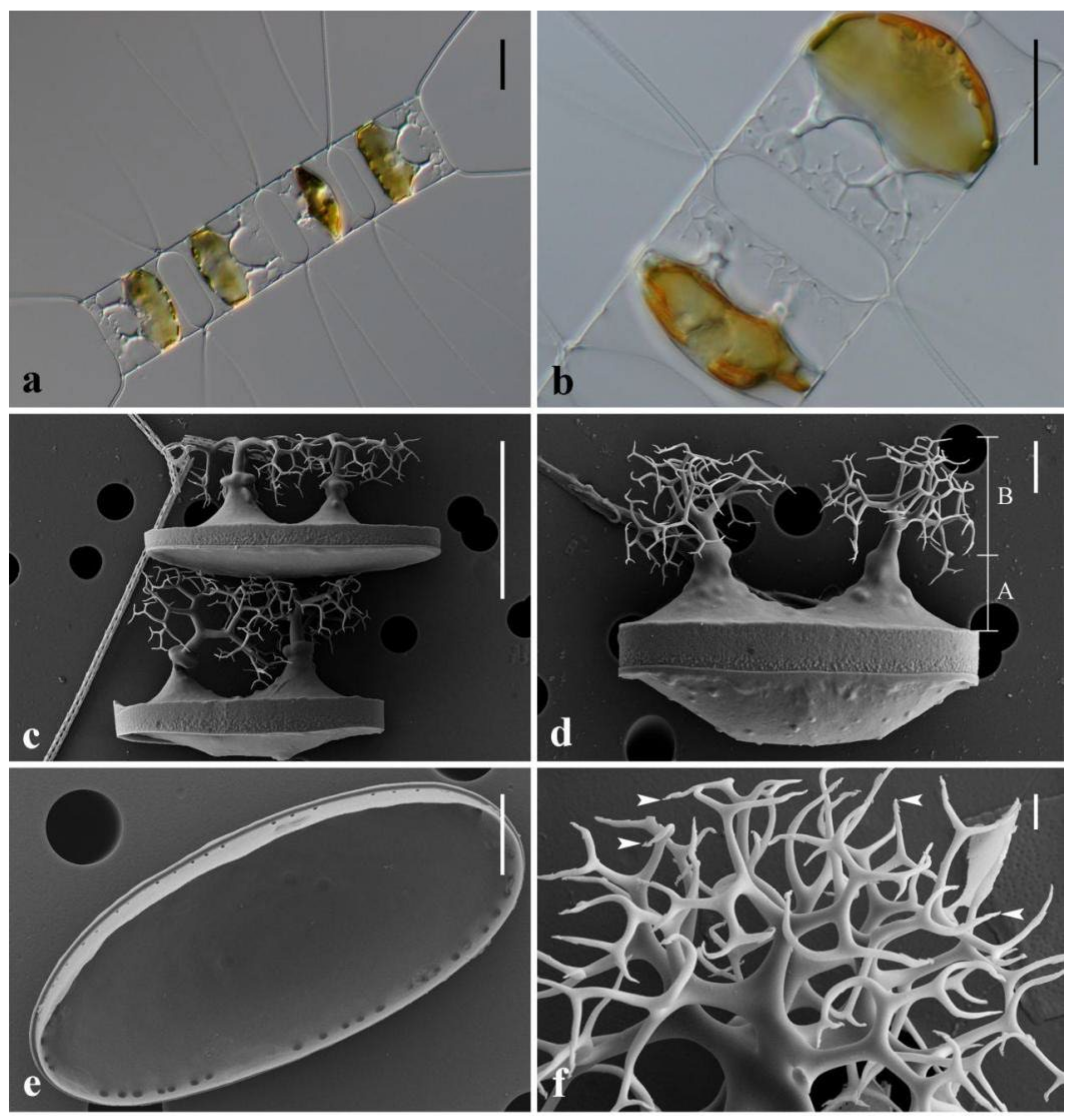
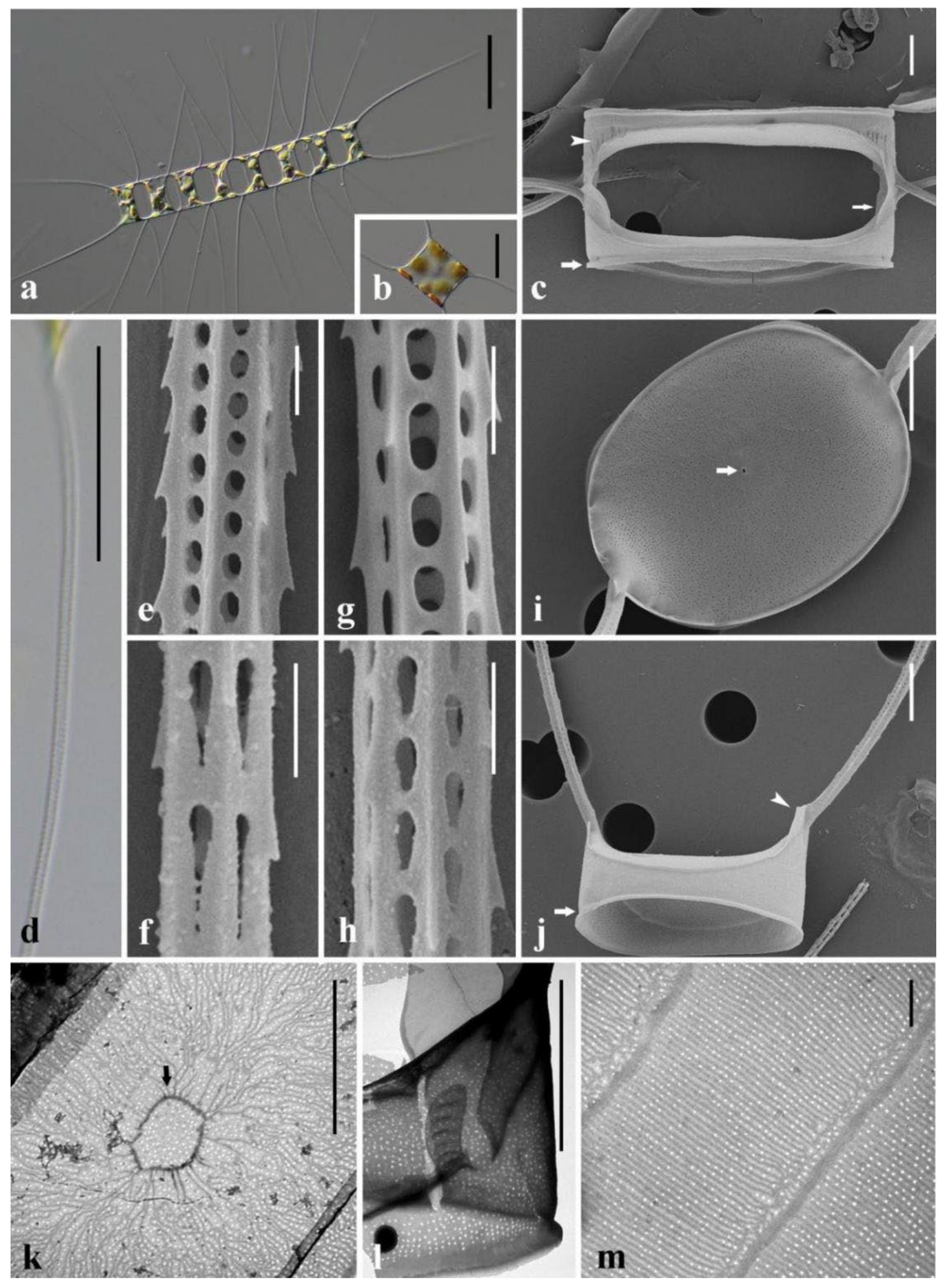
3.1. Morphological Description of Three Chaetoceros Species
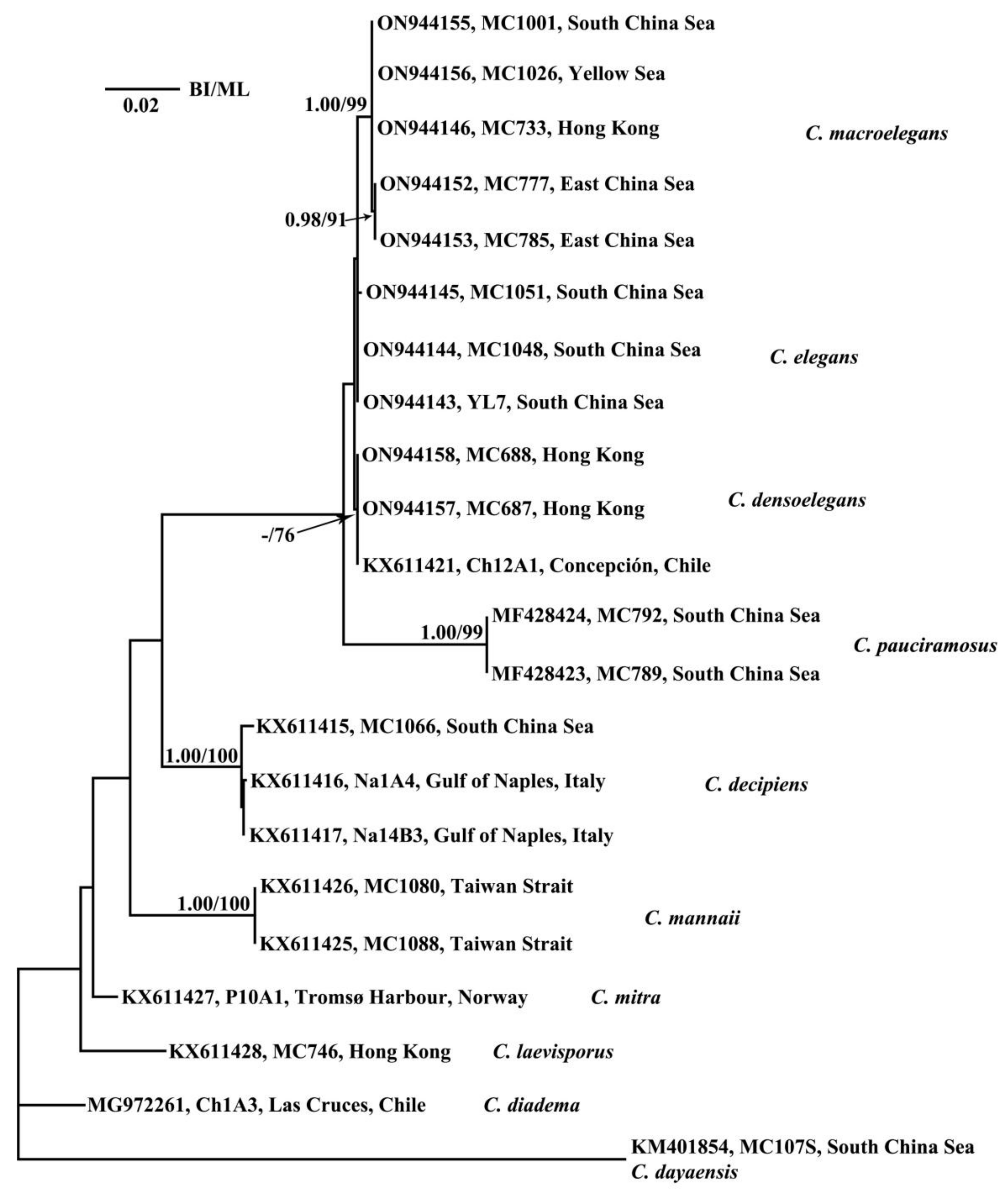
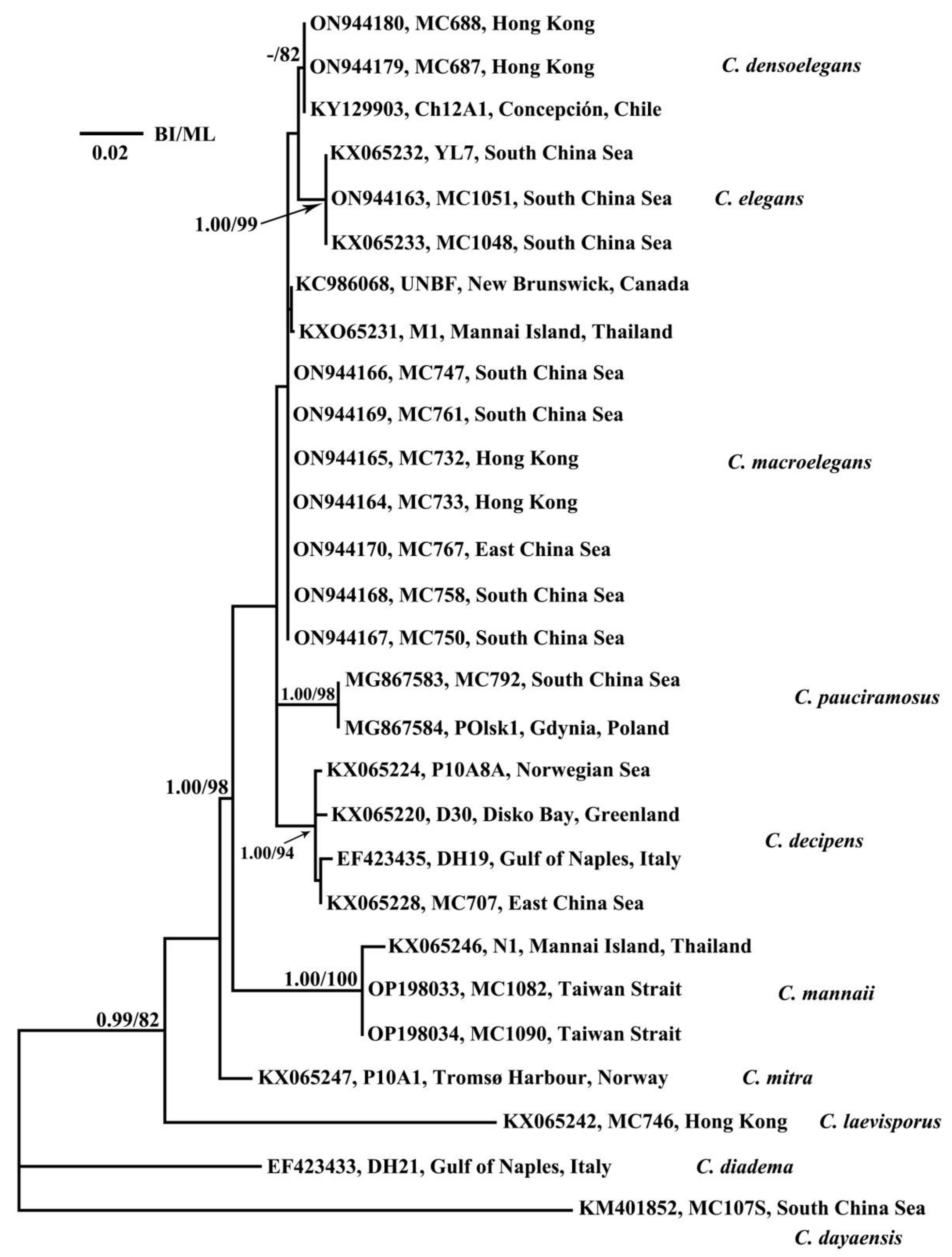
3.2. Phylogenetic Analyses
3.3. CBC and HCBC Analyses
4. Discussion
4.1. Taxonomic Positions of the New Taxa
4.2. Detailed Comparison of the Three Taxa
4.3. Comparing the Three Taxa in This Study with Close Species in the C. lorenzianus Complex
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guiry, M.D.; Guiry, G.M. AlgaeBase. World-Wide Electronic Publication, National University of Ireland, Galway. Available online: http://www.algaebase.org (accessed on 10 January 2022).
- Chamnansinp, A.; Li, Y.; Lundholm, N.; Moestrup, Ø. Global diversity of two widespread, colony-forming diatoms of the marine plankton, Chaetoceros socialis (syn. C. radians) and Chaetoceros gelidus sp. nov. J. Phycol. 2013, 49, 1128–1141. [Google Scholar] [CrossRef] [PubMed]
- Chamnansinp, A.; Moestrup, Ø.; Lundholm, N. Diversity of the marine diatom Chaetoceros (Bacillariophyceae) in Thai waters-revisiting Chaetoceros compressus and Chaetoceros contortus. Phycologia 2015, 54, 161–175. [Google Scholar] [CrossRef]
- Xu, X.J.; Lundholm, N.; Li, Y. A study of Chaetoceros debilis sensu lato species (Bacillariophyceae), with emendation of C. debilis and description of C. galeatus sp. nov. J. Phycol. 2020, 56, 784–797. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lundholm, N.; Moestrup, Ø. Chaetoceros rotosporus sp. nov. (Bacillariophyceae), a species with unusual resting spore formation. Phycologia 2013, 52, 600–608. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, S.; Lundholm, N.; Lü, S. Morphology and molecular phylogeny of Chaetoceros dayaensis sp. nov. (Bacillariophyceae), characterized by two 90° rotations of the resting spore during maturation. J. Phycol. 2015, 51, 469–479. [Google Scholar] [CrossRef]
- Gaonkar, C.C.; Kooistra, W.H.C.F.; Lange, C.B.; Montresor, M.; Sarno, D. Two new species in the Chaetoceros socialis complex (Bacillariophyta): C. sporotruncatus and C. dichatoensis, and characterization of its relatives, C. radicans and C. cinctus. J. Phycol. 2017, 53, 889–907. [Google Scholar] [CrossRef]
- Kaczmarska, I.; Sammanta, B.; Ehrman, J.M.; Porcher, E.M.A. Auxosporulation in Chaetoceros acadianus sp. nov. (Bacillariophyceae), a new member of the section Compressa. Eur. J. Phycol. 2019, 54, 206–221. [Google Scholar] [CrossRef]
- Xu, X.J.; Chen, Z.Y.; Lundholm, N.; Li, Y. Diversity in the section Compressa of the genus Chaetoceros (Bacillariophyceae), with description of two new species from Chinese warm waters. J. Phycol. 2019, 55, 104–117. [Google Scholar] [CrossRef]
- Xu, X.J.; Lundholm, N.; Chen, Z.Y.; Li, Y. Revisiting section Compressa of Chaetoceros (Bacillariophyceae), with descriptions of C. brevispinosus sp. nov. and C. ornatus comb. nov. Phycologia 2019, 58, 614–627. [Google Scholar] [CrossRef]
- Li, Y.; Boonprakob, A.; Gaonkar, C.C.; Kooistra, W.H.C.F.; Lange, C.B.; Hernández-Becerril, D.; Chen, Z.Y.; Moestrup, Ø.; Lundholm, N. Diversity in the globally distributed diatom genus Chaetoceros (Bacillariophyceae): Three new species from warm-temperate waters. PLoS ONE 2017, 12, e0168887. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Lundholm, N.; Moestrup, Ø.; Kownacka, J.; Li, Y. Chaetoceros pauciramosus sp. nov. (Bacillariophyceae), a widely distributed brackish water species in the C. lorenzianus complex. Protist 2018, 169, 615–631. [Google Scholar] [CrossRef] [PubMed]
- De Luca, D.; Sarno, D.; Piredda, R.; Kooistra, W.H.C.F. A multigene phylogeny to infer the evolutionary history of Chaetocerotaceae (Bacillariophyta). Mol. Phylogenet. Evol. 2019, 140, 106575. [Google Scholar] [CrossRef] [PubMed]
- Gaonkar, C.C.; Piredda, R.; Minucci, C.; Mann, D.; Montresor, M.; Sarno, D.; Kooistra, W.H.C.F. Annotated 18S and 28S rDNA reference sequences of taxa in the planktonic diatom family Chaetocerotaceae. PLoS ONE 2018, 13, e0208929. [Google Scholar] [CrossRef] [PubMed]
- Guillard, R.L.; Hargraves, P.E. Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia 1993, 32, 234–236. [Google Scholar] [CrossRef]
- Kraus, D. Daniel’s XL Toolbox Addin for Excel, Version 6 October 2014. 2014. Available online: http://xltoolbox.sourceforge.net (accessed on 7 April 2022).
- Anonymous. Proposals for a standardization of diatom terminology and diagnoses. Nova Hedwig. Beih. 1975, 53, 323–354. [Google Scholar]
- Rines, J.E.B.; Hargraves, P.E. The Chaetoceros Ehrenberg (Bacillariophyceae) flora of Narragansett Bay, Rhode Island, USA. Bibl. Phycol. 1988, 79, 5–196. [Google Scholar]
- Suto, I. Taxonomy of the marine resting spore genera Dicladia Ehrenberg, Monocladia gen. nov. and Synendrum Ehrenberg and their stratigraphic significance in Miocene strata. Diatom Res. 2003, 18, 331–356. [Google Scholar] [CrossRef]
- Ishii, K.I.; Iwataki, M.; Matsuoka, K.; Imai, I. Proposal of identification criteria for resting spores of Chaetoceros species (Bacillariophyceae) from a temperate coastal sea. Phycologia 2011, 50, 351–362. [Google Scholar] [CrossRef]
- Lundholm, N.; Daugbjerg, N.; Moestrup, Ø. Phylogeny of the Bacillariaceae with emphasis on the genus Pseudo-nitzschia (Bacillariophyceae) based on partial LSU rDNA. Eur. J. Phycol. 2002, 37, 115–134. [Google Scholar] [CrossRef]
- Scholin, C.A.; Herzog, M.; Sogin, M.; Anderson, D.M. Identification of group- and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). II. Sequence analysis of a fragment of the LSU rDNA. J. Phycol. 1994, 30, 999–1011. [Google Scholar] [CrossRef]
- Nunn, G.B.; Theisen, B.F.; Christensen, B.; Arctander, P. Simplicity-correlated size growth of the nuclear 28S ribosomal RNA D3 expansion segment in the crustacean order Isopoda. J. Mol. Evol. 1996, 42, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.; Mi, T.Z.; Yu, Z.G. Detection of Phaeocystis globosa using sandwich hybridization integrated with nuclease protection assay (NPA-SH). J. Environ. Sci. 2008, 20, 1481–1486. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for window 95/98/NT. Nucleic Acids Symp. 1999, 41, 95–98. [Google Scholar]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa ad mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Boc, A.; Diallo, A.B.; Makarenkov, V. T-REX: A web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012, 40, 573–579. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Mark, P.V. MrBayes 3.2: Efficient Bayesian inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModeltest v2. Program Distributed by the Author; Evolutionary Biology Center, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Seibel, P.; Müller, T.; Dandekar, T.; Wolf, M. Synchronous visual analysis and editing of RNA sequence and secondary structure alignments using 4SALE. BMC Res. Notes 2008, 1, 91. [Google Scholar] [CrossRef]
- Wolf, M.; Koetschan, C.; Müller, T. ITS2, 18S, 16S or any other RNA-simply aligning sequences and their individual secondary structures simultaneously by an automatic approach. Gene 2014, 546, 145–149. [Google Scholar] [CrossRef]
- Teng, S.T.; Tan, S.N.; Lim, H.C.; Dao, V.H.; Bates, S.S.; Leaw, C.P. High diversity of Pseudo-nitzschia along the northern coast of Sarawak (Malaysian Borneo), with descriptions of P. bipertita sp. nov. and P. limii sp. nov. (Bacillariophyceae). J. Phycol. 2016, 52, 973–989. [Google Scholar] [CrossRef]
- Brunel, J. Orientation of setae in the genus Chaetoceros, in regard to the apical axis. J. Mar. Biol. Ass. India 1972, 14, 315–327. [Google Scholar]
- Chen, Z.Y.; Xu, X.J.; Zhu, S.Y.; Zhai, M.Y.; Li, Y. Species diversity and geographical distribution of the Chaetoceros lorenzianus complex along the coast of China. Biodivers. Sci. 2019, 27, 149–158. [Google Scholar]
- Hasle, G.R.; Syvertsen, E.E. Marine diatoms. In Identifying Marine Phytoplankton; Tomas, C.R., Ed.; Academic Press: San Diego, CA, USA, 1997; pp. 5–385. [Google Scholar]
- Amato, A.; Montresor, M. Morphology, phylogeny and sexual cycle of Pseudo-nitzschia mannii sp. nov. (Bacillariophyceae): A pseudo-cryptic species within the P. pseudodelicatissima complex. Phycologia 2008, 47, 487–497. [Google Scholar] [CrossRef]
- Casteleyn, G.; Chepurnov, V.A.; Leliaert, F.; Mann, D.G.; Bates, S.S.; Lundhulm, N.; Rhodes, L.; Sabbe, K.; Vyverman, W. Pseudo-nitzschia pungens (Bacillariophyceae): A cosmopolitan diatom species? Harmful Algae 2008, 7, 241–257. [Google Scholar] [CrossRef]
- Lee, S.D.; Park, J.S.; Yun, S.M.; Lee, J.H. Critical criteria for identification of the genus Chaetoceros (Bacillariophyceae) based on setae ultrastructure. I. Subgenus Chaetoceros. Phycologia 2014, 53, 174–187. [Google Scholar] [CrossRef]
- Lee, S.D.; Joo, H.M.; Lee, J.H. Critical criteria for identification of the genus Chaetoceros (Bacillariophyceae) based on setae ultrastructure. II. Subgenus Hyalochaete. Phycologia 2014, 53, 614–638. [Google Scholar] [CrossRef]
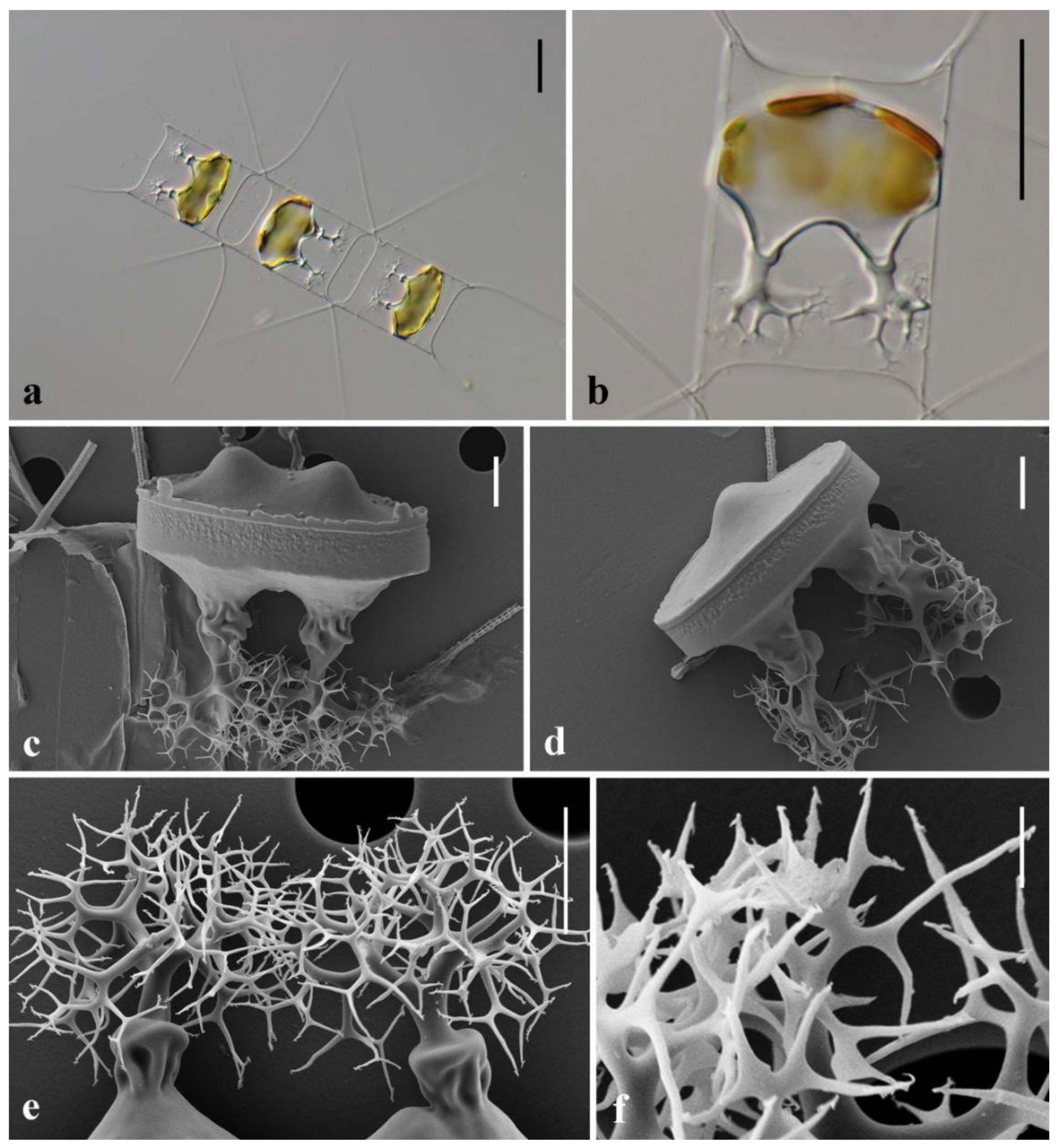

| Species | Strain | Locality | Date | Accession Number | ||
|---|---|---|---|---|---|---|
| LSU | ITS | SSU | ||||
| C. elegans | YL7 * | Dapeng Bay, 22.5933° N, 114.3991° E | 27 August 2010 | KX065232 | ON972673 | ON944143 |
| MC1048 | Dapeng Bay, 22.5953° N, 114.3981° E | 30 September 2015 | KX065233 | ON972674 | ON944144 | |
| MC1051 | Dapeng Bay, 22.5953° N, 114.3981° E | 1 October 2015 | ON944163 | ON972675 | ON944145 | |
| C. macroelegans | MC733 | Hong Kong, 22.2960° N, 114.1840° E | 10 July 2015 | ON944164 | ON972676 | ON944146 |
| MC732 | Hong Kong, 22.2960° N, 114.1840° E | 10 July 2015 | ON944165 | ON972677 | ON944147 | |
| MC747 | Jiangmen, 21.8361° N, 113.1916° E | 5 August 2015 | ON944166 | ON972678 | ON944148 | |
| MC750 | Jiangmen, 21.8361° N, 113.1916° E | 5 August 2015 | ON944167 | / | ON944149 | |
| MC758 | Jiangmen, 21.8361° N, 113.1916° E | 5 August 2015 | ON944168 | / | / | |
| MC761 | Jiangmen, 21.8361° N, 113.1916° E | 5 August 2015 | ON944169 | / | ON944150 | |
| MC767 | Ningbo, 29.8636° N, 121.5611° E | 6 August 2015 | ON944170 | ON972679 | ON944151 | |
| MC777 | Ningbo, 29.8636° N, 121.5611° E | 7 August 2015 | ON944171 | ON972680 | ON944152 | |
| MC785 * | Ningbo, 29.8636° N, 121.5611° E | 7 August 2015 | ON944172 | ON972681 | ON944153 | |
| MC788 | Zhuhai, 22.1614° N, 113.3436° E | 17 August 2015 | KX065236 | ON972682 | / | |
| MC790 | Zhuhai, 22.1614° N, 113.3436° E | 17 August 2015 | ON944173 | ON972683 | / | |
| MC1000 | Zhanjiang, 20.9566° N, 110.3998° E | 26 August 2015 | ON944174 | ON972684 | ON944154 | |
| MC1001 | Zhanjiang, 20.9566° N, 110.3998° E | 26 August 2015 | ON944175 | ON972685 | ON944155 | |
| MC1026 | Qingdao, 36.0329° N, 120.3473° E | 9 September 2015 | ON944176 | / | ON944156 | |
| UNBF | New Brunswick, Canada, 45.00° N, 66.733° W | 7 September 2010 | KC986068 | / | / | |
| M1 | Mannai Island, Thailand, 12.6111° N, 101.6836° E | 2 June 2008 | KXO65231 | / | / | |
| C. densoelegans | MC687 * | Hong Kong, 22.3520° N, 114.1139° E | 3 April 2015 | ON944179 | ON972686 | ON944157 |
| MC688 | Hong Kong, 22.2960° N, 114.1840° E | 3 April 2015 | ON944180 | ON972687 | ON944158 | |
| Ch12A1 | Concepción, Chile, 36.5133° S and 73.1291° W | 29 October 2013 | KY129903 | / | KX611421 | |
| Characters | C. macroelegans | C. densoelegans | C. elegans | |
|---|---|---|---|---|
| Brunel group | Ⅰ | Ⅰ | Ⅰ | |
| Aperture shape | elliptical to hexagonal | elliptical to hexagonal | quadrangular-rectangular | |
| Poroids on valve face | present | present | present | |
| External tube of rimoportula | short | short | short | |
| Basal part of seta | absent or short | absent or short | present, distinct | |
| Seta poroid shape | Terminal seta | tear-shaped, round-oval | tear-shaped, round-oval | tear-shaped |
| Intercalary seta | tear-shaped, round-oval | tear-shaped, round-oval | tear-shaped | |
| Seta poroid size/density (10 µm) | Terminal seta | 0.5–1.3 (0.7 ± 0.2) a (tear-shaped)/ | 0.5–0.7 (0.6 ± 0.1) a (tear-shaped)/ | 0.6–1.2 (0.9 ± 0.2) b/ |
| 6.5–16.0 (11.0 ± 2.2) a (tear-shaped) | 9.3–14.2 (12.3 ± 2.7) a (tear-shaped) | 7.2–12.8 (9.4 ± 1.7) b | ||
| 0.4–0.6 (0.4 ± 0.1) a (round-oval)/ | 0.2–0.5 (0.4 ± 0.1) b (round-oval)/ | |||
| 12.0–19.1 (15.8 ± 2.3) a (round-oval) | 15.0–27.0 (19.5 ± 3.1) b (round-oval) | |||
| Intercalary seta | 0.7– 2.3 (1.2 ± 0.3) a (tear-shaped)/ | 0.6–0.9 (0.7 ± 0.1) b (tear-shaped)/ | 0.4–1.0 (0.7 ± 0.2) b/ | |
| 5.0–13.0 (7.9 ± 1.7) a (tear-shaped) | 8.4–14.0 (11.2 ± 2.1) b (tear-shaped) | 8.0–18.5 (12.0 ± 3.1) b | ||
| 0.4– 0.5 (0.4 ± 0.03) a (round-oval)/ | 0.3–0.5 (0.4 ± 0.1) b (round-oval)/ | |||
| 15.3–18.3 (17.1 ± 1.2) a (round-oval) | 13.5–27.0 (20.0 ± 4.2) b (round-oval) | |||
| Apical axis (µm) | 12.6–39.7 | 25.0–26.5 | 9.2–14.7 | |
| (27.9 ± 10.8) | (25.5 ± 0.6) | (12.5 ± 1.8) | ||
| Pervalvar axis (µm) | 11.3–28.4 | 9.4–15.8 | 9.2–16.4 | |
| (17.5 ± 3.9) | (12.4 ± 2.2) | (12.0 ± 1.7) | ||
| Aperture in pervalvar axis (µm) | 6.3–12.6 | 8.3–14.5 | 7.8–12.9 | |
| (8.9 ± 2.0) | (11.4 ± 1.5) | (10.2 ± 2.0) | ||
| Aperture height/ | 0.2–0.7 | 0.3–0.6 | 0.5–0.9 | |
| pervalvar axis index | (0.4 ± 0.1) a | (0.5 ± 0.1) b | (0.7 ± 0.1) b | |
| Resting spore | two branching processes | two branching processes | unknown | |
| Apical axis (μm) | 20.3–47.1 (33.8 ± 7.2) | 18.3–32.9 (28.6 ± 4.2) | ||
| Pervalvar axis of primary valve (μm) | 8.4–16.3 (11.2 ± 2.2) | 9.1–13.6 (11.4 ± 1.2) | ||
| Height of branching processes (μm) | 9.5–14.9 (11.4 ± 1.3) | 9.6–16.3 (13.3 ± 1.8) | ||
| Height of elevation (μm) | 4.4–10.3 (6.9 ± 1.6) | 6.0–9.6 (7.5 ± 1.0) | ||
| Outer slope of elevation | Acute angle | Acute angle | ||
| Joining of two elevations | Near mantle | Near mantle | ||
| Structure of distal tips | Hook | Hook | ||
| Secondary valve bulges | 1–2 | 1–2 | ||
| Branching times | 5–6 | 5–6 | ||
| Elevations/branching processes | 0.4–0.8 (0.6 ± 0.1) | 0.4–1.0 (0.6 ± 0.1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Chen, Z.; Lundholm, N.; Teng, S.T.; Xu, X.; Li, Y. Semicryptic Diversity around Chaetoceros elegans (Bacillariophyta, Mediophyceae), and the Description of Two New Species. Diversity 2022, 14, 676. https://doi.org/10.3390/d14080676
Chen X, Chen Z, Lundholm N, Teng ST, Xu X, Li Y. Semicryptic Diversity around Chaetoceros elegans (Bacillariophyta, Mediophyceae), and the Description of Two New Species. Diversity. 2022; 14(8):676. https://doi.org/10.3390/d14080676
Chicago/Turabian StyleChen, Xiumei, Zuoyi Chen, Nina Lundholm, Sing Tung Teng, Xiaojing Xu, and Yang Li. 2022. "Semicryptic Diversity around Chaetoceros elegans (Bacillariophyta, Mediophyceae), and the Description of Two New Species" Diversity 14, no. 8: 676. https://doi.org/10.3390/d14080676
APA StyleChen, X., Chen, Z., Lundholm, N., Teng, S. T., Xu, X., & Li, Y. (2022). Semicryptic Diversity around Chaetoceros elegans (Bacillariophyta, Mediophyceae), and the Description of Two New Species. Diversity, 14(8), 676. https://doi.org/10.3390/d14080676







