Taxonomic Discussion on Cyanobacterial Systematics at Family Level, with Special Regards to Phormidiaceae by Using the Strains of Chinese Newly Recorded Genera Ancylothrix and Potamolinea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cyanobacterial Collection and Cultivation
2.2. Morphological Observation
2.3. DNA Extraction and PCR Amplification
2.4. Phylogenetic Analyses
2.5. Analyses of 16S–23S Internal Transcribed Spacer (ITS)
3. Results
3.1. Morphological Description
3.1.1. Phormidium-like sp. 1
3.1.2. Phormidium-like sp. 2
3.1.3. Phormidium-like sp. 3
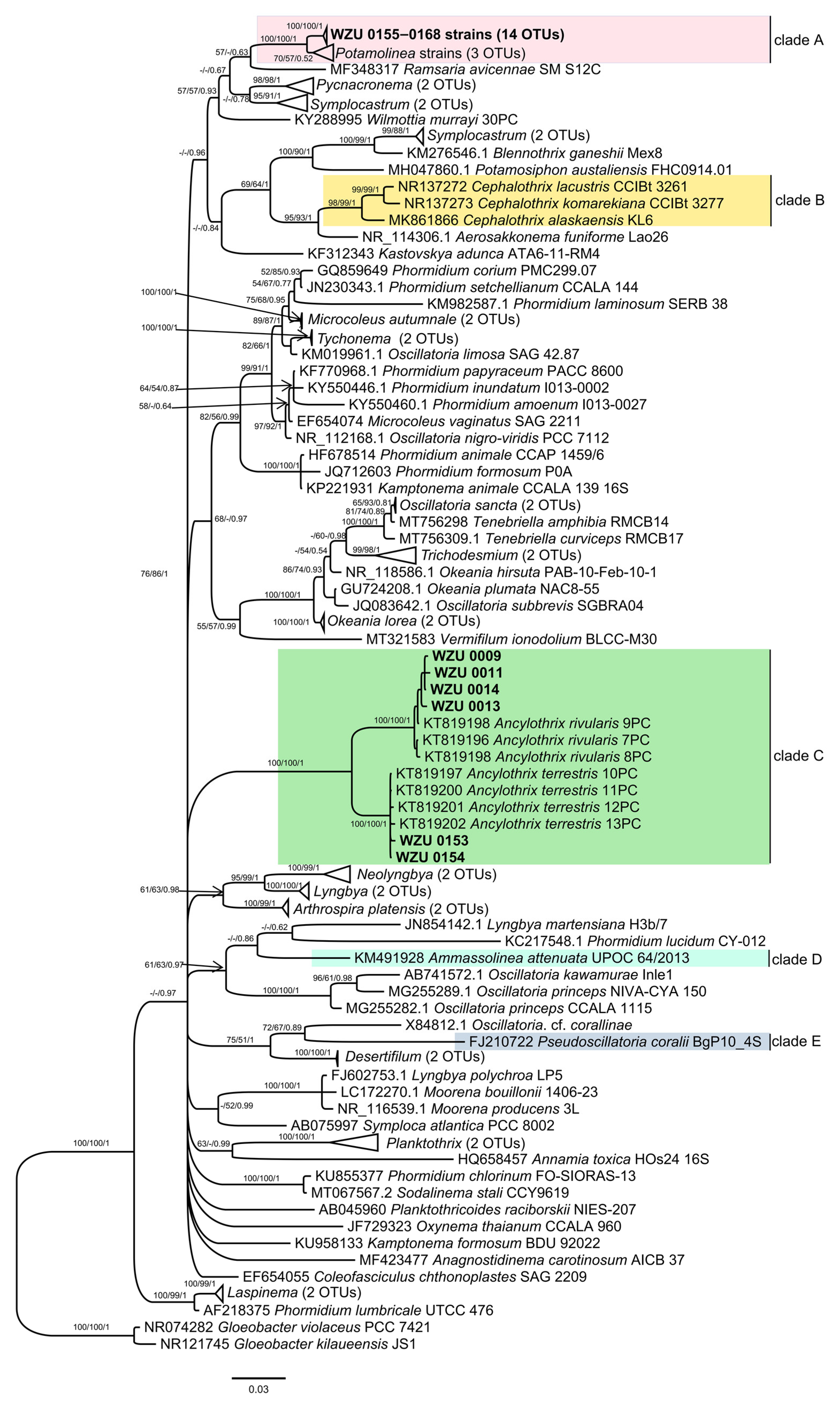
3.2. Molecular and Phylogenetic Analyses
3.3. Analyses of ITS between 16S and 23S rRNA Gene and Secondary Structures
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 1979, 111, 1–61. [Google Scholar] [CrossRef] [Green Version]
- Anagnostidis, K.; Komárek, J. Modern approach to the classification system of cyanophytes. 1–Introduction. Algol. Stud./Arch. Für Hydrobiol. 1985, 38–39, 291–302. [Google Scholar]
- Hoffmann, L.; Komárek, J.; Kaštovský, J. System of cyanoprokaryotes (cyanobacteria)—State in 2004. Algol. Stud. 2005, 117, 95–115. [Google Scholar] [CrossRef]
- Hoffmann, L.; Komárek, J.; Kaštovský, J. Proposal of cyanobacterial system—2004. In Süsswasserflora Von Mitteleuropa 19/2; Büdel, B., Krienitz, L., Gärtner, G., Schagerl, M., Eds.; Elsevier: Heidelberg, Germany, 2005; pp. 657–660. [Google Scholar]
- Fiore, M.F.; Sant’anna, C.L.; Azevedo, M.T.D.P.; Komárek, J.; Kaštovský, J.; Sulek, J.; Lorenzi, A.A.S. The cyanobacterial genus Brasilonema, gen. nov., a molecular and phenotypic evaluation. J. Phycol. 2007, 43, 789–798. [Google Scholar] [CrossRef]
- Bohunická, M.; Pietrasiak, N.; Johansen, J.R.; Gómez, E.B.; Hauer, T.; Gaysina, L.A.; Lukešová, A. Roholtiella, gen. nov.(Nostocales, Cyanobacteria)—A tapering and branching cyanobacteria of the family Nostocaceae. Phytotaxa 2015, 197, 84–103. [Google Scholar] [CrossRef] [Green Version]
- Genuário, D.B.; Vaz, M.G.M.V.; Hentschke, G.S.; Sant’anna, C.L.; Fiore, M.F. Halotia gen. nov., a phylogenetically and physiologically coherent cyanobacterial genus isolated from marine coastal environments. Int. J. Syst. Evol. Microbiol. 2015, 15, 663–675. [Google Scholar] [CrossRef] [Green Version]
- Sciuto, K.; Moro, I. Detection of the new cosmopolitan genus Thermoleptolyngbya (Cyanobacteria, Leptolyngbyaceae) using the 16S rRNA gene and 16S–23S ITS region. Mol. Phylogenetics Evol. 2016, 105, 15–35. [Google Scholar] [CrossRef]
- Martins, M.D.; Rigonato, J.; Taboga, S.R.; Branco, L.H.Z. Proposal of Ancylothrix gen. nov., a new genus of phormidiaceae (Cyanobacteria, Oscillatoriales) based on a polyphasic approach. Int. J. Syst. Evol. Microbiol. 2016, 66, 2396–2405. [Google Scholar] [CrossRef]
- Martins, M.D.; Branco, L.H.Z. Potamolinea gen. nov (Oscillatoriales, Cyanobacteria): A phylogenetically and ecologically coherent cyanobacterial genus. Int. J. Syst. Evol. Microbiol. 2016, 66, 3632–3641. [Google Scholar] [CrossRef]
- Borges, H.L.F.; Branco, L.H.Z.; Martins, M.D.; Lima, C.S.; Barbosa, P.T.; Lira, G.A.S.T.; Bittencourt-Oliveira, M.C.; Molica, R.J.R. Cyanotoxin production and phylogeny of benthic cyanobacterial strains isolated from the northeast of Brazil. Harmful Algae 2015, 43, 46–57. [Google Scholar] [CrossRef]
- Yap-Dejeto, L.G.; Batula, H.S. Bloom of Trichodesmium (Oscillatoriales, Phormidiaceae) and seasonality of potentially harmful phytoplankton in San Pedro Bay, Leyte, Philippines. Rev. De Biol. Trop. 2016, 64, 897–911. [Google Scholar]
- Te, S.H.; Tan, B.F.; Boo, C.Y.; Thompson, J.R.; Gin, K.Y.H. Genomics insights into production of 2-methylisoborneol and a putative cyanobactin by Planktothricoides sp. SR001. Stand. Genom. Sci. 2017, 12, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozwalak, P.; Podkowa, P.; Buda, J.; Niedzielski, P.; Zawierucha, K. Cryoconite—From minerals and organic matter to bioengineered sediments on glacier’s surfaces. Sci. Total Environ. 2022, 807, 150874. [Google Scholar] [CrossRef] [PubMed]
- Hašler, P.; Dvořák, P.; Poulíčková, A.; Casamatta, D.A. A novel genus Ammassolinea gen. nov. (Cyanobacteria) isolated from sub–tropical epipelic habitats. Fottea 2014, 14, 241–248. [Google Scholar] [CrossRef] [Green Version]
- Malone, C.F.d.S.; Rigonato, J.; Laughinghouse, H.D.; Schmidt, É.C.; Bouzon, Z.L.; Wilmotte, A.; Sant’Anna, C.L. Cephalothrix gen. nov. (Cyanobacteria): Towards an intraspecific phylogenetic evaluation by multilocus analyses. Int. J. Syst. Evol. Microbiol. 2015, 65, 2993–3007. [Google Scholar] [CrossRef]
- Rasoulouniriana, D.; Siboni, N.; Ben-Dov, E.; Kramarsky-Winter, E.; Loya, Y.; Kushmaro, A. Pseudoscillatoria coralii gen. nov., sp nov., a cyanobacterium associated with coral black band disease (BBD). Dis. Aquat. Org. 2009, 87, 91–96. [Google Scholar] [CrossRef]
- Ichimura, T. Isolation and culture methods of algae. Methods Phycol. Stud. 1979, 2, 294–305. [Google Scholar]
- Neilan, B.A.; Jacobs, D.; Goodman, A.E. Genetic diversity and phylogeny of toxic cyanobacteria determined by DNA polymorphisms within the phycocyanin locus. Appl. Environ. Microbiol. 1995, 61, 3875–3883. [Google Scholar] [CrossRef] [Green Version]
- Edwards, U.; Rogall, T.; Blöcker, H.; Emde, M.; Böttger, E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar] [CrossRef] [Green Version]
- Gkelis, S.; Rajaniemi, P.; Vardaka, E.; Moustaka–gouni, M.; Lanaras, T.; Sivonen, K. Limnothrix redekei (Van Goor) Meffert (Cyanobacteria) strains from Lake Kastoria, Greece from a separate phylogenetic group. Microb. Ecol. 2005, 49, 176–182. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Page, R.D.M. TreeView: An application to display phylogenetic trees on personal computers. Bioinformatics 1996, 12, 357–358. [Google Scholar] [CrossRef] [Green Version]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Rennie, W.; Kanoria, S.; Liu, C.; Mallick, B.; Long, D.; Wolenc, A.; Ding, Y. STarMirDB: A database of microRNA binding sites. RNA Biol. 2016, 13, 554–560. [Google Scholar] [CrossRef]
- Osorio-Santos, K.; Pietrasiak, N.; Bohunická, M.; Miscoe, L.H.; Kováčik, L.; Martin, M.P.; Johansen, J.R. Seven new species of Oculatella (Pseudanabaenales, Cyanobacteria): Taxonomically recognizing cryptic diversification. Eur. J. Phycol. 2014, 49, 450–470. [Google Scholar] [CrossRef] [Green Version]
- Caires, T.A.; Lyra, G.D.; Hentschke, G.S.; da Silva, A.M.S.; de Araujo, V.L.; Sant’Anna, C.L.; Nunes, J.M.D. Polyphasic delimitation of a filamentous marine genus, Capillus gen. nov. (Cyanobacteria, Oscillatoriaceae) with the description of two Brazilian species. Algae 2018, 33, 291–304. [Google Scholar] [CrossRef]
- Pietrasiak, N.; Mühlsteinová, R.; Siegesmund, M.A.; Johansen, J.R. Phylogenetic placement of Symplocastrum (Phormidiaceae, Cyanophyceae) with a new combination S. californicum and two new species: S. flechtnerae and S. torsivum. Phycologia 2014, 53, 529–541. [Google Scholar] [CrossRef]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (Cyanobacterial genera) 2014, using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Miscoe, L.H.; Johansen, J.R.; Vaccarino, M.A.; Pietrasiak, N.; Sherwood, A.R. The diatom flora and cyanobacteria from caves on Kauai, Hawaii. II. Novel cyanobacteria from caves on Kauai, Hawaii. Bibl. Phycol. 2016, 123, 75–152. [Google Scholar]
- Casamatta, D.; Stanić, D.; Gantar, M.; Richardson, L.L. Characterization of Roseofilum reptotaenium (Oscillatoriales, Cyanobacteria) gen. et sp. nov. isolated from Caribbean black band disease. Phycologia 2012, 51, 489–499. [Google Scholar] [CrossRef]
- Strunecky, O.; Komárek, J.; Šmarda, J. Kamptonema (Microcoleaceae, Cyanobacteria), a new genus derived from the polyphyletic Phormidium on the basis of combined molecular and cytomorphological markers. Preslia 2014, 86, 193–207. [Google Scholar]
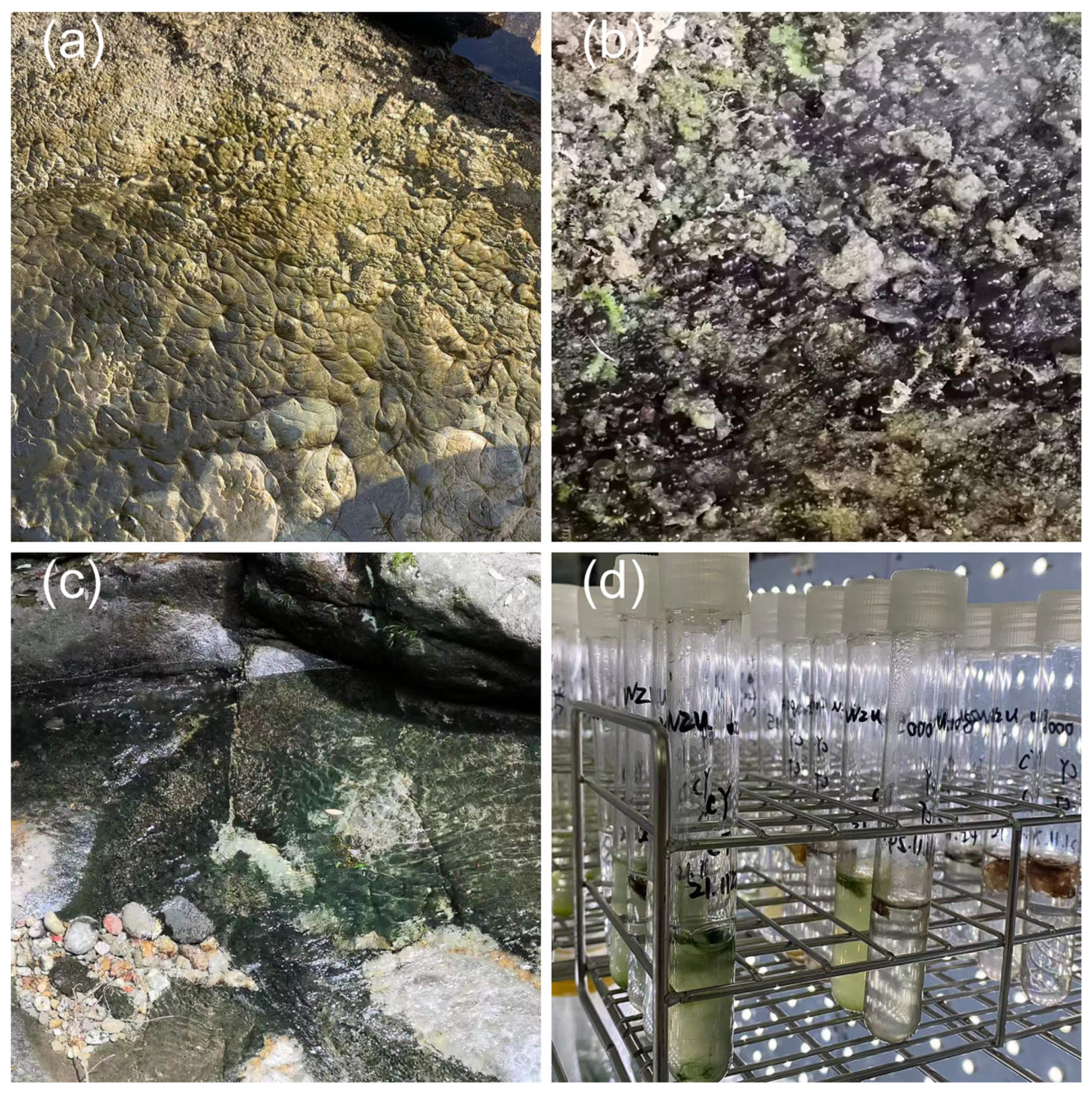
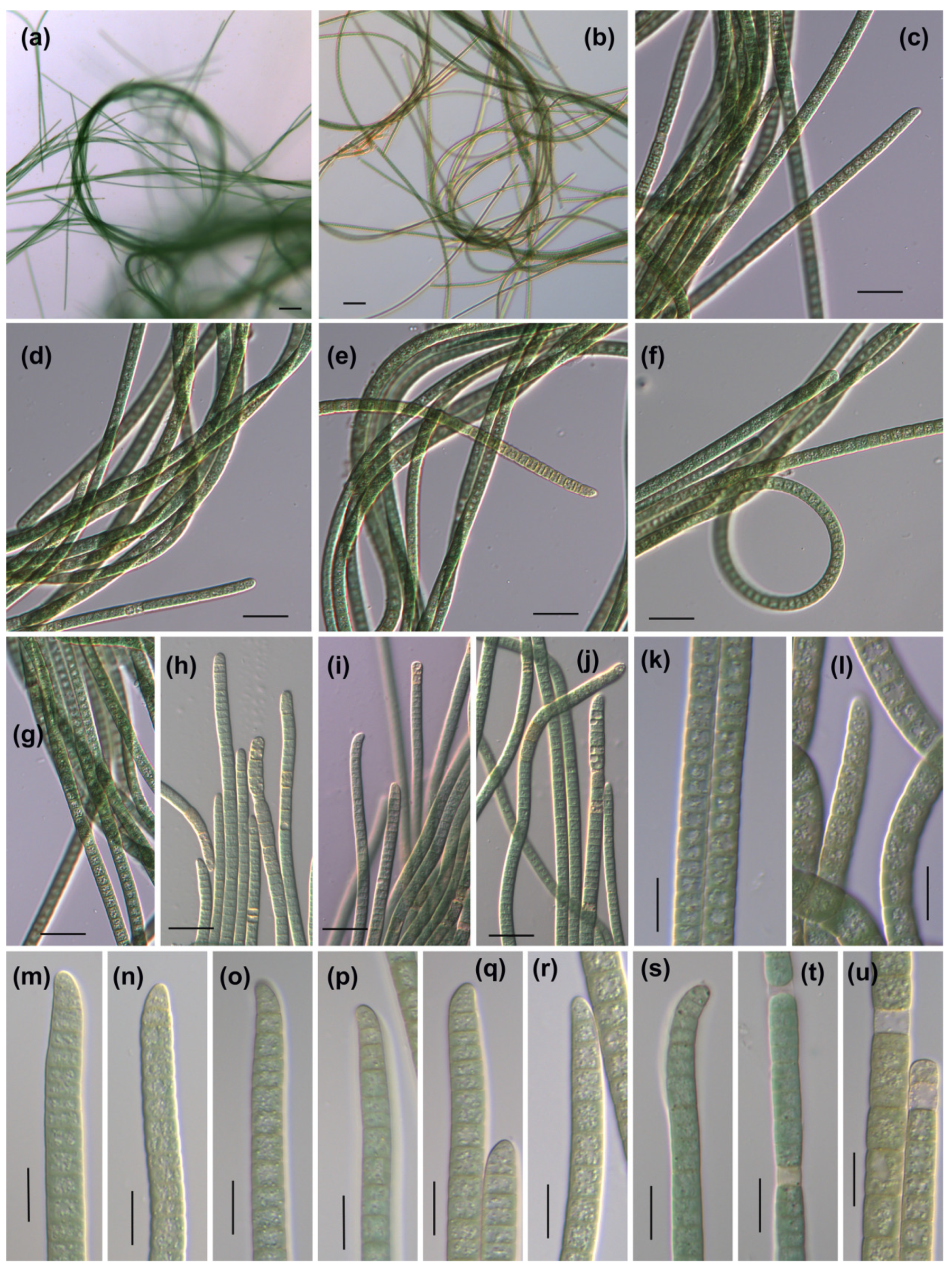
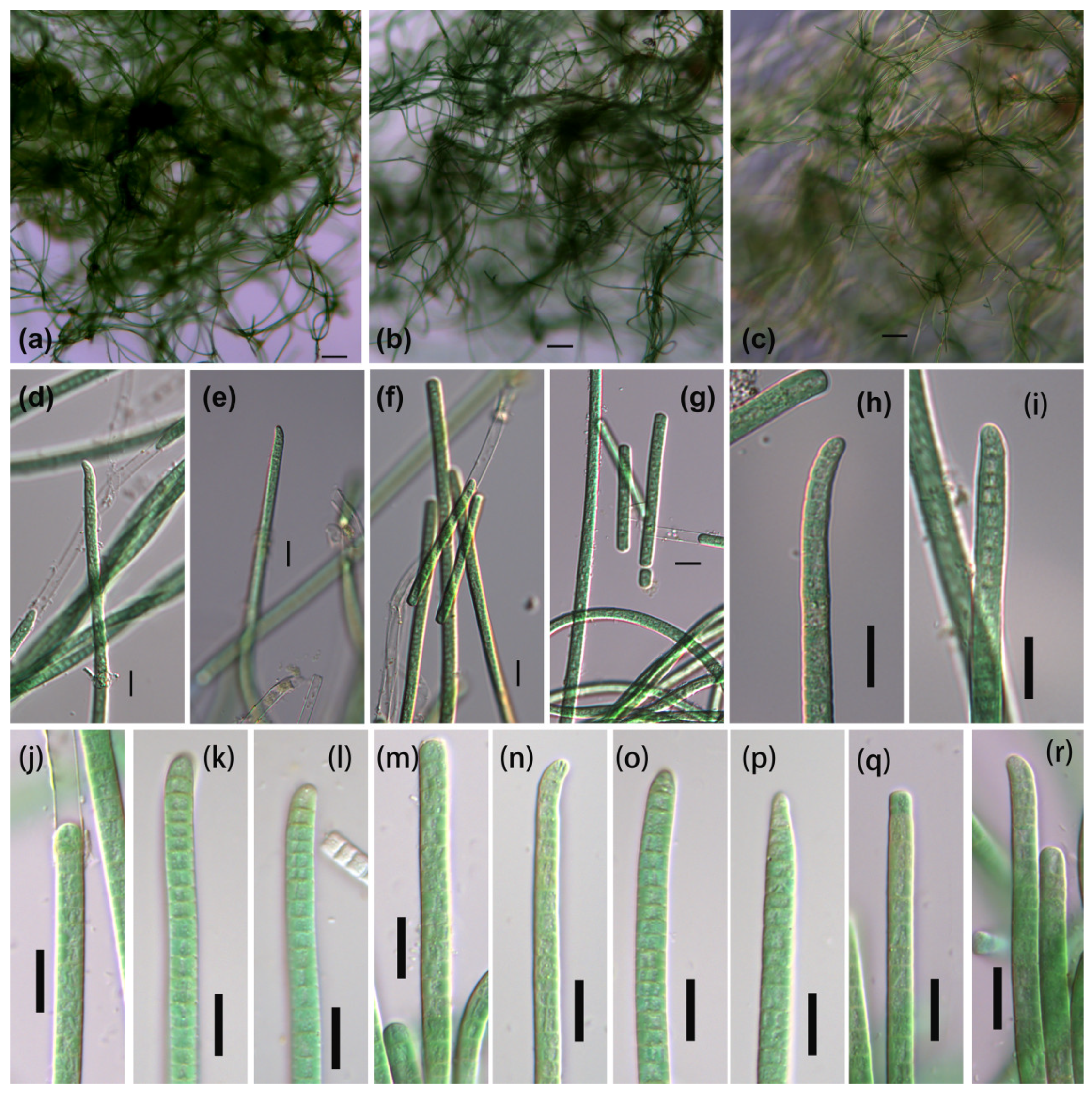

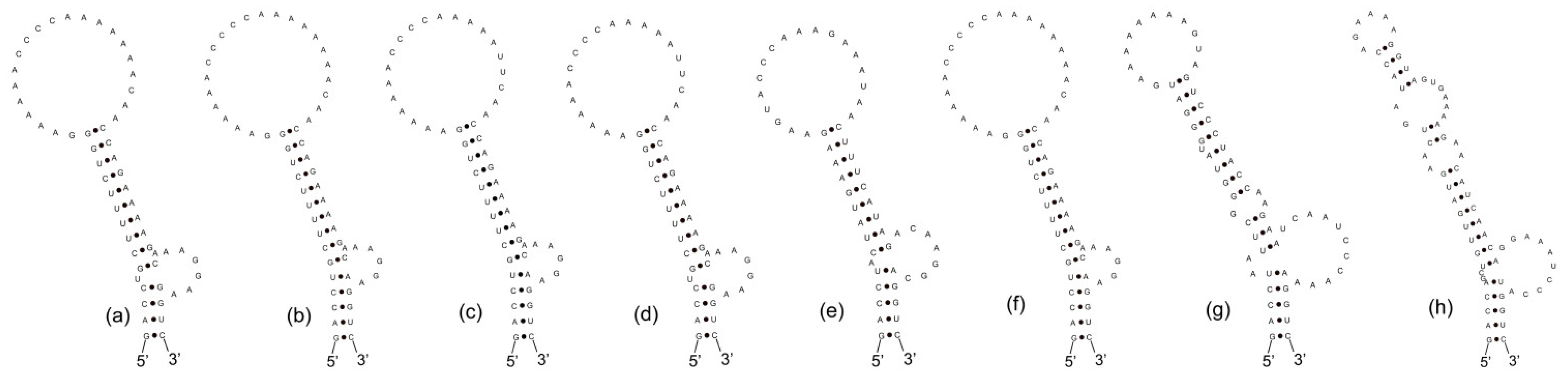


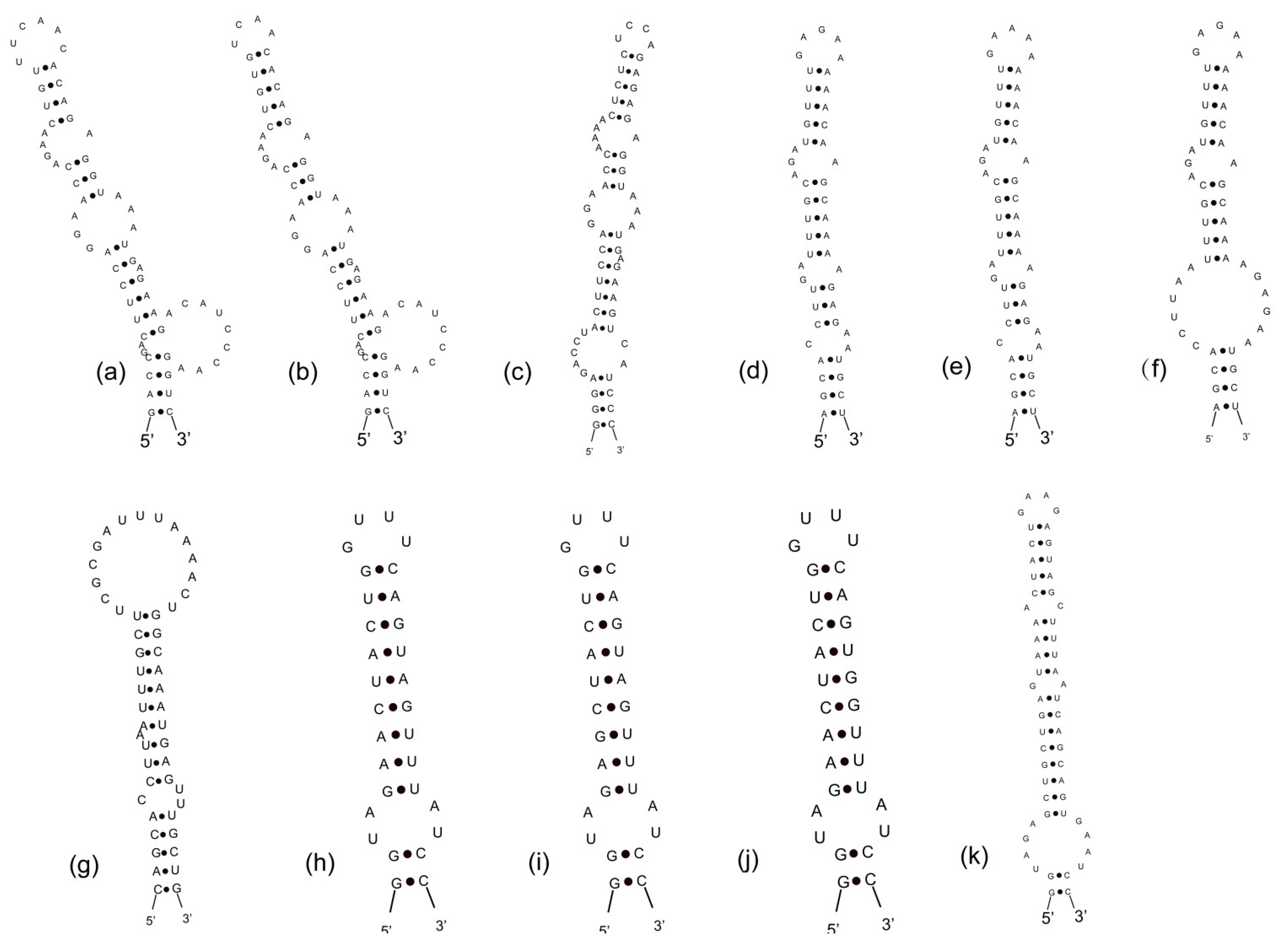
| Strains | Habitat | Locality | Latitude (N) | Longitude (E) |
|---|---|---|---|---|
| WZU 0009, 0011, 0013, 0014 | Benthos | Feiyun River Basin | 27°66′82″ | 120°05′64″ |
| WZU 0153, 0154 | Soil | Dahongyan Scenic Area | 28°81′28″ | 119°66′22″ |
| WZU 0155-0168 | Benthos | Niutou Mountain National Forest Park | 28°66′76″ | 119°49′78″ |
| Strain | GenBank | Complete ITS (nt) | D1–D1′ Helix (nt) | tRNAIle | tRNAAla | Box–B Helix (nt) | Box–A Helix (nt) | D4 | V3 Helix (nt) | D5 |
|---|---|---|---|---|---|---|---|---|---|---|
| WZU 0009 | OL742575 | 441 | 57 | + | + | 39 | 12 | 7 | 43 | 19 |
| WZU 0011 | OL742572 | 441 | 57 | + | + | 39 | 12 | 7 | 43 | 19 |
| WZU 0013 | OL742573 | 445 | 59 | + | + | 39 | 12 | 7 | 43 | 19 |
| WZU 0014 | OL742574 | 441 | 57 | + | + | 40 | 12 | 7 | 43 | 19 |
| 7PC | KT819196 | 437 | 57 | + | + | 40 | 12 | 7 | 43 | 19 |
| 8PC | KT819197 | 438 | 57 | + | + | 40 | 12 | 7 | 43 | 19 |
| 9PC | KT819198 | 436 | 56 | + | + | 40 | 12 | 7 | 43 | 19 |
| 10PC | KT819199 | 515 | 54 | + | + | 38 | 12 | 7 | 54 | 22 |
| 11PC | KT819200 | 364 | 63 | + | - | 39 | 12 | 7 | 12 | 17 |
| 12PC | KT819201 | 409 | 66 | + | - | 33 | 11 | 8 | 44 | 16 |
| 13PC | KT819202 | 516 | 54 | + | + | 38 | 12 | 7 | 54 | 21 |
| WZU 0153 | OM237453 | 511 | 54 | + | + | 36 | 12 | 7 | 54 | 22 |
| WZU 0154 | OM237454 | 511 | 54 | + | + | 36 | 12 | 7 | 54 | 22 |
| Strains | GenBank | Complete ITS (nt) | D1–D1′ Helix (nt) | tRNAIle | tRNAAla | Box–B Helix (nt) | Box–A Helix (nt) | D4 | V3 Helix (nt) | D5 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1PC | KX001786 | 372 | 63 | + | − | 49 | 12 | 7 | 30 | 22 |
| 2PC | KX001787 | 372 | 63 | + | − | 49 | 12 | 7 | 30 | 22 |
| 38PC | KX001788 | 372 | 63 | + | − | 48 | 12 | 7 | 30 | 22 |
| WZU 0155 | OM237439 | 394 | 61 | + | − | 49 | 12 | 7 | 54 | 22 |
| WZU 0156 | OM237440 | 394 | 61 | + | − | 49 | 12 | 7 | 54 | 22 |
| WZU 0157 | OM237441 | 394 | 61 | + | − | 49 | 12 | 7 | 54 | 22 |
| WZU 0158 | OM237442 | 394 | 61 | + | − | 49 | 12 | 7 | 54 | 22 |
| WZU 0159 | OM237443 | 394 | 61 | + | − | 49 | 12 | 7 | 54 | 22 |
| WZU 0160 | OM237444 | 394 | 61 | + | − | 49 | 12 | 7 | 54 | 22 |
| WZU 0161 | OM237445 | 394 | 61 | + | − | 49 | 11 | 8 | 54 | 22 |
| WZU 0162 | OM237446 | 394 | 61 | + | − | 49 | 12 | 7 | 54 | 22 |
| WZU 0163 | OM237447 | 394 | 61 | + | − | 49 | 12 | 7 | 54 | 22 |
| WZU 0164 | OM237448 | 394 | 61 | + | − | 49 | 12 | 7 | 54 | 22 |
| WZU 0165 | OM237449 | 394 | 61 | + | − | 49 | 12 | 7 | 54 | 22 |
| WZU 0166 | OM237450 | 394 | 61 | + | − | 49 | 12 | 7 | 54 | 22 |
| WZU 0167 | OM237451 | 394 | 61 | + | − | 49 | 12 | 7 | 54 | 22 |
| WZU 0168 | OM237452 | 394 | 61 | + | − | 49 | 12 | 7 | 54 | 22 |
| Strains | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. WZU 0009 | ||||||||||||
| 2. WZU 0011 | 99.74 | |||||||||||
| 3. WZU 0013 | 99.66 | 99.57 | ||||||||||
| 4. WZU 0014 | 99.91 | 99.83 | 99.74 | |||||||||
| 5. 7PC | 99.66 | 99.57 | 99.66 | 99.74 | ||||||||
| 6. 8PC | 99.57 | 99.48 | 99.57 | 99.66 | 99.91 | |||||||
| 7. 9PC | 99.74 | 99.66 | 99.74 | 99.83 | 99.91 | 99.83 | ||||||
| 8. 10PC | 95.94 | 95.84 | 96.13 | 96.03 | 96.12 | 96.03 | 96.39 | |||||
| 9. 11PC | 96.03 | 95.94 | 96.13 | 96.12 | 96.22 | 96.12 | 96.39 | 100.00 | ||||
| 10. 12PC | 95.85 | 95.76 | 95.87 | 95.94 | 96.04 | 95.14 | 96.13 | 99.57 | 99.65 | |||
| 11. 13PC | 95.55 | 95.46 | 95.79 | 95.64 | 95.74 | 95.65 | 96.04 | 99.48 | 99.48 | 99.22 | ||
| 12. WZU 0153 | 95.74 | 95.66 | 95.75 | 95.84 | 96.13 | 96.03 | 96.03 | 99.83 | 99.83 | 99.39 | 99.31 | |
| 13. WZU 0154 | 95.93 | 95.84 | 95.75 | 96.03 | 96.12 | 96.03 | 96.03 | 100.00 | 100.00 | 99.57 | 99.48 | 99.83 |
| Strains | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. P. aerugineocaerulea 1PC | ||||||||||||||||
| 2. P. aerugineocaerulea 2PC | 99.93 | |||||||||||||||
| 3. P. aerugineocaerulea 38PC | 99.46 | 99.52 | ||||||||||||||
| 4. WZU 0155 | 98.41 | 98.48 | 98.69 | |||||||||||||
| 5. WZU 0156 | 98.41 | 98.48 | 98.69 | 100 | ||||||||||||
| 6. WZU 0157 | 98.41 | 98.48 | 98.69 | 100 | 100 | |||||||||||
| 7. WZU 0158 | 98.41 | 98.48 | 98.69 | 100 | 100 | 100 | ||||||||||
| 8. WZU 0159 | 98.41 | 98.48 | 98.69 | 100 | 100 | 100 | 100 | |||||||||
| 9. WZU 0160 | 98.41 | 98.48 | 98.69 | 100 | 100 | 100 | 100 | 100 | ||||||||
| 10. WZU 0161 | 98.41 | 98.48 | 98..69 | 100 | 100 | 100 | 100 | 100 | 100 | |||||||
| 11. WZU 0162 | 98.41 | 98.48 | 98.69 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ||||||
| 12. WZU 0163 | 98.41 | 98.48 | 98.69 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |||||
| 13. WZU 0164 | 98.41 | 98.48 | 98.69 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ||||
| 14. WZU 0165 | 98.20 | 98.27 | 98.49 | 99.53 | 99.53 | 99.53 | 99.53 | 99.53 | 100 | 100 | 100 | 100 | 100 | |||
| 15. WZU 0166 | 98.13 | 98.20 | 98.42 | 99.46 | 99.46 | 99.46 | 99.46 | 99.46 | 99.46 | 99.46 | 99.46 | 99.46 | 99.46 | 99.93 | ||
| 16. WZU 0167 | 98.20 | 98.27 | 98.49 | 99.53 | 99.53 | 99.53 | 99.53 | 99.53 | 99.53 | 99.53 | 99.53 | 99.53 | 99.53 | 100 | 99.93 | |
| 17. WZU 0168 | 98.41 | 98.48 | 98.69 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99.53 | 99.46 | 99.53 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Y.; Geng, R.; Shan, L.; Liu, Y.; Zhang, H.; Xiao, P.; Ma, Z.; Li, R. Taxonomic Discussion on Cyanobacterial Systematics at Family Level, with Special Regards to Phormidiaceae by Using the Strains of Chinese Newly Recorded Genera Ancylothrix and Potamolinea. Diversity 2022, 14, 301. https://doi.org/10.3390/d14040301
Cheng Y, Geng R, Shan L, Liu Y, Zhang H, Xiao P, Ma Z, Li R. Taxonomic Discussion on Cyanobacterial Systematics at Family Level, with Special Regards to Phormidiaceae by Using the Strains of Chinese Newly Recorded Genera Ancylothrix and Potamolinea. Diversity. 2022; 14(4):301. https://doi.org/10.3390/d14040301
Chicago/Turabian StyleCheng, Yao, Ruozhen Geng, Liang Shan, Yang Liu, He Zhang, Peng Xiao, Zengling Ma, and Renhui Li. 2022. "Taxonomic Discussion on Cyanobacterial Systematics at Family Level, with Special Regards to Phormidiaceae by Using the Strains of Chinese Newly Recorded Genera Ancylothrix and Potamolinea" Diversity 14, no. 4: 301. https://doi.org/10.3390/d14040301
APA StyleCheng, Y., Geng, R., Shan, L., Liu, Y., Zhang, H., Xiao, P., Ma, Z., & Li, R. (2022). Taxonomic Discussion on Cyanobacterial Systematics at Family Level, with Special Regards to Phormidiaceae by Using the Strains of Chinese Newly Recorded Genera Ancylothrix and Potamolinea. Diversity, 14(4), 301. https://doi.org/10.3390/d14040301









