Ex Situ Conservation of Plant Genetic Resources: An Overview of Chickpea (Cicer arietinum L.) and Lentil (Lens culinaris Medik.) Worldwide Collections
Abstract
1. Introduction
Scope and Focus of This Review
2. Assembly and Management of Chickpea and Lentil Collections
3. The Passport Data of the Collections
4. Non-Omic and Omic Relative to the Two Collections
5. Conclusions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ladizinsky, G. The origin of lentil and its wild gene pool. Euphytica 1979, 28, 179–187. [Google Scholar] [CrossRef]
- Lev-Yadun, S.; Gopher, A.; Abbo, S. The cradle of agriculture. Science 2000, 288, 1062–1063. [Google Scholar] [CrossRef] [PubMed]
- Bellon, M.R. Conceptualizing interventions to support on-farm genetic resources conservation. World Dev. 2004, 32, 159–172. [Google Scholar] [CrossRef]
- Zander, P.; Amjath-Babu, T.S.; Preissel, S.; Reckling, M.; Bues, A.; Schlafke, N.; Kuhlman, T.; Bachinger, J.; Uthes, S.; Stoddard, F.; et al. Grain legume decline and potential recovery in European agriculture: A review. Agron. Sust. Develop. 2016, 36, 26. [Google Scholar] [CrossRef]
- Pinheiro de Carvalho, M.A.A.; Bebel, P.J.; Bettencourt, E.; Costa, G.; Dias, S.; Dos Santos, T.M.M.; Slaski, J.J. Cereal landraces genetic resources in worldwide GeneBanks. A review. Agron. Sust. Develop. 2013, 33, 177–203. [Google Scholar] [CrossRef]
- Bellemou, D.; Millan, T.; Gil, J.; Abdelguerfi, A.; Laouar, M. Genetic diversity and population structure of Algerian chickpea (Cicer arietinum L.) genotypes: Use of agro-morphological traits and molecular markers linked or not linked to the gen or QTL of interest. Crop Pasture Sci. 2020, 71, 155–170. [Google Scholar] [CrossRef]
- Piergiovanni, A.R.; Margiotta, B. On farm survival of Apulian legume and cereal landraces in relation to land cover/land use changes. A case study. Ital. J. Agron. 2021, 16, 67–75. [Google Scholar] [CrossRef]
- Thanopoulos, R.; Chatzigeorgiou, T.; Argyropoulou, K.; Kostouros, N.M.; Bebeli, P.J. State of crop landraces in Arcadia (Greece) and in-situ conservation potential. Diversity 2021, 13, 558. [Google Scholar] [CrossRef]
- FAO. Second Global Plan of Action for Plant Genetic Resources for Food and Agriculture; Food and Agriculture Organization of the United Nations: Rome, Italy, 2011; p. 51. [Google Scholar]
- Van Treuren, R.; Van Hintum, T.J.L. Next-generation genebanking: Plant genetic resources management and utilization in the sequencing era. Plant Genet. Resour. 2014, 12, 298–307. [Google Scholar] [CrossRef]
- Engels, J.M.M.; Ebert, A.W. A Critical Review of the Current Global Ex Situ Conservation System for Plant Agrobiodiversity. I. History of the Development of the Global System in the Context of the Political/Legal Framework and Its Major Conservation Components. Plants 2021, 10, 1557. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Gowda, C.L.L.; Sastry, D.V.S.S.R. Plant genetic resources management: Collection, characterization, conservation and utilization. J. SAT Agric. Res. 2008, 6, 16. [Google Scholar]
- Barulina, E.I. Lentils of the USSR and other countries. Bull. Appl. Bot. Genet. Plant Breed. 1930, 40, 265–304. [Google Scholar]
- Tullu, A.; Diederichsen, A.; Suvorova, G.; Vadenberg, A. Genetic and genomic resources of lentil: Status; use and prospects. Plant Genet. Resour. Charact. Util. 2011, 9, 19–29. [Google Scholar] [CrossRef]
- Petrova, S.; Angelova, S. Status of the national chickpea collection in Bulgaria. Bull. Transilv. Univ. Brasov. Med. Sci. Ser. VI 2011, 4, 73–80. [Google Scholar]
- Casals, J.; Casaňas, F.; Simό, J. Is It Still Necessary to Continue to Collect Crop Genetic Resources in the Mediterranean Area? A Case Study in Catalonia. Econ. Bot. 2017, 71, 330–341. [Google Scholar] [CrossRef]
- Saker, A.; Bayaa, B.; Erskine, W. Registration of six lentil germplasm lines with resistance to vascular wilt. Crop Sci. 2001, 41, 1655. [Google Scholar] [CrossRef]
- El-Ashkar, F.; Sarker, A.; Erskine, W.; Bayaa, B.; El-Hassan, H.; Kadah, N.; Karim, B.A. Registration of ‘Idlib-4’ lentil. Crop-Sci. 2004, 44, 2261–2262. [Google Scholar] [CrossRef]
- El-Ashkar, F.; Sarker, A.; Erskine, W.; Bayaa, B.; El-Hassan, H.; Kadah, N.; Karim, B.A. Registration of ‘Idlib-3’ lentil. Crop-Sci. 2004, 44, 2261. [Google Scholar] [CrossRef]
- Siddique, K.H.M.; Regan, K.L. Registration of ‘Kimberley Large’ kabuli chickpea. Crop-Sci. 2005, 45, 1659–1660. [Google Scholar] [CrossRef]
- Bibi, N.; Khattak, A.B.; Khattak, G.S.S.; Mehmood, Z.; Ihsanullah, I. Quality and consumer acceptability studies and their inter-relationship of newly evolved desi type chickpea genotypes (Cicer arietinum L.). Int. J. Food Sci. Technol. 2007, 42, 528–534. [Google Scholar] [CrossRef]
- Halewood, M.; Jamora, N.; Lopez Noriega, I.; Anglin, N.L.; Wenzl, P.; Payne, T.; Ndjiondjop, M.N.; Guarino, L.; Lava Kumar, P.; Yazbek, M.; et al. Germplasm Acquisition and Distribution by CGIAR Genebanks. Plants 2020, 9, 1296. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.H.D. Core collections: A practical approach to genetic resources management. Genome 1989, 31, 818–824. [Google Scholar] [CrossRef]
- Pande, S.; Sharma, M. Resistance to Ascochyta blight in chickpea (Cicer arietinum L.). Legume Perspect. 2014, 3, 20–22. [Google Scholar]
- Upadhyaya, H.D.; Ortiz, R. A mini core subset for capturing diversity and promoting utilization of chickpea genetic resources in crop improvement. Theor. Appl. Genet. 2001, 102, 1292–1298. [Google Scholar] [CrossRef]
- Archak, S.; Tyagi, R.K.; Harer, P.N.; Mahase, L.B.; Singh, N.; Dahiya, O.P.; Nizar, M.A.; Singh, M.; Tilekar, V.; Kumar, V.; et al. Characterization of chickpea germplasm conserved in the Indian National Genebank and development of a core set using qualitative and quantitative trait data. Crop J. 2016, 4, 417–424. [Google Scholar] [CrossRef]
- Tripathi, K.; Kumari, J.; Gore, P.G.; Mishra, D.C.; Singh, A.K.; Mishra, G.P.; Gayacharan, C.; Dikshit, H.K.; Singh, N.; Semwal, D.P.; et al. Agro-morphological characterization of lentil germplasm of Indian National genebank and development of a core set for efficient utilization in lentil improvement programs. Front. Plant Sci. 2022, 12, 751429. [Google Scholar] [CrossRef]
- Wambugu, P.W.; Ndjiondjop, M.N.; Henry, R.J. Role of genomics in promoting the utilization of plant genetic resources in genebanks. Brief. Funct. Genom. 2018, 17, 198–206. [Google Scholar] [CrossRef]
- Fayaz, H.; Mir, A.H.; Tyagi, S.; Wani, A.A.; Jan, N.; Yasin, M.; Mir, J.I.; Mondal, B.; Khan, M.A.; Mir, R.R. Assessment of molecular genetic diversity of 384 chickpea genotypes and development of core set of 192 genotypes for chickpea improvement programs. Genet. Res. Crop Evol. 2022, 69, 1193–1205. [Google Scholar] [CrossRef]
- Muehlbauer, F.J.; Sarker, A. Economic Importance of Chickpea: Production, Value, and World Trade. In The Chickpea Genome; Varshney, R.K., Thudi, M., Muehlbauer, F., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 5–12. [Google Scholar]
- Jha, U.C.; Nayyar, H.; Jha, R.; Nath, C.P.; Datta, D. Chickpea Breeding for Abiotic Stress: Breeding Tools and ‘Omics’ Approaches for Enhancing Genetic Gain. In Accelerated Plant Breeding; Gosal, S.S., Wani, S.H., Eds.; Springer: Cham, Switzerland, 2020; Volume 3. [Google Scholar]
- Singh, N.; Wu, S.; Raupp, W.J.; Sehgal, S.; Arora, S.; Tiwari, V.; Vikram, P.; Singh, S.; Chhuneja, P.; Gill, B.S.; et al. Efficient curation of genebanks using next generation sequencing reveals substantial duplication of germplasm accessions. Sci. Rep. 2019, 9, 650. [Google Scholar] [CrossRef]
- Weise, S.; Lohwasser, U.; Oppermann, M. Document or Lose It—On the Importance of Information Management for Genetic Resources Conservation in Genebanks. Plants 2020, 9, 1050. [Google Scholar] [CrossRef]
- Davidson, L.A.; Kimberly, D. Digital Object Identifiers: Promise and Problems for Scholarly Publishing. J. Electron. Publ. 1998, 4. [Google Scholar] [CrossRef]
- Varshney, R.K.; Roorkiwal, M.; Sun, S.; Bajaj, P.; Chitikineni, A.; Thudi, M.; Singh, N.P.; Du, X.; Upadhyaya, H.D.; Khan, A.W.; et al. A chickpea genetic variation map based on the sequencing of 3366 genomes. Nature 2021, 599, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Villa, T.C.; Maxted, N.; Scholten, M.A.; Ford-Lloyd, B.V. Defining and identifying crop landraces. Plant Genet. Resour. 2005, 3, 373–384. [Google Scholar] [CrossRef]
- Casaňas, F.; Simό, J.; Casals, J.; Prohens, J. Toward an evolved concept of landrace. Front. Plant Sci. 2017, 8, 145. [Google Scholar] [CrossRef]
- Ceccarelli, S. Landraces: Importance and use in breeding and environmentally friendly agronomic systems. In Agrobiodiversity Conservation: Securing the Diversity of Crop Wild Relatives and Landraces; Maxted, N., Dulloo, M.E., Ford-Lloyd, B.V., Frese, L., Iriondo, J.M., de Carvalho, M.A.A.P., Eds.; CAB International: Wallingford, UK, 2012; pp. 103–117. [Google Scholar]
- Singh, A.K.; Varaprasad, K.S.; Venkateswaran, K. Conservation costs of plant genetic resources for food and agriculture: Seed genebanks. Agric. Res. 2012, 1, 223–239. [Google Scholar] [CrossRef]
- Ebert, A.W.; Engels, J.M.M. Plant Biodiversity and Genetic Resources Matter! Plants 2020, 9, 1706. [Google Scholar] [CrossRef]
- Gaur, P.M.; Samineni, S.; Thudi, M.; Tripathi, S.; Sajja, S.B.; Jayalakshmi, V.; Mannur, D.M.; Vijayakumar, A.G.; Rao, N.; Ojiewo, C.; et al. Integrated breeding approaches for improving drought and heat adaptation in chickpea (Cicer arietinum L.). Plant Breed. 2019, 138, 389–400. [Google Scholar] [CrossRef]
- Brink, M.; van Hintum, T. Practical consequence of digital sequence information (DSI) definitions and access and benefit sharing scenarios from a plant genebank’s perspective. Plants People Planet 2021, 4, 23–32. [Google Scholar] [CrossRef]
- Akinlade, O.J.; Voss-Fels, K.; Costilla, R.; Kholova, J.; Choudhary, S.; Varshney, R.K.; Hickey, L.T.; Smith, M.R. Designing chickpea for a hotter dried world. Euphytica 2022, 218, 100. [Google Scholar] [CrossRef]
- Gupta, S.; Kumar, T.; Verma, S.; Bharadwaj, C.; Bhatia, S. Development of gene-based markers for use in construction of the chickpea (Cicer arietinum L.) genetic linkage map and identification of QTLs associated with seed weight and plant height. Mol. Biol. Rep. 2015, 42, 1571–1580. [Google Scholar] [CrossRef]
- Li, Y.; Ruperao, P.; Batley, J.; Edwards, D.; Davidson, J.; Hobson, K.; Sutton, T. Genome analysis identified novel candidate genes for ascochyta blight resistance in chickpea using whole genome resequencing data. Front. Plant Sci. 2017, 8, 359. [Google Scholar] [PubMed]
- Verma, P.; Goyal, R.; Chahota, R.K.; Sharma, T.R.; Abdin, M.Z.; Bhatia, S. Construction of a genetic linkage map and identification of QTLs for seed weight and seed size traits in lentil (Lens culinaris Medik.). PLoS ONE 2015, 10, e0139666. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, K.; Coyne, C.J.; Zheng, P.; Saha, G.; Main, D.; Amin, N.; Ma, Y.; Kisha, T.; Bett, K.E.; McGeea, R.J.; et al. Genetic diversity and GWAS of agronomic traits using ICARDA lentil (Lens culinaris Medik.) reference plus collection. Plant Genet. Resour. Charact. Util. 2021, 19, 279–288. [Google Scholar] [CrossRef]
- Sokolkova, A.; Bulyntsev, S.V.; Chang, P.L.; Carrasquilla-Garcia, N.; Igolkina, A.A.; Noujdina, N.V.; von Wettberg, E.; Vishnyakova, M.A.; Cook, D.R.; Nuzhdin, S.V.; et al. Genomic analysis of Vavilov’s historic chickpea landraces reveals footprints of environmental and human selection. Int. J. Mol. Sci. 2020, 21, 3952. [Google Scholar] [CrossRef]
- Khazaei, H.; Caron, C.T.; Fedoruk, M.; Diapari, M.; Vandenberg, A.; Coyne, C.J.; McGee, R.; Bett, K.E. Genetic diversity of cultivated lentil (Lens culinaris Medik.) and its relation to the world’s agro-ecological zones. Front. Plant Sci. 2016, 7, 1093. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, M.; Materne, M.; Cogan, N.O.I.; Rodda, M.; Daetwyler, H.D.; Slater, A.T.; Forster, J.W.; Kaur, S. Assessment of genetic variation within a global collection of lentil (Lens culinaris Medik.) cultivars and landraces using SNP markers. BMC Genet. 2014, 15, 150. [Google Scholar] [CrossRef]
- Idrissi, O.; Piergiovanni, A.R.; Toklu, F.; Houasli, C.; Udupa, S.M.; De Keyser, E.; Van Damme, P.; De Riek, J. Molecular variance and population structure of lentil (Lens culinaris Medik.) landraces from Mediterranean countries as revealed by simple sequence repeat DNA markers: Implications for conservation and use. Plant Genet. Resour. Charact. Util. 2018, 16, 249–259. [Google Scholar] [CrossRef]
- Li, Y.; Ruperao, P.; Batley, J.; Edwards, D.; Martin, W.; Hobson, K.; Sutton, T. Genomic prediction of preliminary yield trials in chickpea: Effect of functional annotation of SNPs and environment. Plant Genome 2021, 15, e20166. [Google Scholar] [CrossRef]
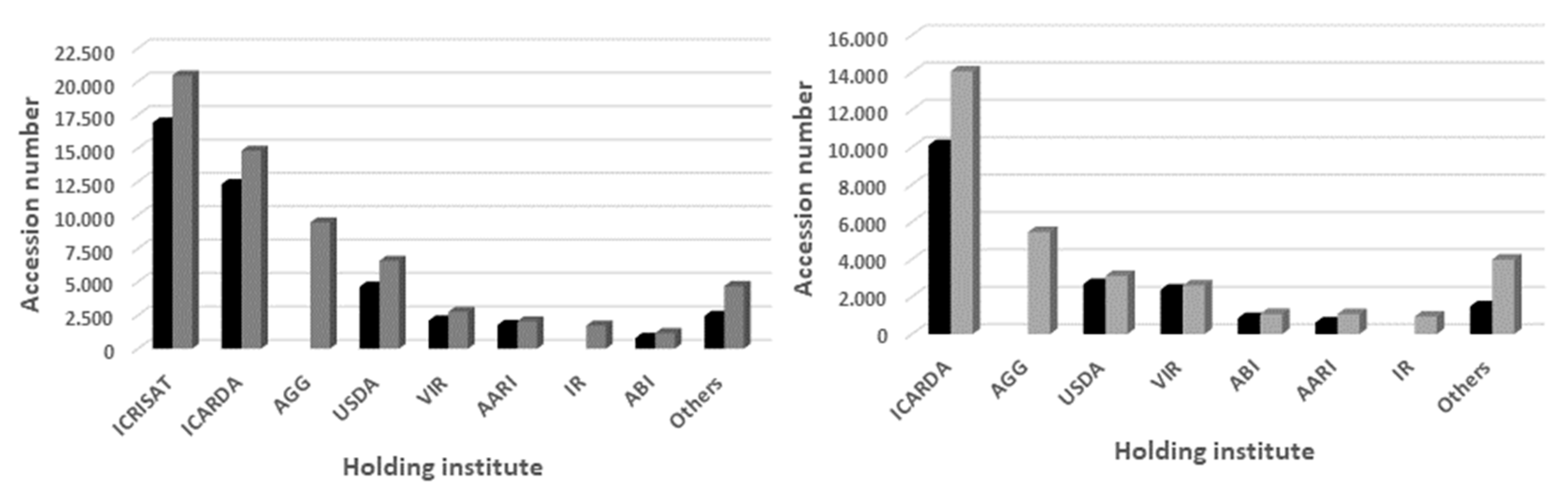
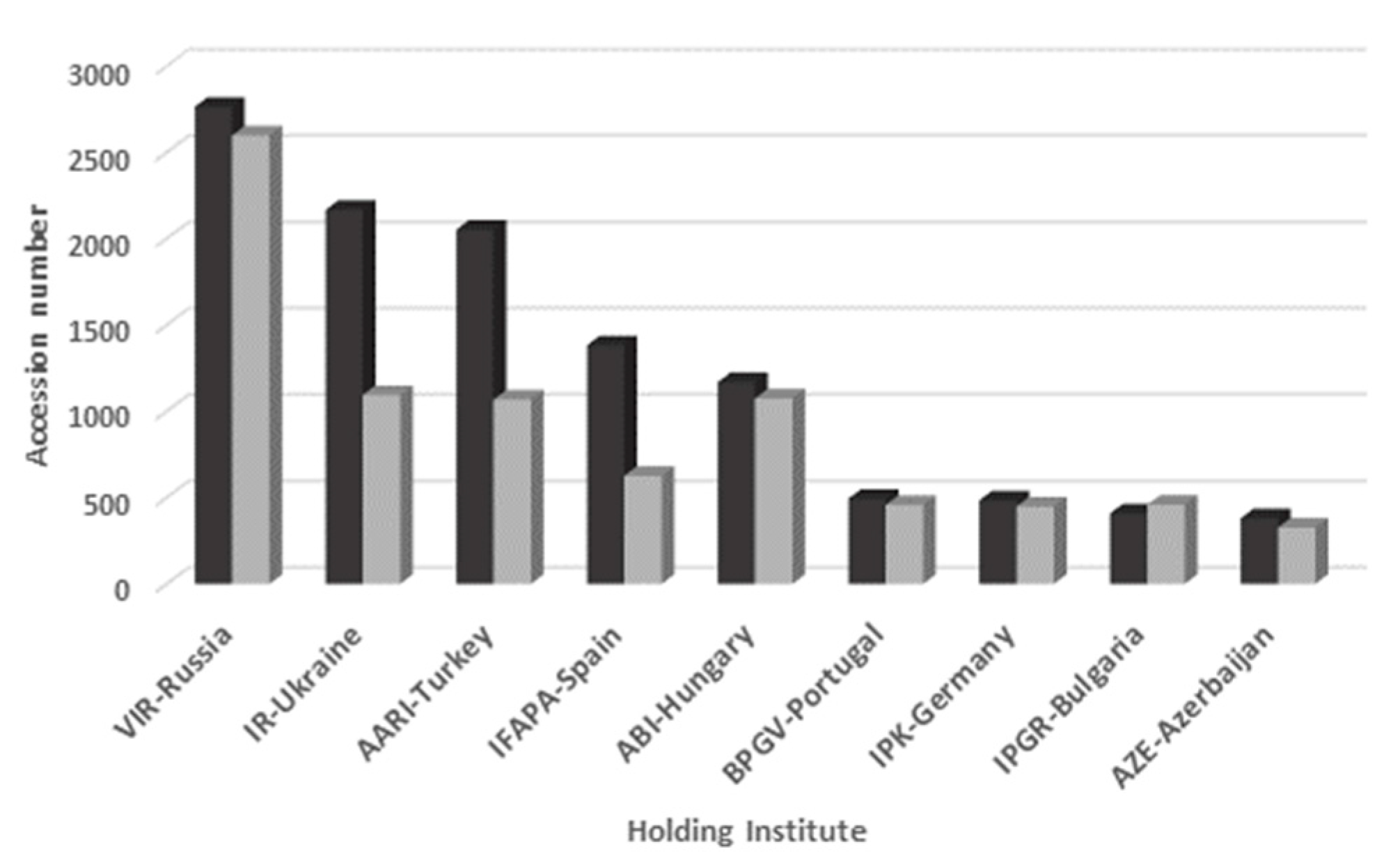
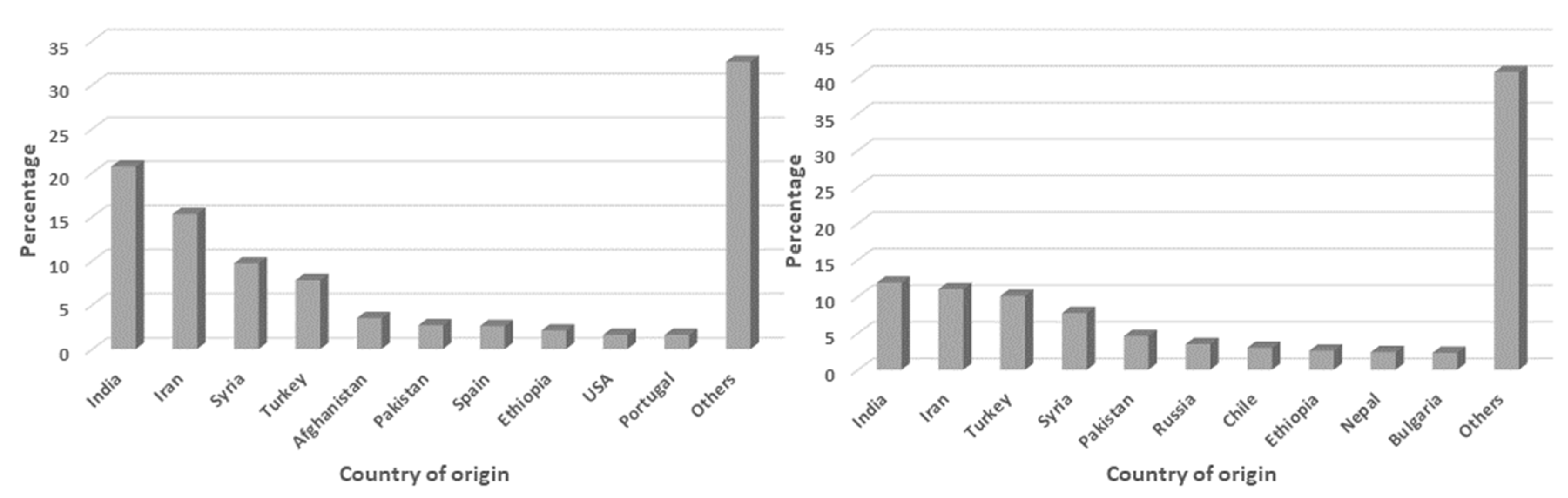
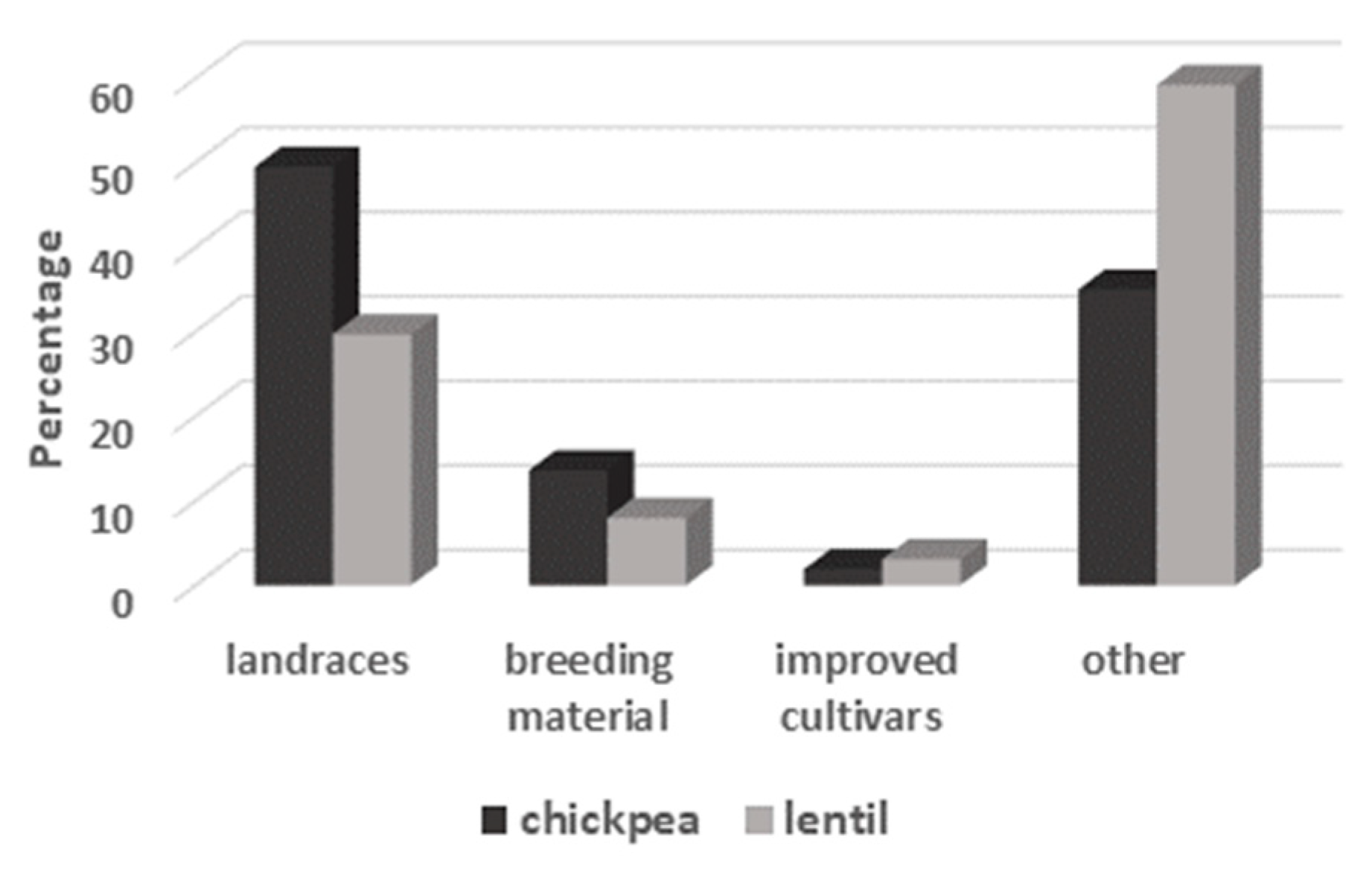
| Genebank Name | No. of Accessions | |
|---|---|---|
| Chickpea | Lentil | |
| International Crops Research Institute for the Semi-Arid Tropics (ICRISAT, India) | 16,928 | ----- |
| International Center for Agricultural Research in the Dry Areas, (ICARDA, Lebanon) | 12,331 | 10,113 |
| United States Department of Agriculture (USDA, USA) | 4652 | 2672 |
| N.I. Vavilov Institute of Plant Industry (VIR, Russia) | 2113 | 2376 |
| Aegean Agricultural Research Institute (AARI, Turkey) | 1790 | 615 |
| Institute for AgroBotany (ABI, Hungary) | 815 | 857 |
| Others | 2467 | 1481 |
| TOTAL | 41,096 | 18,114 |
| Institute Code | GRC005 | CYP004 | ROM007 | ARM059 | ALB026 |
|---|---|---|---|---|---|
| Lentil accessions | |||||
| Total | 97 | 19 | 38 | 34 | 2 |
| Domestic | 97 | 19 | 24 | 22 | 2 |
| Chickpea accessions | |||||
| Total | 177 | 28 | 127 | 42 | 9 |
| Domestic | 177 | 28 | 22 | 36 | 9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piergiovanni, A.R. Ex Situ Conservation of Plant Genetic Resources: An Overview of Chickpea (Cicer arietinum L.) and Lentil (Lens culinaris Medik.) Worldwide Collections. Diversity 2022, 14, 941. https://doi.org/10.3390/d14110941
Piergiovanni AR. Ex Situ Conservation of Plant Genetic Resources: An Overview of Chickpea (Cicer arietinum L.) and Lentil (Lens culinaris Medik.) Worldwide Collections. Diversity. 2022; 14(11):941. https://doi.org/10.3390/d14110941
Chicago/Turabian StylePiergiovanni, Angela Rosa. 2022. "Ex Situ Conservation of Plant Genetic Resources: An Overview of Chickpea (Cicer arietinum L.) and Lentil (Lens culinaris Medik.) Worldwide Collections" Diversity 14, no. 11: 941. https://doi.org/10.3390/d14110941
APA StylePiergiovanni, A. R. (2022). Ex Situ Conservation of Plant Genetic Resources: An Overview of Chickpea (Cicer arietinum L.) and Lentil (Lens culinaris Medik.) Worldwide Collections. Diversity, 14(11), 941. https://doi.org/10.3390/d14110941







