Genome-Wide Genetic Diversity and Population Structure of Local Sudanese Sheep Populations Revealed by Whole-Genome Sequencing
Abstract
1. Introduction
2. Materials and Methods
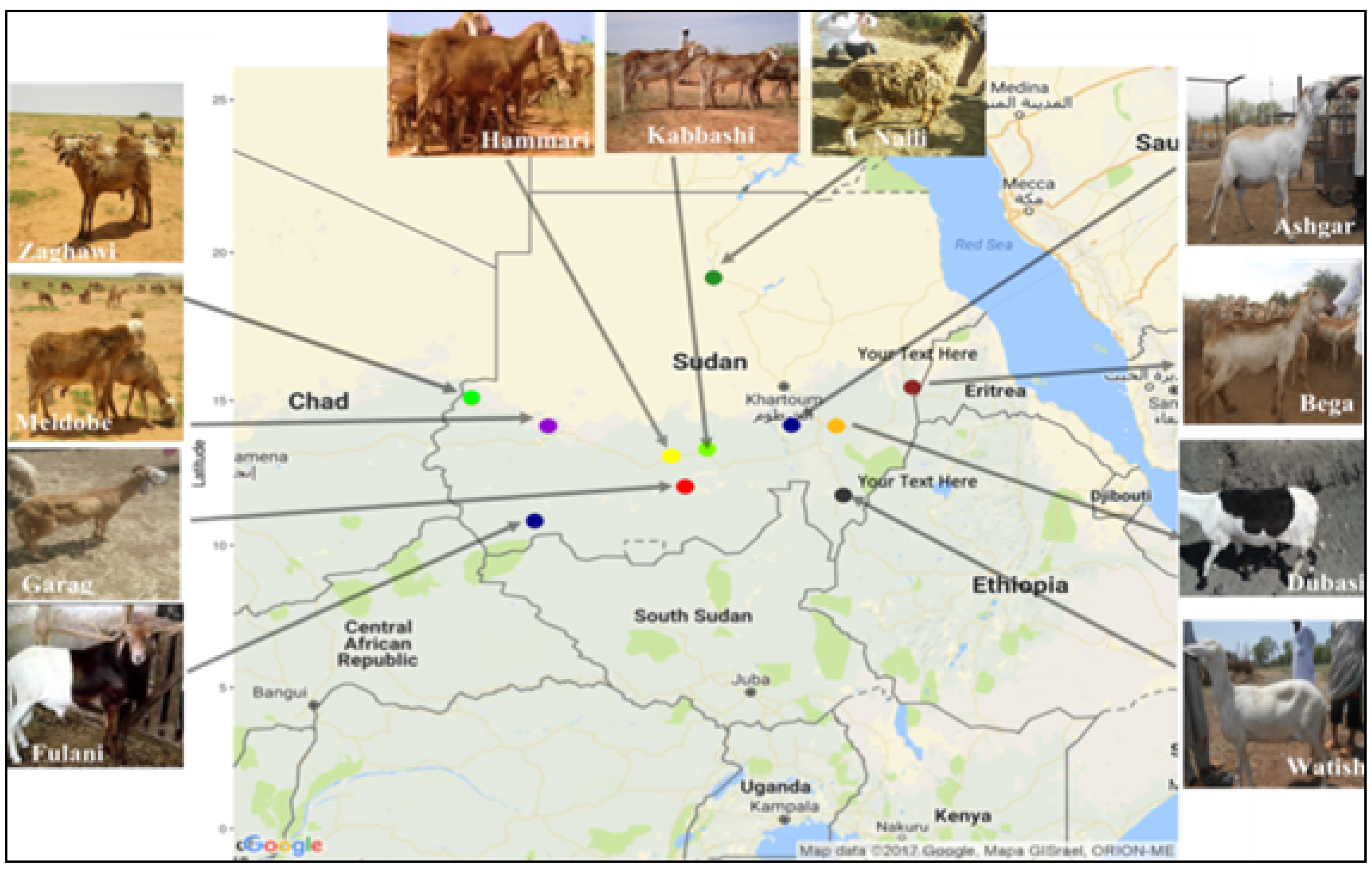
2.1. Sheep Populations, Sample Collection, and DNA Extraction
| Origin | Population | N | Latitude | Longitude | Altitude | Fiber Type | Tail Type | Type |
|---|---|---|---|---|---|---|---|---|
| Sudan | Hammari | 10 | 13.09 | 29.22 | 621.45 | Hairy | Long, thin tail | Desert |
| Kabbashi | 10 | 13.09 | 29.22 | 621.45 | Hairy | Long, thin tail | Desert | |
| Meidobe | 11 | 14.13 | 25.55 | 945.91 | Hairy | Long, thin tail | Desert | |
| Ashgar | 7 | 14.16 | 32.79 | 418.51 | Hairy | Long, thin tail | Desert | |
| Dubasi | 10 | 14.13 | 34.13 | 413.44 | Hairy | Long, thin tail | Desert | |
| Watish | 2 | 11.74 | 34.34 | 484.37 | Hairy | Long, thin tail | Desert | |
| Bega | 8 | 15.45 | 36.39 | 505.42 | Hairy | Long, thin tail | Desert | |
| Zaghawi | 10 | 15.09 | 23.27 | 727.35 | Hairy | Long, thin tail | Arid-upland | |
| Fulani | 10 | 10.85 | 25.15 | 473.85 | Hairy | Short, thin tail | West African | |
| Garag | 7 | 12.04 | 29.63 | 679.42 | Hairy | Short, thin tail | Fused | |
| Naili | 5 | 19.16 | 30.48 | 227.37 | Wooly | Long, thin tail | Reverin | |
| Ethiopia | Kefis | 13 | 9.5 | 40.17 | 890 | Hairy | Fat tail | - |
| Gafera | 10 | 11.52 | 36.90 | 2500 | Hairy | Fat rump | - | |
| Doyogena | 10 | 7.35 | 37.78 | 2324 | Hairy | Fat tail | - | |
| Libya | Barbary | 12 | 32.52 | 15.15 | < 100 | Hairy | Fat tail | - |
| Total | 134 | |||||||
2.2. Whole-Genome Sequencing, Variant Detection, and Annotation
2.3. Data Management and Quality Control
2.4. Genetic Diversity and Differentiation
2.5. Genetic Population Structure Analyses
3. Results
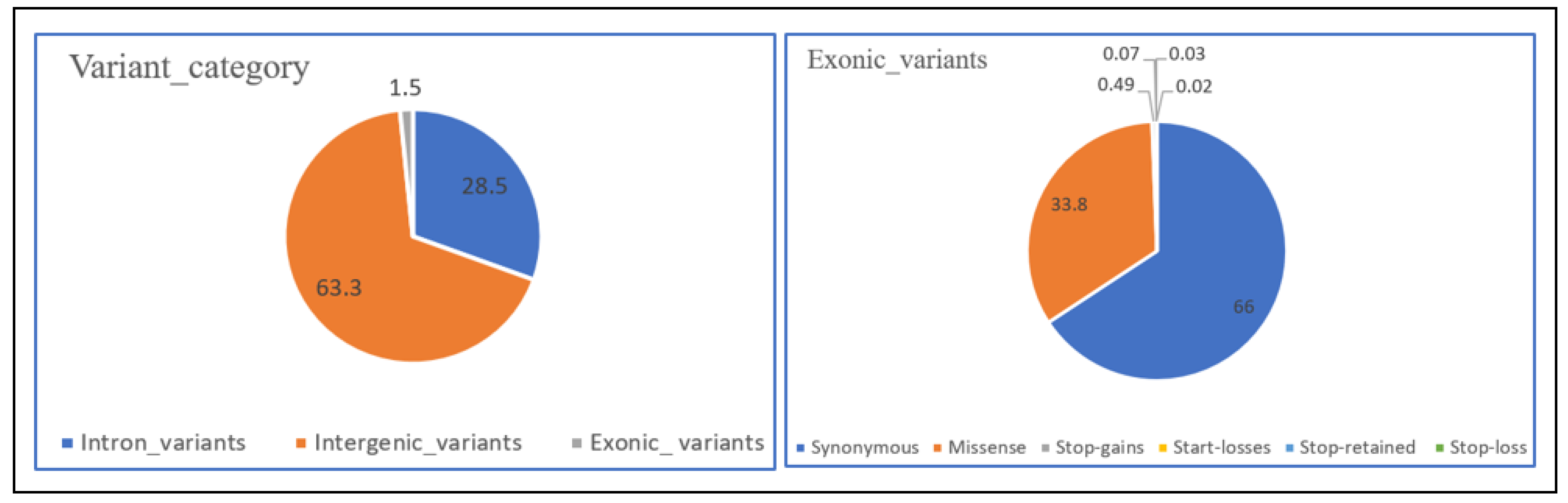
3.1. Variant Detection and Annotation
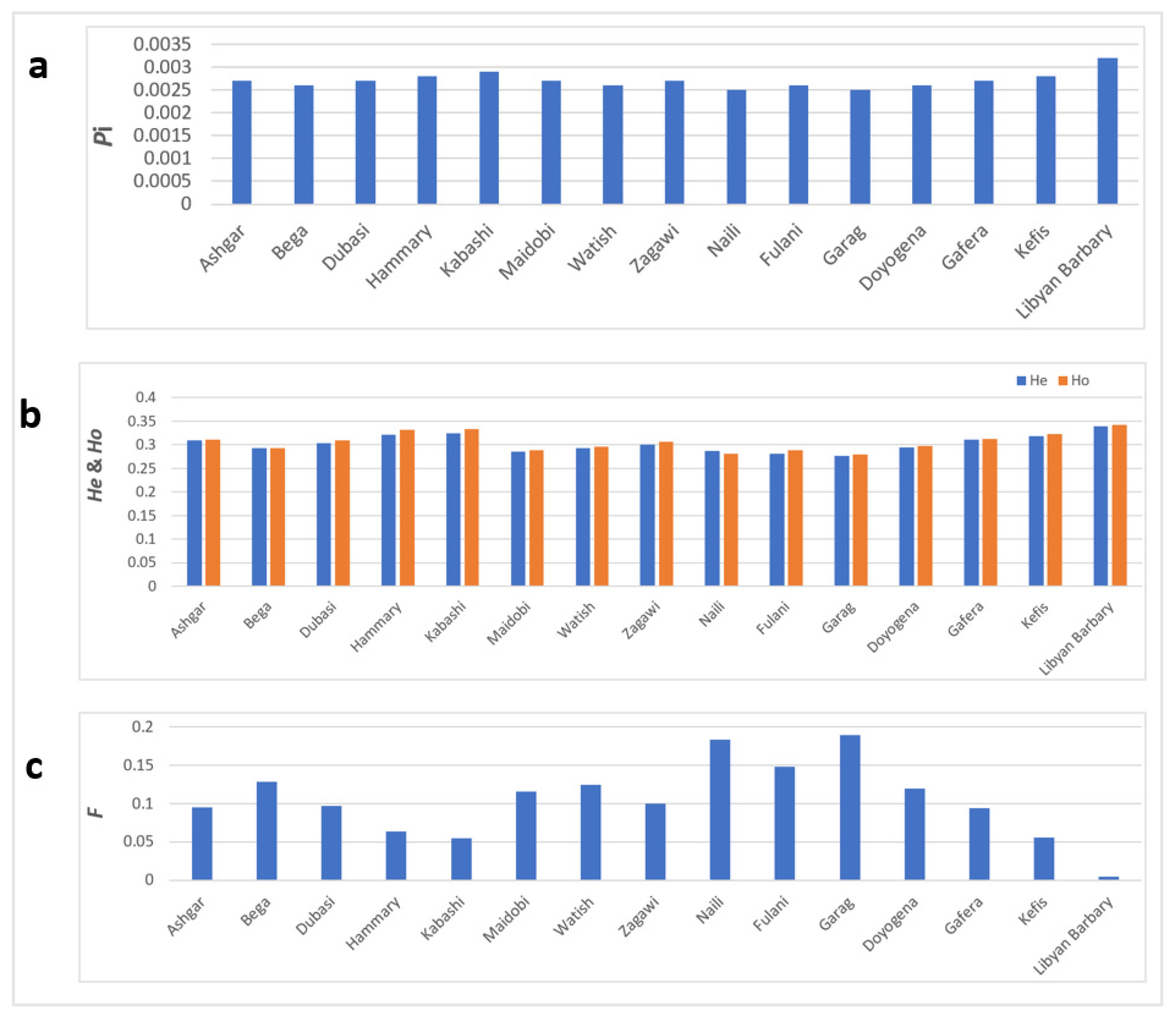
3.2. Estimation of Genetic Diversity
3.3. Population Divergence and Relationships
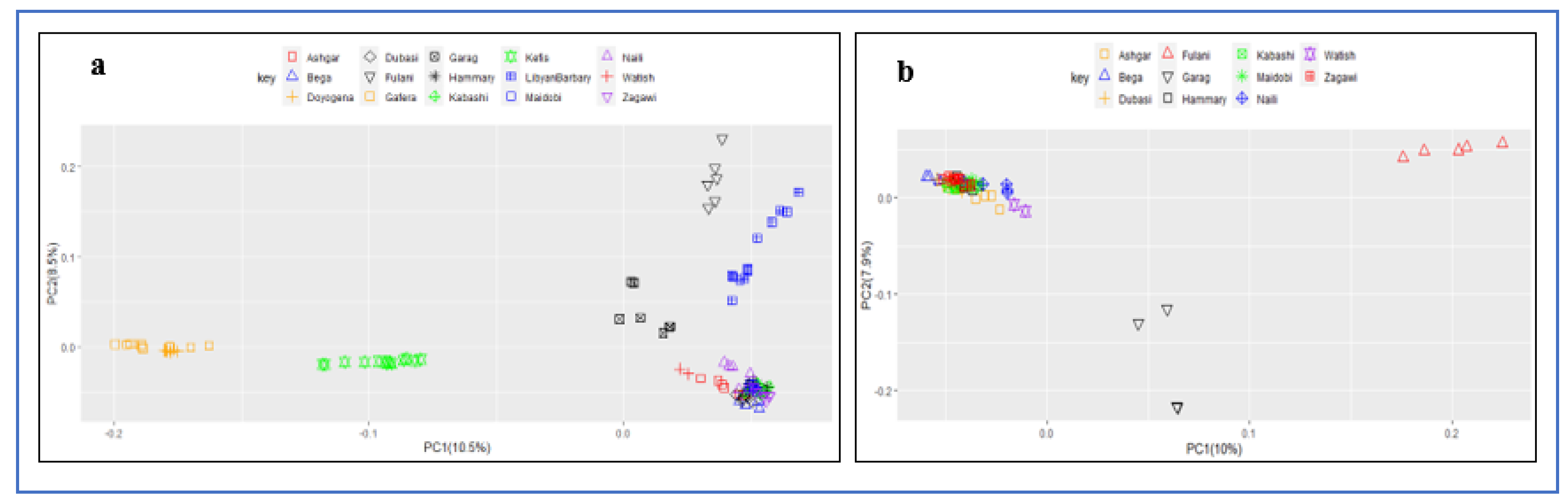
3.4. Principal Component Analysis
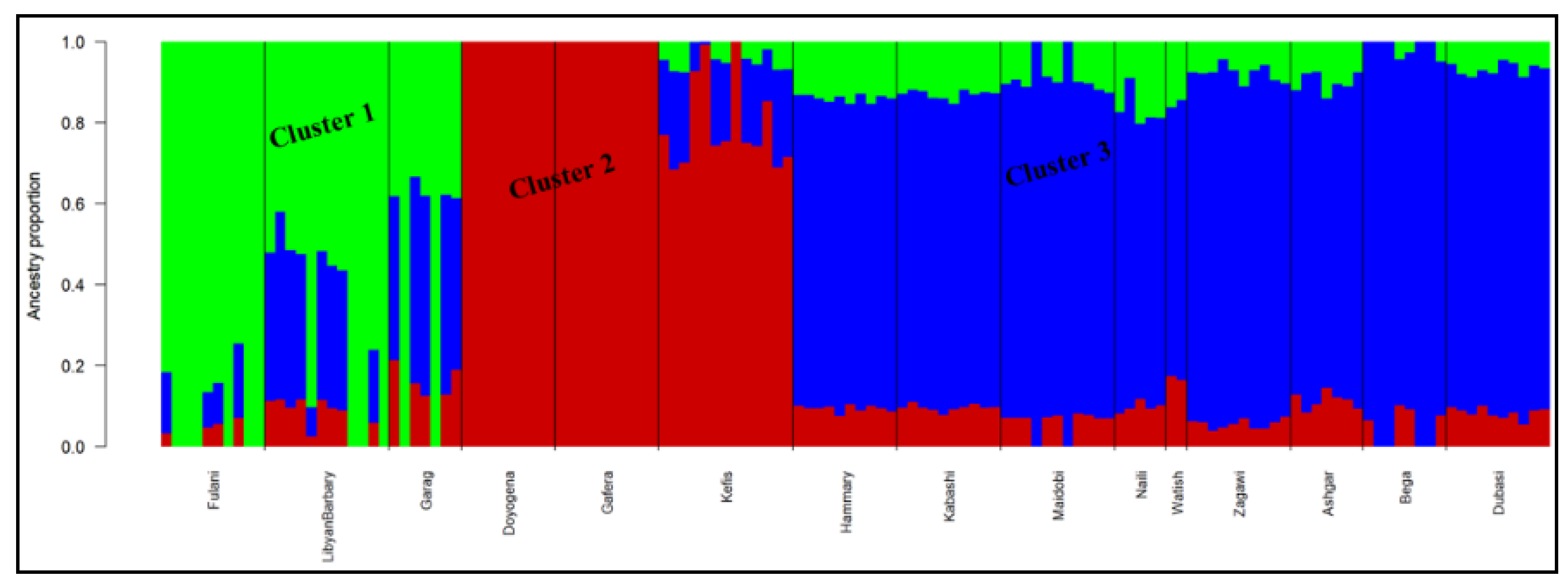
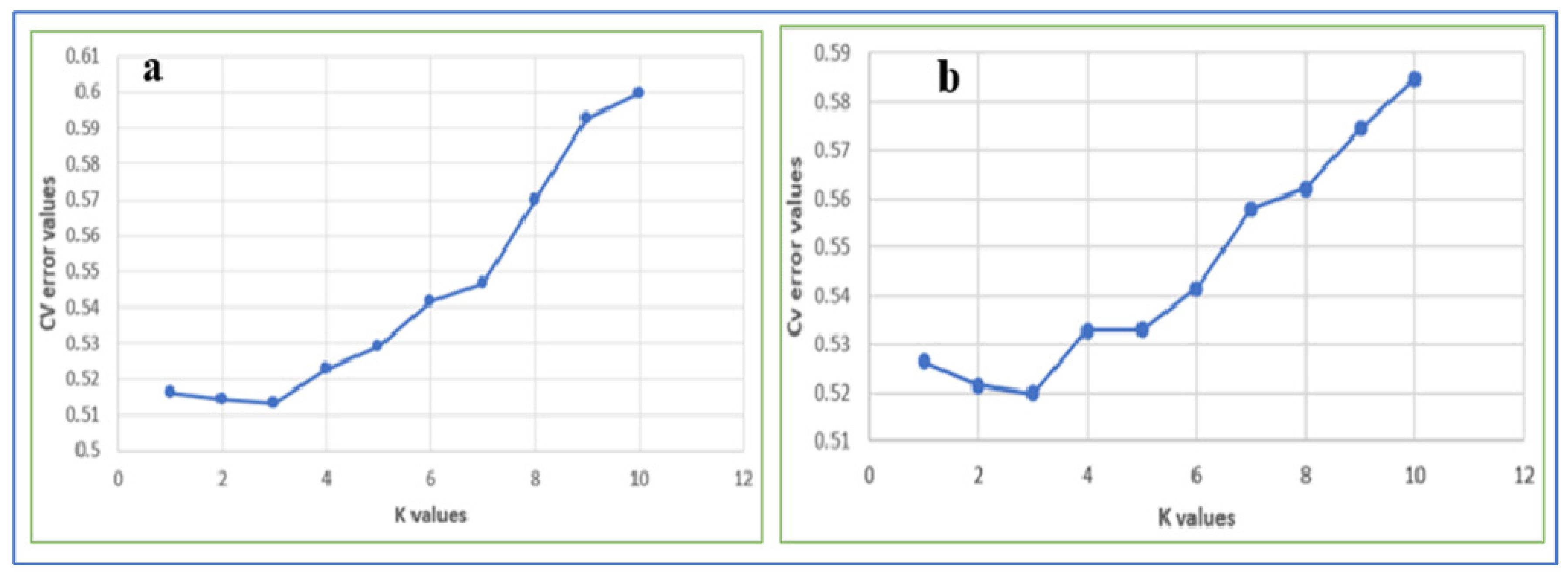
3.5. Genetic Population Structure Analysis
4. Discussion
4.1. Variant Detection and Annotation
4.2. Genetic Diversity, Population Divergence and Relationships
4.3. Genetic Population Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gornas, N.; Weimann, C.; El Hussien, A.; Erhardt, G. Genetic characterization of local Sudanese sheep breeds using DNA markers. Small Rumin. Res. 2011, 95, 7–33. [Google Scholar] [CrossRef]
- FAO. FAO Stat Data; Food and Agriculture Organization: Rome, Italy, 2018. [Google Scholar]
- Central Bank of Sudan. Annual Report. Khartoum, Sudan. 2009. Available online: https://cbos.gov.sd/sites/default/files/annual_09_0_0.pdf (accessed on 6 September 2022).
- MARF. Statistical Bulletin for Animal Resources; Ministry of Animal Resources and Fisheries: Khartoum, Sudan, 2009; No. 19. [Google Scholar]
- Bennett, S.C.J.; John, E.R.; Hewison, J.W. Animal Husbandry, Agriculture in Sudan; Oxford University Press: London, UK, 1948; pp. 663–667. [Google Scholar]
- Wilson, R.T. Livestock in the Republic of the Sudan: Policies, production, problems, possibilities, Bartridge House, Umberleigh, UK. Anim. Husb. Dairy Vet. Sci. 2018, 2, 1–12. [Google Scholar] [CrossRef]
- Alvarez, I.; Traore, A.; Tamboura, H.H.; Kabore, A.; Royo, L.J.; Fernandez, I.; Ouedraogo-Sanou, G.; Sawadogo, L.; Goyache, F. Microsatellite analysis characterizes Burkina Faso as a genetic contact zone between Sahelian and Djallonke sheep. Anim. Biotechnol. 2009, 20, 47–57. [Google Scholar] [CrossRef]
- Abied, A.; Ahbara, A.M.; Berihulay, H.; Xu, L.; Islam, R.; El-Hag, F.M.; Rekik, M.; Haile, A.; Han, J.-L.; Ma, Y.; et al. Genome Divergence and Dynamics in the Thin-Tailed Desert Sheep from Sudan. Front. Genet. 2021, 12, 659507. [Google Scholar] [CrossRef]
- McLeroy, G.B. The sheep of the Sudan, an extensive survey and system of classification. Sudan J. Vet. Anim. Husb. 1961, 2, 19–35. [Google Scholar]
- Wilson, R.T.; Clarke, S.E. Studies on the livestock of Southern Darfur, Sudan. The ecology and livestock resources of the area. Trop. Anim. Health Prod. 1975, 7, 165–187. [Google Scholar]
- Gizaw, S.; Komen, H.; Hanotte, O.; Van Arendonk, J.A. Indigenous sheep resources of Ethiopia: Types, production systems and farmers preferences. Anim. Genet. Resour. 2008, 43, 25–39. [Google Scholar] [CrossRef]
- Amane, A.; Belay, G.; Nasser, Y.; Kyalo, M.; Dessie, T.; Kebede, A.; Tarekegn, G.M. Genome-wide insights of Ethiopian indigenous sheep populations reveal the population structure related to tail morphology and phylogeography. Genes Genom. 2020, 42, 1169–1178. [Google Scholar] [CrossRef]
- Abualazayium, M. Animal Wealth and Animal Production in Sudan, 3rd ed.; Khartoum University Press: Khartoum, Sudan, 2004; pp. 50–51. [Google Scholar]
- Devendra, C.; McLeroy, G. Goat and Sheep Production in the Tropics; Longman Group UK Ltd.: Essex, UK, 1982; p. 271. [Google Scholar]
- Tapio, I.; Tapio, M.; Grislis, Z.; Holm, L.E.; Jeppsson, S.; Kantanen, J.; Miceikiene, I.; Olsaker, I.; Viinalass, H.; Eythorsdottir, E. Unfolding of population structure in Baltic sheep breeds using microsatellite analysis. Heredity 2005, 94, 448–456. [Google Scholar] [CrossRef]
- Kardos, M.; Luikart, G.; Bunch, R.; Dewey, S.; Edwards, W.; McWilliam, S.; Kijas, J. Whole-genome resequencing uncovers molecular signatures of natural and sexual selection in wild bighorn sheep. Mol. Ecol. 2015, 24, 5616–5632. [Google Scholar] [CrossRef]
- Yang, J.I.; Li, W.R.; Lv, F.H.; He, S.G.; Tian, S.L.; Peng, W.F.; Liu, M.J. Whole-genome sequencing of native sheep provides insights into rapid adaptations to extreme environments. Mol. Biol. Evol. 2016, 33, 2576–2592. [Google Scholar] [CrossRef]
- Daetwyler, H.D.; Capitan, A.; Pausch, H.; Stothard, P.; Van Binsbergen, R.; Brøndum, R.F.; Hayes, B.J. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 2014, 46, 858–865. [Google Scholar] [CrossRef]
- FAO. Molecular Genetic Characterization of Animal Genetic Resources; FAO Animal Production and Health Guidelines: Rome, Italy, 2011; No. 9. [Google Scholar]
- Ahbara, A.; Bahbahani, H.; Almathen, F.; Al Abri, M.; Agoub, M.O.; Abeba, A.; Kebede, A.; Musa, H.H.; Mastrangelo, S.; Pilla, F.; et al. Genome-wide variation, candidate regions and genes associated with fat deposition and tail morphology in Ethiopian indigenous sheep. Front. Genet. 2019, 9, 699. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Sham, P.C. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Yuan, Z.; Liu, E.; Liu, Z.; Kijas, J.W.; Zhu, C.; Hu, S.; Ma, X.; Zhang, L.; Du, L.; Wang, H.; et al. Selection signature analysis reveals genes associated with tail type in Chinese indigenous sheep. Anim. Genet. 2017, 48, 55–66. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinform 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Use R!); Springer: New York, NY, USA, 2009; ISBN 0387981403. [Google Scholar]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Davenport, K.M.; Hiemke, C.; McKay, S.D.; Thorne, J.W.; Lewis, R.M.; Taylor, T.; Murdoch, B.M. Genetic structure and admixture in sheep from terminal breeds in the United States. Anim. Genet. 2020, 51, 284–291. [Google Scholar] [CrossRef]
- Wiener, P.; Robert, C.; Ahbara, A.; Salavati, M.; Abebe, A.; Kebede, A.; Wragg, D.; Friedrich, J.; Vasoya, D.; Hume, D.A.; et al. Whole-Genome Sequence Data Suggest Environmental Adaptation of Ethiopian Sheep Populations. Genome Biol. Evol. 2021, 13, evab014. [Google Scholar] [CrossRef]
- Park, S.D.; Magee, D.A.; McGettigan, P.A.; Teasdale, M.D.; Edwards, C.J.; Lohan, A.J.; MacHugh, D.E. Genome sequencing of the extinct Eurasian wild aurochs, Bos primigenius, illuminates the phylogeography and evolution of cattle. Genome Biol. 2015, 16, 234. [Google Scholar] [CrossRef]
- Mufarrih, M.E. Sudan desert sheep: Their origin, ecology, and production potential. World Anim. Rev. 1991, 66, 23–31. [Google Scholar]
- Edea, Z.; Dadi, H.; Dessie, T.; Kim, K.S. Genomic signatures of high-altitude adaptation in Ethiopian sheep populations. Genes Genom. 2019, 41, 973–981. [Google Scholar] [CrossRef]
- Peter, C.; Bruford, M.; Perez, T.; Dalamitra, S.; Hewitt, G.; Erhardt, G.; Econogene Consortium. Genetic diversity and subdivision of 57 European and Middle Eastern sheep populations. Anim. Genet. 2007, 38, 37–44. [Google Scholar] [CrossRef]
- MARF. Estimates of Animal Populations, Annual Report. Federal Ministry of Animal Resources and Fisheries; Khartoum, Sudan, 2013; Available online: https://openjicareport.jica.go.jp/pdf/12233656_02.pdf (accessed on 6 September 2022).
- Madrigal, L.; Ware, B.; Miller, R.; Saenz, G.; Chavez, M.; Dykes, D. Ethnicity, gene flow, and population subdivision in Limon, Costa Rica. Am. J. Phys. Anthropol. 2001, 114, 99–108. [Google Scholar] [CrossRef]
- Gizaw, S.; Van Arendonk, J.A.; Komen, H.; Windig, J.; Hanotte, O. Population structure, genetic variation, and morphological diversity in indigenous sheep of Ethiopia. Anim. Genet. 2007, 38, 621–628. [Google Scholar] [CrossRef]
- McLeroy, G.B. The sheep of the Sudan. 2. Eco-types and tribal breeds. Sudan J. Vet. Sci. Anim. Husb. 1961, 2, 101–151. [Google Scholar]
- Gaouar, S.B.S.; Kdidi, S.; Ouragh, L. Estimating population structure and genetic diversity of five Moroccan sheep breeds by microsatellite markers. Small Rumin. Res. 2016, 144, 23–27. [Google Scholar] [CrossRef]
- Muigai, A.W.; Hanotte, O. The origin of African sheep: Archaeological and genetic perspectives. Afr. Archaeol. Rev. 2013, 30, 39–50. [Google Scholar] [CrossRef]
- Gifford-Gonzalez, D.; Hanotte, O. Domesticating animals in Africa: Implications of genetic and archaeological findings. J. World Prehistory 2011, 24, 1–23. [Google Scholar] [CrossRef]
- Wilson, R.T. Small ruminant production and the small ruminant genetic resource in tropical Africa. In Food and Agriculture Organization of The United Nations; Springer: Rome, Italy, 1991; ISBN 0387981403. [Google Scholar]
- Ryder, M.L. Sheep. In Evolution of Domesticated Animals; Mason, I.L., Ed.; Longman: New York, NY, USA, 1984; pp. 63–84. [Google Scholar]
- Hanotte, O.; Bradley, D.G.; Ochieng, J.W.; Verjee, Y.; Hill, E.W.; Rege, J.E.O. African pastoralism: Genetic imprints of origins and migrations. Science 2002, 296, 336–339. [Google Scholar] [CrossRef]
- Muigai, A.W.T. Characterization and Conservation of Indigenous Animal Genetic Resources: The Fat-Tailed and Thin-Tailed Sheep of Africa. Ph.D. Thesis, Jomo Kenyatta University of Agriculture and Technology, Juja, Kenya, 2003. [Google Scholar]
- Aswani, O.B. Genetic Diversity and Relationships of African Sheep: A Y Chromosome Perspective. Master’s Thesis, University of Egerton, Njoro, Kenya, 2007. [Google Scholar]
- Moradi, M.H.; Nejati-Javaremi, A.; Moradi-Shahrbabak, M.; Dodds, K.G.; McEwan, J.C. Genomic scan of selective sweeps in thin and fat tail sheep breeds for identifying of candidate regions associated with fat deposition. BMC Genet. 2012, 13, 10. [Google Scholar] [CrossRef]
- Moioli, B.; Pilla, F.; Ciani, E. Signatures of selection identify loci associated with fat tail in sheep. J. Anim. Sci. 2015, 93, 4660–4669. [Google Scholar] [CrossRef]
- Abdulkarim, A. Small ruminant contribution to meat production in Libya. Egypt. J. Sheep Goat Sci. 2015, 10, 13–18. [Google Scholar]


| Population | Ashgar | Bega | Dubasi | Fulani | Garag | Hama | Kaba | Maid | Naili | Zagawi | Doyo | Kefis | Gafera |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bega | 0.027 | ||||||||||||
| Dubasi | 0.004 | 0.026 | |||||||||||
| Fulani | 0.060 | 0.085 | 0.063 | ||||||||||
| Garag | 0.052 | 0.078 | 0.055 | 0.081 | |||||||||
| Hama | 0.002 | 0.024 | 0.002 | 0.058 | 0.049 | ||||||||
| Kabba | 0.001 | 0.026 | 0.003 | 0.060 | 0.051 | 0.004 | |||||||
| Maid | 0.009 | 0.030 | 0.009 | 0.062 | 0.056 | 0.005 | 0.004 | ||||||
| Naili | 0.018 | 0.042 | 0.019 | 0.070 | 0.064 | 0.013 | 0.014 | 0.021 | |||||
| Zagawi | 0.012 | 0.033 | 0.012 | 0.065 | 0.058 | 0.005 | 0.006 | 0.010 | 0.024 | ||||
| Doyo | 0.065 | 0.091 | 0.0680 | 0.105 | 0.108 | 0.060 | 0.062 | 0.070 | 0.080 | 0.073 | |||
| Kefis | 0.052 | 0.075 | 0.055 | 0.095 | 0.101 | 0.048 | 0.049 | 0.056 | 0.065 | 0.059 | 0.066 | ||
| Gafera | 0.069 | 0.093 | 0.070 | 0.107 | 0.112 | 0.063 | 0.063 | 0.072 | 0.083 | 0.075 | 0.042 | 0.069 | |
| LB | 0.044 | 0.067 | 0.047 | 0.072 | 0.091 | 0.039 | 0.041 | 0.048 | 0.048 | 0.050 | 0.085 | 0.067 | 0.082 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amane, A.; Belay, G.; Tijjani, A.; Dessie, T.; Musa, H.H.; Hanotte, O. Genome-Wide Genetic Diversity and Population Structure of Local Sudanese Sheep Populations Revealed by Whole-Genome Sequencing. Diversity 2022, 14, 895. https://doi.org/10.3390/d14110895
Amane A, Belay G, Tijjani A, Dessie T, Musa HH, Hanotte O. Genome-Wide Genetic Diversity and Population Structure of Local Sudanese Sheep Populations Revealed by Whole-Genome Sequencing. Diversity. 2022; 14(11):895. https://doi.org/10.3390/d14110895
Chicago/Turabian StyleAmane, Agraw, Gurja Belay, Abdulfatai Tijjani, Tadelle Dessie, Hassan H. Musa, and Olivier Hanotte. 2022. "Genome-Wide Genetic Diversity and Population Structure of Local Sudanese Sheep Populations Revealed by Whole-Genome Sequencing" Diversity 14, no. 11: 895. https://doi.org/10.3390/d14110895
APA StyleAmane, A., Belay, G., Tijjani, A., Dessie, T., Musa, H. H., & Hanotte, O. (2022). Genome-Wide Genetic Diversity and Population Structure of Local Sudanese Sheep Populations Revealed by Whole-Genome Sequencing. Diversity, 14(11), 895. https://doi.org/10.3390/d14110895






