Abstract
Dongxiang wild rice (Oryza rufipogon Griff.) germplasm is a precious resource for the improvement of agronomic traits in rice. Rice seeds also harbor a diverse endophytic bacterial community, and their interactions with their hosts and each other can influence plant growth and adaptability. Here, we investigated the community composition of cultivable endophytic bacteria obtained from the surface-sterilized seeds of Dongxiang wild rice and screened them for plant growth-promoting traits. Phylogenetic analysis of 16S rRNA gene sequences indicated that the 47 isolates were affiliated with five classes and 13 discrete genera, and Bacillus and Microbacterium predominated. Evaluations of plant growth promoting (PGP) traits showed that 45 endophytic bacteria isolates produced between 3.37 and 90.11 μg mL−1 of Indole-3-acetic acid (IAA), with the highest yield of 90.11 μg mL−1 (Fse28). Further, 37 of the isolates were able to solubilize mineral phosphate, while 28 other isolates had the ability of N2-fixation, 17 isolates possessed 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity with the highest yield of 20.72 μmol mg−1 protein h−1 (Fse35), and 17 isolates were also able to produce siderophores. The two strains Fse28 and Fse35 had multiple PGP traits that significantly improved the agronomic traits (root length, shoot length, dry matter, and chlorophyll content) of cultivated rice seedlings. Our results illustrate the rich diversity of seed endophytic bacteria in Dongxiang wild rice and their potential for developing novel efficient bioinoculants to enhance soil fertility and favor seedling growth.
1. Introduction
Plants can establish relationships with other members of their ecosystem, enjoying benefits to their growth and development while providing an ecological niche for thriving microorganisms [1,2]. Endophytes are microorganisms that inhabit the various tissues and organs of some or all stages of healthy plants and do not cause substantial damage to the host plant [3]. Researchers have successfully classified and reported more than 200 genera of endophytic bacteria from different plant tissues [4]. Endophytic bacteria sometimes provide multiple benefits to host plants, such as stimulation of plant growth by promoting biological nitrogen fixation, phosphate solubilization, phytohormone production (indole-3-acetic acid (IAA) and gibberellins), ACC deaminase activity, and enhancing antagonism against plant pathogens [5,6,7,8]. As a new agricultural resource, the study of endophytic bacteria and their interactions with plants has attracted much attention recently in the fields of plant science, agronomy, thermology, and ecology [9,10].
Seeds are as much an important plant organ as roots, leaves, and flowers, which have had to evolve in association with diverse microbial communities [11,12,13]. Previous studies have reported that seed microbes may originate from the earliest stages of seed development on the parent plant and then are able to establish early in soil grown seedlings [14,15]. Evidence has also indicated that vertical or seed transmission can significantly contribute to the plant microbiome [16,17]. Indeed, endophytic seed microbes have been previously shown to have potential to promote plant growth or control plant diseases [11,18,19]. Some examples of these bacteria are Bacillus, Enterococcus, Paenibacillus, and Methylobacterium, which were isolated from Eucalyptus species seeds [20]. The common bacterial genera reported in seeds of different plant species are Paenibacillus, Micrococcus, Staphylococcus, Pantoea, Acinetobacter, Bacillus, and Pseudomonas [21]. Some seed endophytes are involved in plant growth promotion and can protect the host plant against pathogens. For example, Khalaf et al. [22] reported that Bacillus and Paenibacillus isolated from cucurbit seeds emit volatile organic compounds (VOCs) and secrete extracellular ribonucleases which could suppress fungal pathogens. Jeong et al. [23] observed that the colonization of seed endophytic bacterium Kosakonia cowanii GG1 into Arabidopsis thaliana resulted in the stimulation of plant growth under drought conditions. Xu et al. [24] found a seed-borne Bacillus strain that improved the root and shoot growth of tomato, probably through the production of ACC deaminase and nitrogen fixation. These results illustrate how seed endophytic bacteria may express functions beneficial to host plants and suggest they have potential for biotechnological applications.
Dongxiang wild rice (DXWR, Oryza rufipogon Griff.), the northernmost wild rice in Jiangxi Province, China and even the world (28°14′ N), is endemic to Dongxiang County with an annual average temperature of 17.7 °C and low temperature average in winter of −12.8 °C [25]. The ecological environment is surrounded by low hills, marshes, ditches, and ponds [26]. The presence in DXWR of genes related to high grain yield, disease and insect resistance, fertility restoration, as well as cold and drought tolerance have been extensively studied [27,28]. As an example, the NBS-LRR resistance gene of DXWR was cloned based on the conserved motif of NBS, and sequence analysis revealed that some amino acids were deleted [29]. Zhou et al. [30] found that, compared with cultivated rice, Dongxiang wild rice had a very high survival rate under salt stress, which was associated with large differences in its leaf and root transcriptomes. Microbes associated with plants may also play important roles in affecting significant plant properties. Previous studies have reported a number of endophytic bacteria (including Phytobacter diazotrophicus) with high nitrogen fixing enzyme activity and probiotic potential that were isolated from wild rice [31,32]. A study revealed a hitherto unreported endophytic bacterium from wild rice germplasm, Microbacterium laevaniformans, with high production of indole acetic acid and gibberellic acid [33]. Our earlier study described rhizobacteria [34], endophytic bacteria [35], fungi [36], and actinomycetes [37] from different tissues (root, stem and leaf) of DXWR, which showed high diversity. Zeng et al. [38] reported that Pantoea agglomerans T21 isolated from rhizosphere with multiple PGP traits could stimulate the growth of cultivated rice. However, the diversity and PGP effects of seed-colonizing endophytic bacteria from DXWR remain poorly understood. Therefore, assessing the community and beneficial functions of seed-colonizing endophytic bacteria will extend our knowledge of plant and microbial interactions.
In the present study, we investigated the community composition of cultivable endophytic bacteria obtained from the surface-sterilized seeds of Dongxiang wild rice using five isolation media, including nutrient agar (NA), 10-fold diluted nutrient agar (TDNA), Reasoner’s 2A agar (R2A), Tryptone soy agar (TSA), and Baird-Parker agar (BPA). To further investigate the function of the cultivatable seed endophytic bacteria community, we measured nitrogen fixation, phosphorus solubilization, indole acetic acid (IAA) production, iron production carrier, and ACC deaminase activity of endophytic bacteria. We also screened for their plant growth promotion potential, inoculating them on domesticated rice seeds to evaluate their effect on seedling germination and growth. Our results show that DXWR seeds possess many cultivatable strains of bacterial endophytes that have potential for agricultural applications.
2. Materials and Methods
2.1. Isolation of Endophytic Bacteria
Healthy seeds of DXWR were collected from natural populations in a natural reserve located in Dongxiang County, Jiangxi Province, China (28°14′ N latitude and 116°30′ E longitude). All seeds were stored in an icebox and immediately brought to the laboratory. The tissues of the samples were screened for endophytic bacteria within 24 h.
Surface sterilization involved shaking seeds in autoclaved distilled water thrice followed by 5 min rinsing in sodium hypochlorite (5%), followed by draining of the bleach. Seeds were then rinsed with autoclaved distilled water, before being washed with 95% ethanol for 5 min. After draining the ethanol, the seeds were rinsed three times with autoclaved distilled water. To verify that the surface sterilization was adequate, 200 μL of the last wash were plated and cultured on five different types of agar media: nutrient agar (NA), 10-fold diluted nutrient agar (TDNA), Reasoner’s 2A agar (R2A), Tryptone soy agar (TSA), and Baird-Parker agar (BPA) and these plates were incubated for 3 days at 30 °C.
Once seed surface sterility was confirmed, 15 seeds per gram were grounded gently in an autoclaved mortar using 0.5 mL of 50 mM Na2HPO4 buffer. The ground seed suspension (100 μL) was used for microbial culturing: 10-fold serial dilutions in 50 mM Na2HPO4 (10×, 100× and 1000×) were streaked on NA, TDNA, R2A, TSA, and BPA media followed by incubation for 1–7 days at 30 °C. Morphologically unique bacterial colonies from each plate were selected, streaked on fresh plates to purify, and finally cultured in LB broth (10 g/L tryptone, 5 g/L yeast extract and 5 g/L NaCl, pH 7.2) for glycerol stock and DNA extraction.
2.2. DNA Extraction, 16S rRNA Gene Amplification, Sequencing, and Strain Identification
Endophytic bacteria were cultured in LB broth at 37 °C with constant shaking for 24 h. For the taxonomic indentation of bacterial endophytes, their genomic DNA was extracted using an EZNA bacterial DNA kit (D3350-01, Omega, Norcross, GA, USA) following the manufacturer’s instruction. The 16S rRNA gene was amplified using 27F (5’-AGAGTTTGATCCTG GCTCAG-3’) and 1492R (5’-TACGGCTACC TTGTTACGACTT-3’) universal primers [39]. Approximately 2 μL of total DNA (50 ng genomic DNA) was added to a PCR mixture containing 20 μL of rTaq PCR Mastermix (Takara Bio, Beijing, China), 2 μL of each primer (10 μM working stock), and H2O to a final volume of 40 μL. Amplification was set at the following conditions in a DNA Thermal Cycler (S1000, BioRad, Hercules, CA, USA): denaturation at 95 °C for 2 min, followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 1.5 min and final extension at 72 °C for 5 min. As a negative control, the template DNA was replaced by sterile double-distilled water. The PCR amplified products were resolved in 1.2% (w/v)) agarose gel and documented using the Gel documentation system (Gel Doc XR+, BioRad). Amplicons of about 1500 bp were selected and gel purified using a Gel Extraction Kit (Omega). The purification and sequencing of the PCR products was performed by Shanghai Invitrogen Company Ltd. (Shanghai, China).
The bacterial 16S rRNA sequences were matched against the nucleotide database using the Basic Local Alignment Search Tool of the US National Centre for Biotechnology Information (NCBI) for the final identification of the endophytic bacteria. Sequences were deposited in GenBank under accession numbers KJ733853~KJ733899. The most appropriate relative sequences were selected based on maximum identity and habitat in the NCBI database and imported into MEGA 5.0 [40]. To reveal the genetic distances between different bacterial strains intuitively, we constructed a rooted phylogenetic tree by using the neighbor-joining method combined with bootstrap analysis with 1000 replicates.
2.3. Assay for Plant Growth-Promoting Activities
Bacterial isolates were screened for N-fixation ability by observing the growth on nitrogen-free semisolid medium (BAz) [41]. Production of siderophore was estimated on chrome-azurol S-agar medium by observing the development of orange color around the bacterial colony [42]. To test the ability of bacteria to solubilize inorganic phosphate, isolates were inoculated in duplicate to the NBRIP plate at 30 °C for 10 days [43]. The P content in the supernatant was tested by a spectrophotometrical method involving a 96-well microplate [44]. The production of IAA was determined by the colorimetric methodology described by Ribeiro and Cardoso [45] with some modifications. The ability of the isolates to produce ACC deaminase was also screened on minimal media containing ACC as their sole nitrogen source, as described by Penrose and Glick [46]. All the experiments were performed in triplicate and repeated three times.
2.4. Evaluation of Plant Growth Promotion in Rice Treated with Endophytic Bacteria
Based on the performance of PGPB in the experiments, two selected isolates, Fse28 and Fse35 were put through seed germination and seedling growth assays conducted in a greenhouse. Rice seeds were surface sterilized as previously described in this paper. Five sterilized seeds were placed on nutrient agar to confirm the surface sterilization. The sterilized seeds were inoculated with each bacterial suspension (approximately 108 CFU mL−1) or sterile water (uninoculated control) for 12 h, and then germinated on filter paper moistened with distilled water in a dark incubator at 30 °C. Evaluations of the length (cm) of the root and shoot were made after 7 days.
After seeds were germinated in the dark for 7 days, seedlings were transferred to pots (diameter, 15 cm; depth, 9 cm) that were filled with sterile silica sand. Hence, 10 seedlings were planted in each pot and three replicates were used for each treatment. The growth chamber experiment was conducted with a photoperiod cycle of 14 h light/10 h dark at 28 °C. Hoagland’s nutrient solution (Hoagland’s composition (mg L−1): KNO3 607, Ca(NO3)2·4H2O 945.0, MgSO4·7H2O 493.0, NH4H2PO4 115.0, H3BO3 2.86, MnCl2·4H2O 2.13, ZnSO4·7H2O 0.22, CuSO4·5H2O 0.08, H2MoO·4H2O 0.02, FeSO4·7H2O 2.78, Na2-EDTA 3.72) was watered every two days. After 10 and 20 days, plants were carefully removed from the pots and length data were recorded. The roots and shoots were separated and washed in distilled water, then dried and weighed.
For measuring chlorophyll content, 100 mg of finely chopped fresh leaves were placed in a capped measuring tube containing 25 mL of 80% acetone, and placed inside a refrigerator (4–8°C) for 28 h. The chlorophyll content was measured at 646.6 and 663.6 nm in a spectrophotometer and calculated using the equation of Porra [47].
2.5. Statistical Analysis
All results represent the means based on three or more independent replicates. Differences were compared with the one-way analysis of variance test and means were compared using Duncan’s multiple range test, where p < 0.05 was considered to indicate a significant difference. Results were expressed as the mean ± SD.
3. Results
3.1. Diversity and Community Composition of Seed Cultivable Endophytic Bacteria
The surface disinfection of seeds was efficient at eliminating epiphytic bacteria because the seed imprint on nutrient agar yielded no bacterial growth. A total 165 endophytic bacteria were isolated from the seed of DXWR. Based on their morphological characteristics, 47 apparently distinct isolates were selected for further study. NA and TDNA mediums yielded in the highest number of isolates and highest diversity with eight different genera. The R2A medium obtained the next highest diversity with six different genera, while TSA and BPA medium obtained only five different genera each. Bacillus spp. Were recovered from all five medium. Meanwhile, the strains Facklamia spp., Acinetobacter spp., and Burkholderia spp. could be only isolated from NA. Methylobacterium spp. was only isolated from TDNA. Micrococcus spp. was only isolated from TSA (Table 1).

Table 1.
The numbers and genera of endophytic bacteria recovered on the five different isolation media from DXWR seeds.
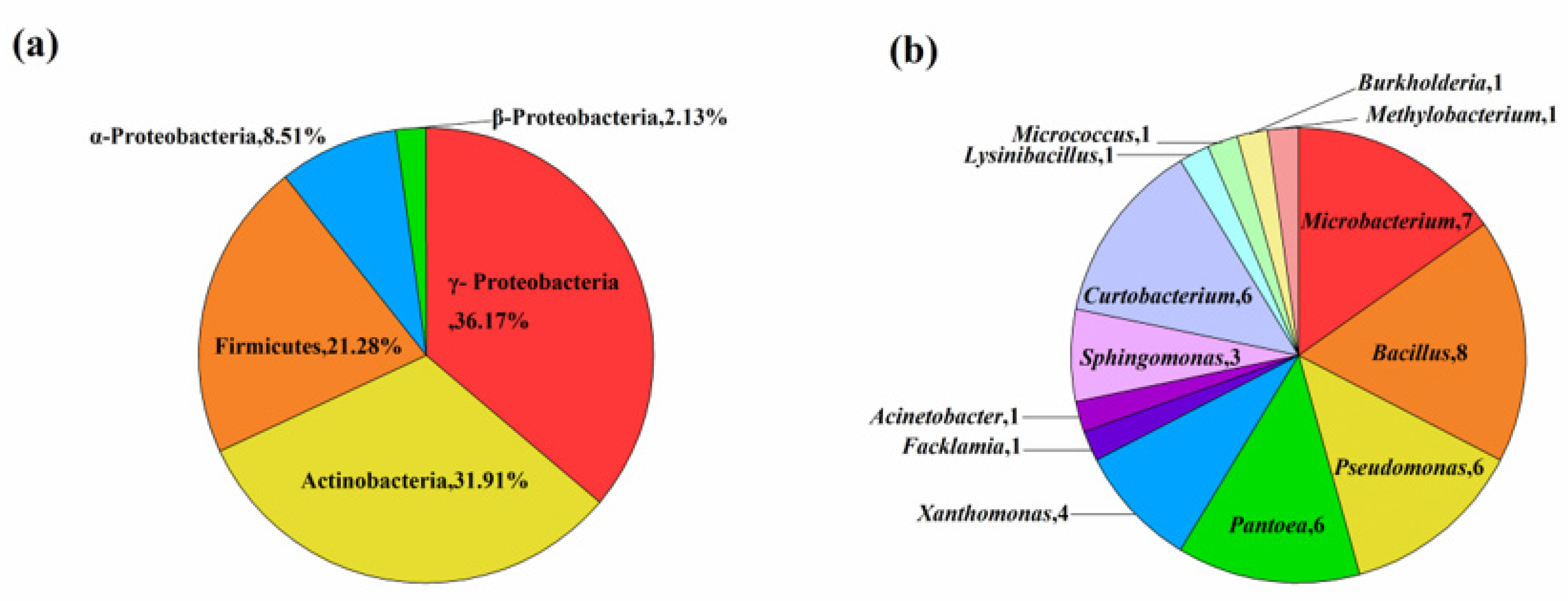
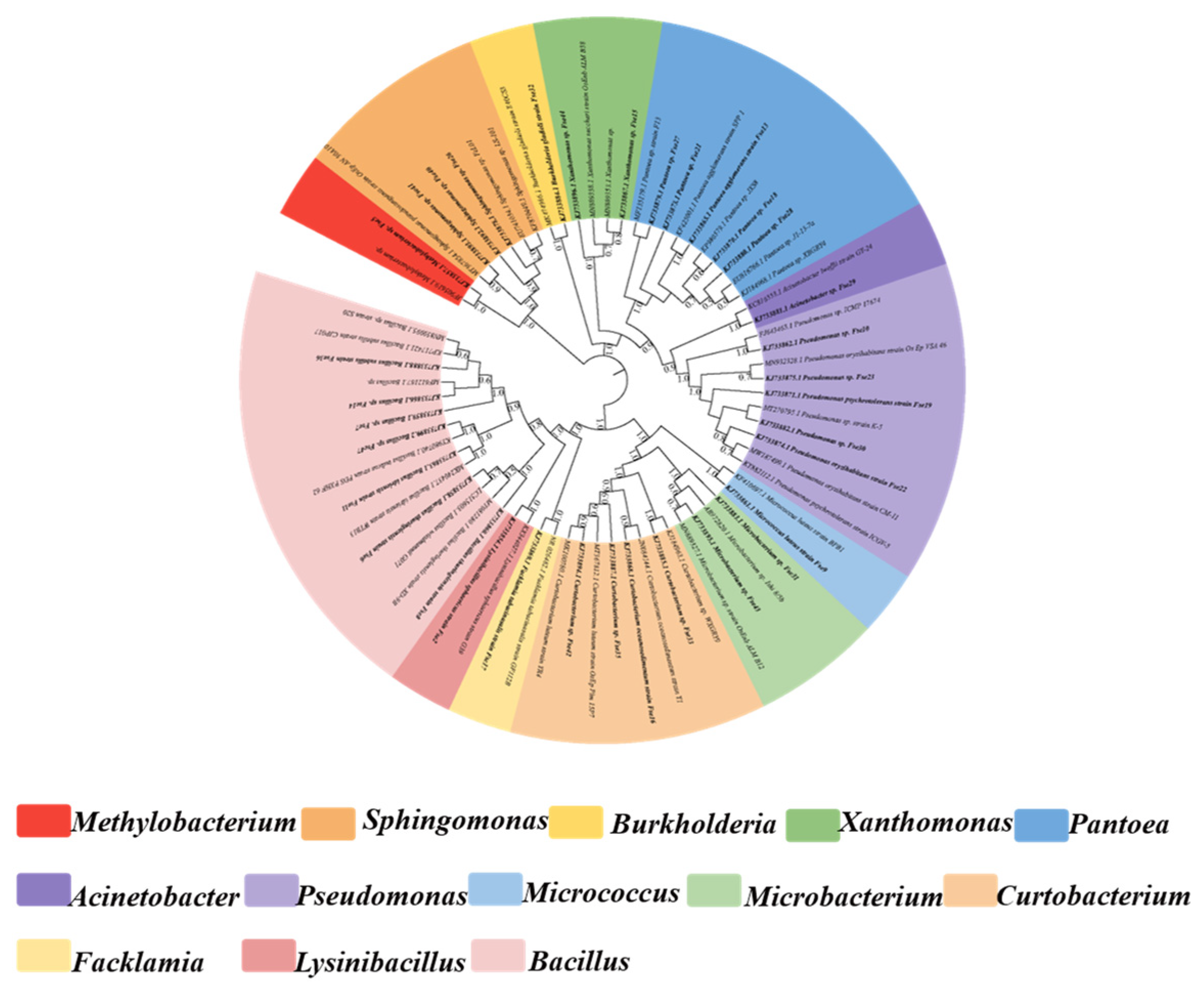
Analysis of 16S rDNA sequence from the 47 isolates indicated significant genetic diversity in to three different classes (α-, β- and γ-proteobacteria, Firmicutes and Actinobacteria) and 13 discrete genera (Bacillus, Microbacterium, Curtobacterium, Pseudomonas, Pantoea, Xanthomonas, Sphingomonas, Methylobacterium, Burkholderia, Acinetobacter, Lysinibacillus, Facklamia and Micrococcus) (Table 2, Figure 1). Members of the class γ-Proteobacteria were predominant in composition (n = 17, 36.2%), followed by Actinobacteria (n = 15, 31.9%), Firmicutes (n = 10, 21.3%), α-Proteobacteria (n = 4, 8.5%), and β-Proteobacteria (n = 1, 2.1%) (Figure 1a). Among 47 bacterial endophytes, Bacillus (n = 8, 17%) and Microbacterium (n = 8, 17%) were the predominant taxa, followed by Curtobacterium (n = 7, 14%), whereas six strains each of Pseudomonas and Pantoea, four strains of Xanthomonas, three strains of Sphingomonas, and one strain each of Methylobacterium, Burkholderia, Acinetobacter, Lysinibacillus, Facklamia, and Micrococcus were also identified (Figure 1b). The phylogenetic tree constructed based on 16S rDNA gene sequences for the 47 isolates are shown in Figure 2. The phylogenetic dendrogram indicates a division of the endophytic bacteria into thirteen distinct clades with different colors representing discrete genera (Figure 2). Overall, these results demonstrate the presence of abundant and diverse endophytic bacteria within seeds of DXWR.
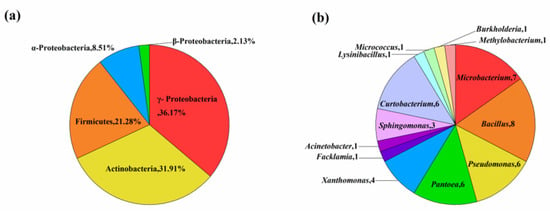
Figure 1.
Taxonomic diversity of the obtained representative bacterial isolates. (a) Composition percentages of the isolates at the phylum level. (b) Numbers of isolates at the genus level.
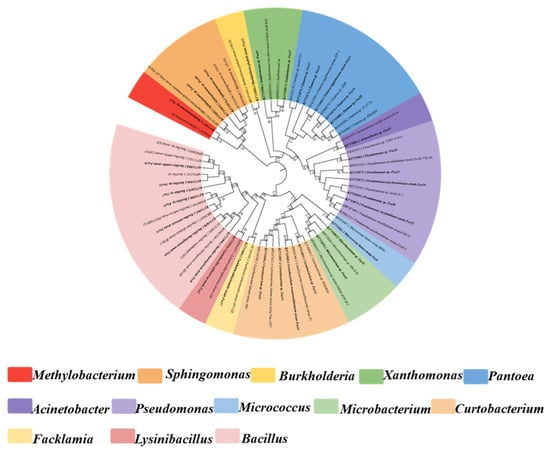
Figure 2.
Phylogenetic tree of representative strains of each species of endophytic bacteria in DXWR and relative strains in GenBank database based on 16S rDNA sequences.
3.2. Screening for PGP Traits of Isolates from Oryza Rufipogon Griff. In Vitro
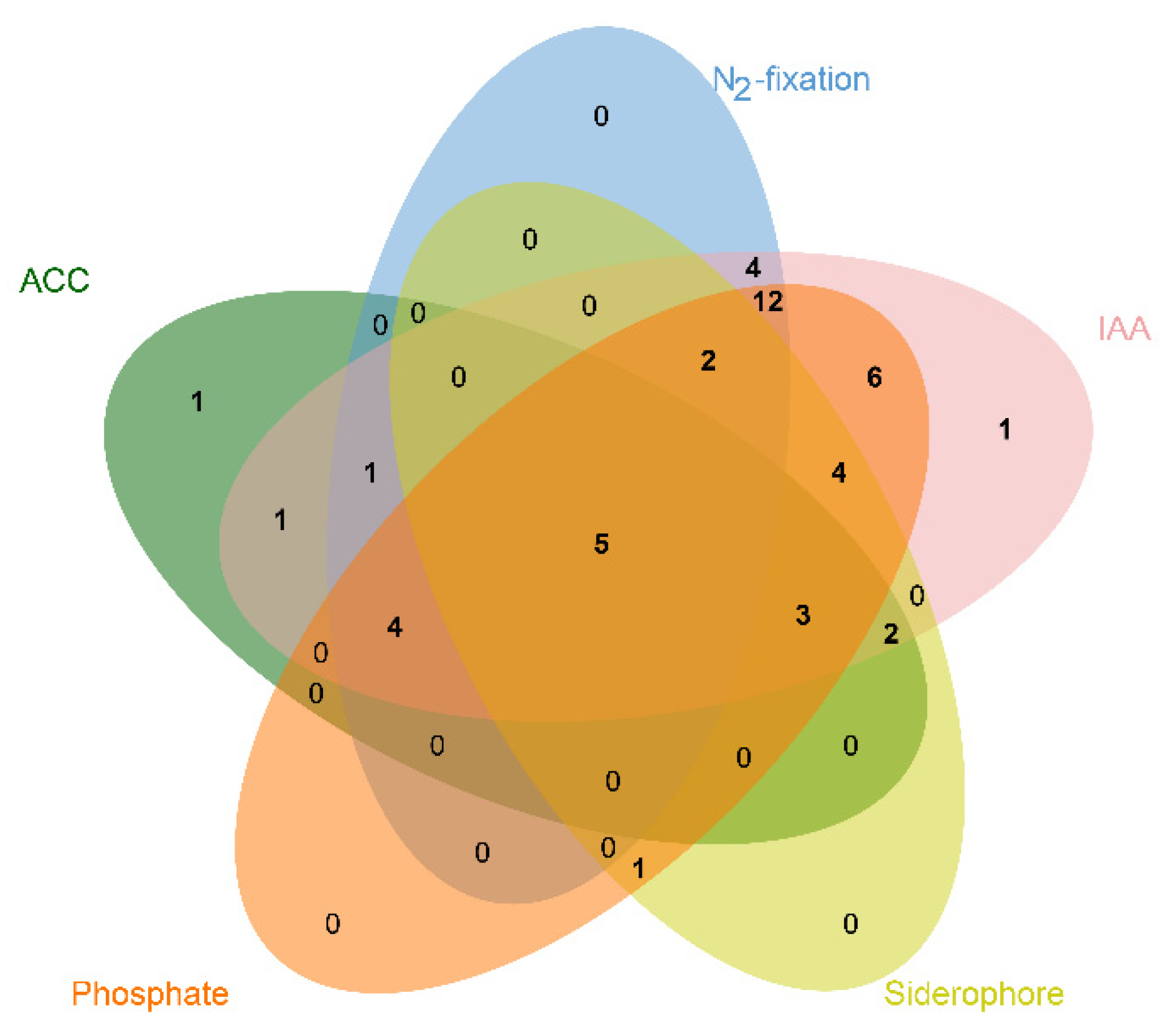
The potential of the isolates for plant growth promotion in this study was evaluated by screening for N2 fixation, IAA production, ACC deaminase activity, siderophore production and mineral phosphate solubilization. The differential PGP traits identified in all of 47 isolates were summarized in Table 2. The numbers of endophytic bacteria isolated from seeds with different PGP traits is shown in Figure 3. Among the 47 isolates, a higher percentage of the isolates showed IAA production (95.74%), followed by phosphate solubilization (78.72%), nitrogen fixation (59.57%), siderophore production (36.17%), and ACC deaminase activity (36.17%), respectively. All the strains except for Fse11 and Fse47 possessed the ability to produce IAA at different efficiencies. The highest levels of IAA production (90.11 ± 5.90 μg mL−1) were observed for Pantoea sp. strain Fse28. On the basis of the phosphate solubilization assay, 37 bacterial isolates were able to mobilize calcium phosphate, with Methylobacterium sp. strain Fse5 showing the highest activity of phosphate solubilization (576.34 ± 24.63 μg mL−1). Seventeen strains produced ACC deaminase with activity ranging from 2.12 μmol mg−1 protein h−1 (Curtobacterium sp. strain Fse42) to 20.72 μmol mg−1 protein h−1 (Curtobacterium sp. strain Fse35). Further, 17 bacteria were positive for siderophore production with a higher proportion of Bacilli than Actinobacteria. Twenty-eight isolates grew in a N-free medium, indicative of their ability for N-fixation, however none of these were Bacilli. Looking for strains with multiple plant growth promoting traits, two strains (Fse28 and Fse35) were selected for further evaluation in seed germination and potted plant growth experiments.

Table 2.
Summary of the endophytic bacteria isolated from DXWR seeds together with their respective strain codes, GenBank accession numbers, closest affiliations among the representative isolates in GenBank according to 16S rRNA sequence analysis, and PGP traits.
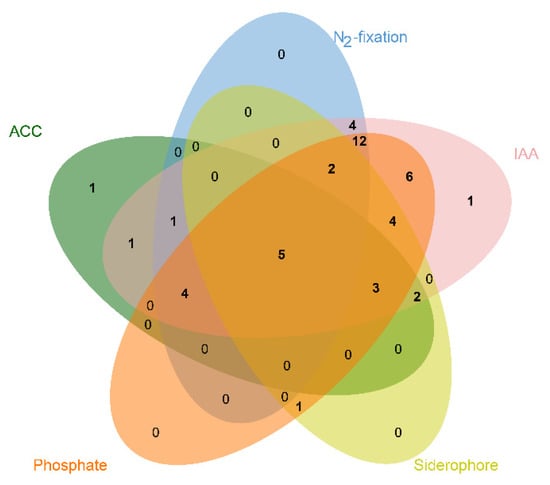
Figure 3.
Venn diagram showing the distribution of seed-colonizing endophytic bacteria with different PGP traits isolated from DXWR seeds.
3.3. Effect of Bio-Inoculum Treatment on Rice Growth-Promoting under Growth Chamber
The effect of the two multi-functional PGP strains (Fse28 and Fse35) on seed germination and plant growth is shown in Table 3. The tested strains enhanced seed germination percentage, resulting in 75.67% germination when rice seeds were treated with the isolate Fse28 in comparison to the negative control 71.67%. Seeds inoculated with Fse28 and Fse35 presented higher results than the uninoculated seeds in terms of shoot and root length as well. Inoculation with Pantoea sp. Fse28 significantly (p ≤ 0.05) increased shoot height and root length as compared to uninoculated plants, with a growth rate of 41.6% and 47.6%, respectively.

Table 3.
Effects of inoculation with two putative PGPE strains on rice seed germination.
After 10 and 20 days of the bacteria and host–plant association, the plant growth index was different between inoculated and uninoculated seedlings (Table 4, Figure 4). After 10 days, bacteria-inoculated rice seedlings showed significant increases in plant shoot length, dry weight, and chlorophyll content as compared to the uninoculated controls, although there was no significant difference in root length. Inoculation with Fse28 caused a strong increase in shoot length (18.84 cm), dry weight (28.34 mg), and chlorophyll content (1.82 mg/g), which was 31.3%, 50.7%, and 219.3% higher than the control, respectively. Data on root length, shoot length, dry weight, and chlorophyll content indicate that inoculation with Fse28 enhanced growth parameters significantly (p = 0.05) in comparison with the control after 20 days of development, and the seedling inoculated with Fse35 also had significant increases in shoot length and chlorophyll content. In summary, Fse28 was the most effective of the isolates tested in enhancing rice root, shoot, dry weight, and chlorophyll content.

Table 4.
Effects of inoculation with two PGPE strains on different growth parameters in rice after transplantation for 10 and 20 days.

Figure 4.
Effects of inoculating with PGPE bacteria on the growth of shoots and roots in rice seedlings after treatment for 10 and 20 days. Representative images are shown of rice seedlings with or without inoculation using endophytic bacteria for 10 and 20 days ((A) and (B), respectively).
4. Discussion
Plant seeds harbor complex and variable microbial communities that may be an important source of endophytic bacteria [11]. Most of the publications on seed endophytic bacteria have reported members belonging to Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes [21]. Strains assigned to Pantoea, Bacillus, Methylobacterium, Rhizobiumr, Xanthomonas, Sphingomonas, and Microbacterium have been found inside both the hybrid and cultivated rice seed [48,49]. In the present study, the molecular characterization of 47 strains revealed a significant taxonomic diversity, including members of α-, β-, and γ-proteobacteria, Firmicutes, and Actinobacteria, representing 13 different genera (Table 2 and Figure 1). The dominant genus observed in this study was Bacillus (at least eight out of 47 isolates), which is consistent with previous reports [50]. Apart from Bacillus, the Actinobacterial Microbacterium and Curtobacterium were the prevalent genera (Table 2), as also reported for mature seeds of rice varieties cultivated in the Philippines and Japan [51,52]. The core microbiome of Bacillus and Microbacterium from seeds is consistent with our previous results from Dongxiang wild rice [35], and is supported by the recent finding that most seed transmitted bacteria are part of a core plant microbiome [17]. Some seed-borne pathogenic endophytes, such as Burkholderia spp. and Xanthomonas spp., were identified in our study (Figure 1b). However, Cottyn et al. [53] found that many Burkholderia spp. and Xanthomonas spp. isolated from rice seed fail to cause disease symptoms in common cultivated rice, suggesting they are in fact not pathogenic. Interestingly, we obtained strains of Lysinibacillus sphaericus and Facklamia tabacinasalis, which have not been obtained previously identified in other rice seed studies, suggesting that the study of different plant varieties, using different isolation protocols and distinct growth media can yield differences in the cultivatable population of bacterial endophytes species [54,55].
IAA is a plant hormone which stimulates the development of root systems and promotes plant-microbe interactions [56,57]. Almost all the isolates in the present study are capable of producing IAA, with the strain Pantoea sp. Fse28 showing the highest IAA production (90.11 ± 5.90 mg L−1) (Table 2 and Figure 3), which representhigher levels of IAA production than typically found in other seed endophytic bacteria [24,58,59]. The solubilization of insoluble phosphates by PGP bacteria could increase the availability of the limiting nutrient phosphorus to the host [60]. Previous experiments have shown that endophytic bacteria possess the capacity to solubilize immobilized mineral phosphates [61], suggesting that during initial colonization, endophytic bacteria could enhance phosphate availability to the host plant. About 79% of bacteria have the trait of CaP-Solubilization, three of which strains were shown to result in high levels of soluble phosphorus release (>500 μg ml−1) (Table 2 and Figure 3). Similarly high numbers of phosphate solubilizing bacteria were observed in rice cultivars grown in acidic P-limited soil, as previously documented by Hameed [62]. The abundance of phosphate solubilizing bacteria in Dongxiang wild rice seeds may be related to its long-term growth in low phosphate soil, where phosphate solubilizing bacteria could assist the plant to obtain more available phosphate from the red soil for the host plant to grow.
Plants form symbiotic relationships with microbes, including endophytes, in order to facilitate biological nitrogen fixation via the conversion of atmospheric nitrogen gas into a usable form of nitrogen [5,63]. In this study, 59.57% of the seed-associated microbes (Sphingomonas, Pseudomonas, Pantoea, and Microbacterium) could grow on N-free media (Table 2; Figure 3), which suggests that they were capable of either nitrogen fixation or N-scavenging [64,65,66]. The ability to produce siderophores is a trait that facilitates bacteria-plant associations and contributes to the colonization of the plant [67]. Some siderophore-producing bacteria have been reported to be found in rice, with the main genera being Pantoea, Bacillus, Pseudomonas, and Burkholderia [68,69]. Our findings showed 17 bacterial endophytes affiliated to 11 different genera (Table 2; Figure 3). Moreover, the diversity of the siderophore-producing bacteria was significantly higher in DXWR seeds than other studies [70,71]. It was noteworthy that we identified high siderophore production capacity in F. tabacinasalis for the first time.
Bacteria can help plants to withstand stress by reducing the level of the stress hormone ethylene through the activity of the enzyme ACC deaminase which hydrolyses ACC into α-ketobutyrate and ammonia instead of ethylene [72]. Organisms with ACC deaminase activiy of 0.062–2.664 μmol α-ketobutyrate mg−1 protein h−1 or higher can promote host plant growth [73]. In our study, among the 47 isolates, 17 strains were identified with ACC deaminase activity of 2.12–20.72 μmol α-ketobutyrate mg−1 protein h−1, where Fse28 and Fse35 had the highest ACC deaminase activities of 19.51 ± 1.64 and 20.72 ± 1.89 μmol α-ketobutyrate mg−1 protein h−1, respectively (Table 2). In addition, Fse28 and Fse35 have five PGP traits, such as IAA production, siderophore-producing, nitrogen fixation, phosphorus solubilization, and ACC deaminase activities. PGP validation experiments found that Fse28 (Pantoea sp.) and Fse35 (Curtobacterium sp.) significantly improved the agronomic traits of cultivated rice seedlings (Figure 4). Pantoea spp. has been reported to enhance plant development and improve the photosynthetic efficiency of crops [74], and Curtobacterium spp. could improve the drought resistance and metal tolerance of plants [75,76]. These studies suggested that the endophytic bacteria Fse28 and Fse35 could potentially be used to stimulate tolerance to environmental stress and promote plant growth.
5. Conclusions
Endophytic bacteria community structure was analyzed in the seeds of DXWR using culture-dependent methods. The seeds of DXWR harbored an abundant and diverse culturable endophytic bacteria community, some of which were capable of promoting plant growth. Most of the isolated endophytic bacteria were capable of producing IAA and solubilizing phosphate, and some could also produce siderophores, exhibit ACC deaminase activities, and fix nitrogen. The inoculation of rice seedlings with strains Fse28 and Fse35 exhibited a significant increase in root length, shoot length, dry matter, and chlorophyll content compared with the controls also grown under growth chamber conditions. We think that the mechanisms these microbes use to stimulate rice germination and growth work by increasing the nutrient availability of nitrogen, phosphorous, and iron as well as the production of phytohormones. In future studies, we will investigate the dynamics of plant nitrogen, phosphorus, and potassium after inoculation to further understand the mechanisms used by these plant growth-promoting bacteria to benefit their host.
Author Contributions
Conceptualization, D.Z.; data curation, Y.J.; formal analysis, R.Y.; funding acquisition, D.Z.; investigation, Z.Z.; methodology, T.L., X.Z., and Y.W.; resources, X.Z. and J.X.; validation, Y.W.; visualization, T.L. and Y.J.; writing—original draft, Z.Z.; writing—review & editing, D.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31760160, 31960078).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Sequences were deposited in GenBank under accession numbers KJ733853~KJ733899.
Acknowledgments
We would like to thanks Key Laboratory of Protection and Utilization of Subtropical Plant Resources of Jiangxi Province for technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cordovez, V.; Dini-Andreote, F.; Carrion, V.J.; Raaijmakers, J.M. Ecology and evolution of plant microbiomes. Annu. Rev. Microbiol. 2019, 73, 69–88. [Google Scholar] [CrossRef]
- Compant, S.; Cambon, M.C.; Vacher, C.; Mitter, B.; Samad, A.; Sessitsch, A. The plant endosphere world—Bacterial life within plants. Environ. Microbiol. 2021, 23, 1812–1829. [Google Scholar] [CrossRef]
- Santoyo, G.; Moreno-Hagelsieb, G.; Orozco-Mosqueda, M.D.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef]
- Ali, M.; Ali, Q.; Sohail, M.A.; Ashraf, M.F.; Saleem, M.H.; Hussain, S.; Zhou, L. Diversity and taxonomic distribution of endophytic bacterial community in the rice plant and its prospective. Int. J. Mol. Sci. 2021, 22, 10165. [Google Scholar] [CrossRef] [PubMed]
- Rana, K.L.; Kour, D.; Kaur, T.; Devi, R.; Yadav, A.N.; Yadav, N.; Dhaliwal, H.S.; Saxena, A.K. Endophytic microbes: Biodiversity, plant growth-promoting mechanisms and potential applications for agricultural sustainability. Antonie Van Leeuwenhoek 2020, 113, 1075–1107. [Google Scholar] [CrossRef]
- Hu, J.; Wei, Z.; Weidner, S.; Friman, V.-P.; Xu, Y.-C.; Shen, Q.-R.; Jousset, A. Probiotic Pseudomonas communities enhance plant growth and nutrient assimilation via diversity-mediated ecosystem functioning. Soil. Biol. Biochem. 2017, 113, 122–129. [Google Scholar] [CrossRef]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef]
- Shahzad, R.; Khan, A.L.; Bilal, S.; Waqas, M.; Kang, S.-M.; Lee, I.-J. Inoculation of abscisic acid-producing endophytic bacteria enhances salinity stress tolerance in Oryza sativa. Environ. Exp. Bot. 2017, 136, 68–77. [Google Scholar] [CrossRef]
- Orozco-Mosqueda, M.d.C.; Flores, A.; Rojas-Sánchez, B.; Urtis-Flores, C.A.; Morales-Cedeño, L.R.; Valencia-Marin, M.F.; Chávez-Avila, S.; Rojas-Solis, D.; Santoyo, G. Plant growth promoting bacteria as bioinoculants: Attributes and challenges for sustainable crop improvement. Agronomy 2021, 11, 1167. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Babalola, O.O. Roles of plant endosphere microbes in agriculture—A review. J. Plant Growth Regul. 2021, 1–18. [Google Scholar] [CrossRef]
- Samreen, T.; Naveed, M.; Nazir, M.Z.; Asghar, H.N.; Khan, M.I.; Zahir, Z.A.; Kanwal, S.; Jeevan, B.; Sharma, D.; Meena, V.S.; et al. Seed associated bacterial and fungal endophytes: Diversity, life cycle, transmission, and application potential. Appl. Soil Ecol. 2021, 168, 104191. [Google Scholar] [CrossRef]
- Mitter, B.; Pfaffenbichler, N.; Flavell, R.; Compant, S.; Antonielli, L.; Petric, A.; Berninger, T.; Naveed, M.; Sheibani-Tezerji, R.; von Maltzahn, G.; et al. A new approach to modify plant microbiomes and traits by Introducing beneficial bacteria at flowering into progeny seeds. Front. Microbiol. 2017, 8, 11. [Google Scholar] [CrossRef] [Green Version]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial endophyte colonization and distribution within plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, E.B. The seed microbiome: Origins, interactions, and impacts. Plant Soil 2018, 422, 7–34. [Google Scholar] [CrossRef]
- Rodríguez, C.E.; Mitter, B.; Barret, M.; Sessitsch, A.; Compant, S. Commentary: Seed bacterial inhabitants and their routes of colonization. Plant Soil 2017, 422, 129–134. [Google Scholar] [CrossRef]
- Johnston-Monje, D.; Lundberg, D.S.; Lazarovits, G.; Reis, V.M.; Raizada, M.N. Bacterial populations in juvenile maize rhizospheres originate from both seed and soil. Plant Soil 2016, 405, 337–355. [Google Scholar] [CrossRef] [Green Version]
- Johnston-Monje, D.; Gutiérrez, J.P.; Lopez-Lavalle, L.A.B. Seed-Transmitted Bacteria and Fungi Dominate Juvenile Plant Microbiomes. Front. Microbiol. 2021, 12, 737616. [Google Scholar] [CrossRef] [PubMed]
- Shade, A.; Jacques, M.A.; Barret, M. Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr. Opin. Microbiol. 2017, 37, 15–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, M.M.; Flory, E.; Koyro, H.W.; Abideen, Z.; Schikora, A.; Suarez, C.; Schnell, S.; Cardinale, M. Consistent associations with beneficial bacteria in the seed endosphere of barley (Hordeum vulgare L.). Syst. Appl. Microbiol. 2018, 41, 386–398. [Google Scholar] [CrossRef]
- Ferreira, A.; Quecine, M.C.; Lacava, P.T.; Oda, S.; Azevedo, J.L.; Araujo, W.L. Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol. Lett. 2008, 287, 8–14. [Google Scholar] [CrossRef] [Green Version]
- Truyens, S.; Weyens, N.; Cuypers, A.; Vangronsveld, J. Bacterial seed endophytes: Genera, vertical transmission and interaction with plants. Environ. Microbiol. Rep. 2015, 7, 40–50. [Google Scholar] [CrossRef]
- Khalaf, E.M.; Raizada, M.N. Bacterial seed endophytes of domesticated cucurbits antagonize fungal and oomycete pathogens Including powdery mildew. Front. Microbiol. 2018, 9, 42. [Google Scholar] [CrossRef]
- Jeong, S.; Kim, T.M.; Choi, B.; Kim, Y.; Kim, E. Invasive Lactuca serriola seeds contain endophytic bacteria that contribute to drought tolerance. Sci. Rep. 2021, 11, 13307. [Google Scholar] [CrossRef]
- Xu, M.; Sheng, J.; Chen, L.; Men, Y.; Gan, L.; Guo, S.; Shen, L. Bacterial community compositions of tomato (Lycopersicum esculentum Mill.) seeds and plant growth promoting activity of ACC deaminase producing Bacillus subtilis (HYT-12-1) on tomato seedlings. World J. Microb. Biot. 2014, 30, 835–845. [Google Scholar] [CrossRef]
- Xie, J.; Agrama, H.A.; Kong, D.; Zhuang, J.; Hu, B.; Wan, Y.; Yan, W. Genetic diversity associated with conservation of endangered Dongxiang wild rice (Oryza rufipogon). Genet. Resour. Crop Evol. 2010, 57, 597–609. [Google Scholar] [CrossRef]
- Qiu, B.Y.; Xiong, Y.Z.; Yu, L.Q.; Hu, B.L.; Zhang, Z.; Xie, J.K. Cluster analysis of plant morphological characters in population of Dong Xiang wild rice. Acta Agric. Jiangxi 2006, 18, 1–5. [Google Scholar] [CrossRef]
- Li, L.; Chen, H.; Mao, D. Pyramiding of rapid germination loci from Oryza Sativa cultivar ‘Xieqingzao B’ and cold tolerance loci from Dongxiang wild rice to increase climate resilience of cultivated rice. Mol. Breed. 2019, 39, 85. [Google Scholar] [CrossRef]
- Qi, W.D.; Chen, H.P.; Yang, Z.Z.; Hu, B.L.; Luo, X.D.; Ai, B.; Luo, Y.; Huang, Y.; Xie, J.K.; Zhang, F.T. Systematic characterization of long non-coding RNAs and their responses to drought stress in Dongxiang wild rice. Rice Sci. 2020, 27, 21–31. [Google Scholar] [CrossRef]
- Cui, L.L.; Li, L.; Han, X.X.; Xing, J.J.; Li, L.L.; Yang, Z.G.; Luo, X.J.; Li, D.; Xie, L.L.; Cao, M.L. Cloning and analysis of candidate RPS2 in Dongxiang wild rice. J. Hunan Agric. Univ. 2009, 35, 111–115. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, P.; Cui, F.; Zhang, F.; Luo, X.; Xie, J. Transcriptome Analysis of Salt Stress Responsiveness in the Seedlings of Dongxiang Wild Rice (Oryza rufipogon Griff.). PLoS ONE. 2016, 11, e0146242. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.X.; Peng, G.X.; Wang, E.T.; Yan, H.; Yuan, Q.H.; Zhang, W.; Lou, X.; Wu, H.; Tan, Z.Y. Diverse endophytic nitrogen-fixing bacteria isolated from wild rice Oryza rufipogon and description of Phytobacter diazotrophicus gen. nov. sp. nov. Arch. Microbiol. 2008, 189, 431–439. [Google Scholar] [CrossRef]
- Chaudhary, H.J.; Peng, G.; Hu, M.; He, Y.; Yang, L.; Luo, Y.; Tan, Z. Genetic diversity of endophytic diazotrophs of the wild rice, Oryza alta and identification of the new diazotroph, Acinetobacter oryzae sp. nov. Microb. Ecol. 2012, 63, 813–821. [Google Scholar] [CrossRef]
- Borah, M.; Das, S.; Bora, S.S.; Boro, R.C.; Barooah, M. Comparative assessment of multi-trait plant growth-promoting endophytes associated with cultivated and wild Oryza germplasm of Assam, India. Arch. Microbiol. 2021, 203, 2007–2028. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Wang, Y.; Zeng, Q.; Yan, R.; Zhang, Z.; Zhu, D. Diversity and plant growth promoting activities of the cultivable rhizobacteria of Dongxiang wild rice (Oryza rufipogon). Biodivers. Sci. 2011, 19, 476–484. [Google Scholar]
- Chen, Z.Y.; Liu, J.; Yang, X.P.; Liu, M.; Wang, Y.; Zhang, Z.B.; Zhu, D. Community composition and diversity of cultivable endophytic bacteria isolated from Dongxiang wild rice. Biodivers. Sci. 2019, 27, 1320–1329. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Gao, B.L.; Li, X.X.; Zhang, Z.B.; Yan, R.M.; Yang, H.L.; Zhu, D. Phylogenetic diversity of culturable endophytic fungi in Dongxiang wild rice (Oryza rufipogon Griff), detection of polyketide synthase gene and their antagonistic activity analysis. Fungal Biol. 2015, 119, 1032–1045. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.B.; Deng, Y.M.; Xiong, Y.Y.; Wang, Y.; Yan, R.M.; Zhu, D. Isolation of endophytic actinomycetes from dongxiang wild rice (oryza rufipogon) and analysis of secondary metabolite of active strain s123. Microbiol. China 2015, 42, 1662–1670. [Google Scholar]
- Zeng, Q.G.; Luo, F.; Zhang, Z.B.; Yan, R.M.; Zhu, D. Phosphate solubilizing rhizospherebacterial t21 isolated from Dongxiang wild rice species promotes cultivated rice growth. Appl. Mech. Mater. 2011, 108, 167–175. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evolut. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estrada-De los Santos, P.; Bustillos-Cristales, R.; Caballero-Mellado, J. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 2001, 67, 2790–2798. [Google Scholar] [CrossRef] [Green Version]
- Arora, N.K.; Verma, M. Modified microplate method for rapid and efficient estimation of siderophore produced by bacteria. 3 Biotech 2017, 7, 381. [Google Scholar] [CrossRef] [Green Version]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Mehta, S.; Nautiyal, C.S. An efficient method for qualitative screening of phosphate-solubilizing bacteria. Curr. Microbiol. 2001, 43, 51–56. [Google Scholar] [CrossRef]
- Ribeiro, C.M.; Cardoso, E.J.B.N. Isolation, selection and characterization of root-associated growth promoting bacteria in Brazil Pine (Araucaria angustifolia). Microbiol. Res. 2012, 167, 69–78. [Google Scholar] [CrossRef]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plantarum. 2003, 118, 10–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porra, R.J. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 2002, 73, 149–156. [Google Scholar] [PubMed]
- Zhang, J.; Zhang, C.W.; Yang, J.; Zhang, R.J.; Gao, J.S.; Zhao, X.; Zhao, J.J.; Zhao, D.F.; Zhang, X.X. Insights into Endophytic Bacterial Community Structures of Seeds Among Various Oryza sativa L. Rice Genotypes. J. Plant Growth Regul. 2019, 38, 93–102. [Google Scholar]
- Liu, Y.; Xu, P.; Yang, F.; Li, M.; Yan, H.; Li, N.; Zhang, X.; Wang, W. Composition and diversity of endophytic bacterial community in seeds of super hybrid rice ‘Shenliangyou 5814’ (Oryza sativa L.) and its parental lines. Plant Growth Regul. 2018, 87, 257–266. [Google Scholar] [CrossRef]
- Okunishi, S.; Sako, K.; Mano, H.; Imamura, A.; Morisaki, H. Bacterial flora of endophytes in the maturing seed of cultivated rice (Oryza sativa). Microbes Environ. 2005, 20, 168–177. [Google Scholar] [CrossRef] [Green Version]
- Mano, H.; Tanaka, F.; Watanabe, A.; Kaga, H.; Okunishi, S.; Morisaki, H. Culturable surface and endophytic bacterial flora of the maturing seeds of rice plants (Oryza sativa) cultivated in a paddy field. Microbes Environ. 2006, 21, 86–100. [Google Scholar] [CrossRef] [Green Version]
- Cottyn, B.; Debode, J.; Regalado, E.; Mew, T.W.; Swings, J. Phenotypic and genetic diversity of rice seed-associated bacteria and their role in pathogenicity and biological control. J. Appl. Microbiol. 2009, 107, 885–897. [Google Scholar] [CrossRef]
- Cottyn, B.; Regalado, E.; Lanoot, B.; De Cleene, M.; Mew, T.W.; Swings, J. Bacterial populations associated with rice seed in the tropical environment. Phytopathology 2001, 91, 282–292. [Google Scholar] [CrossRef] [Green Version]
- Rosenblueth, M.; Martinez-Romero, E. Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 2006, 19, 827–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, G.; Kanaujia, N.; Bafana, A. Functional and phylogenetic diversity of root-associated bacteria of Ajuga bracteosa in Kangra valley. Microbiol. Res. 2012, 167, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Duca, D.; Lorv, J.; Patten, C.L.; Rose, D.; Glick, B.R. Indole-3-acetic acid in plant-microbe interactions. Anton. Leeuw. Int. J. Gen. 2014, 106, 85–125. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.; Xu, J.; Acosta, K.; Poulev, A.; Lebeis, S.; Lam, E. Bacterial production of indole related compounds reveals their role in association between duckweeds and endophytes. Front. Chem. 2018, 6, 265. [Google Scholar] [CrossRef] [Green Version]
- Ruiza, D.; Agaras, B.; de Werrab, P.; Wall, L.G.; Valverde, C. Characterization and screening of plant probiotic traits of bacteria isolated from rice seeds cultivated in Argentina. J. Microbiol. 2011, 49, 902–912. [Google Scholar] [CrossRef]
- Shahzad, R.; Waqas, M.; Khan, A.L.; Al-Hosni, K.; Kang, S.M.; Seo, C.W.; Lee, I.J. Indoleacetic acid production and plant growth promoting potential of bacterial endophytes isolated from rice (Oryza sativa L.) seeds. Acta Biol. Hung. 2017, 68, 175–186. [Google Scholar] [CrossRef] [Green Version]
- Rawat, P.; Das, S.; Shankhdhar, D.; Shankhdhar, S.C. Phosphate-solubilizing microorganisms: Mechanism and their role in phosphate solubilization and uptake. J. Soil Sci. Plant Nut. 2020, 21, 49–68. [Google Scholar] [CrossRef]
- Varga, T.; Hixson, K.K.; Ahkami, A.H.; Sher, A.W.; Barnes, M.E.; Chu, R.K.; Battu, A.K.; Nicora, C.D.; Winkler, T.E.; Reno, L.R.; et al. Endophyte-promoted phosphorus solubilization in Populus. Front. Plant Sci. 2020, 11, 567918. [Google Scholar] [CrossRef]
- Hameed, A.; Yeh, M.W.; Hsieh, Y.T.; Chung, W.C.; Lo, C.T.; Young, L.S. Diversity and functional characterization of bacterial endophytes dwelling in various rice (Oryza sativa L.) tissues, and their seed-borne dissemination into rhizosphere under gnotobiotic P-stress. Plant Soil 2015, 394, 177–197. [Google Scholar] [CrossRef]
- Nag, P.; Shriti, S.; Das, S. Microbiological strategies for enhancing biological nitrogen fixation in nonlegumes. J. Appl. Microbiol. 2020, 129, 186–198. [Google Scholar] [CrossRef] [Green Version]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, S.H.; Gururani, M.A.; Chun, S.-C. Isolation and characterization of plant growth promoting endophytic diazotrophic bacteria from Korean rice cultivars. Microbiol. Res. 2014, 169, 83–98. [Google Scholar] [CrossRef]
- Walitang, D.I.; Kim, K.; Madhaiyan, M.; Kim, Y.K.; Kang, Y.; Sa, T. Characterizing endophytic competence and plant growth promotion of bacterial endophytes inhabiting the seed endosphere of rice. BMC Microbiol. 2017, 17, 209. [Google Scholar] [CrossRef]
- Ferreira, M.J.; Silva, H.; Cunha, A. Siderophore-producing rhizobacteria as a promising tool for empowering plants to cope with iron limitation in saline soils: A review. Pedosphere 2019, 29, 409–420. [Google Scholar] [CrossRef]
- Yang, J.H.; Liu, H.X.; Zhu, G.M.; Pan, Y.L.; Xu, L.P.; Guo, J.H. Diversity analysis of antagonists from rice-associated bacteria and their application in biocontrol of rice diseases. J. Appl. Microbiol. 2008, 104, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Loaces, I.; Ferrando, L.; Scavino, A.F. Dynamics, diversity and function of endophytic siderophore-producing bacteria in rice. Microb. Ecol. 2011, 61, 606–618. [Google Scholar] [CrossRef]
- Sultana, S.; Alam, S.; Karim, M.M. Screening of siderophore-producing salt-tolerant rhizobacteria suitable for supporting plant growth in saline soils with iron limitation. J. Agric. Food Res. 2021, 4, 100150. [Google Scholar] [CrossRef]
- Ghavami, N.; Alikhani, H.A.; Pourbabaei, A.A.; Besharati, H. Effects of two new siderophore-producing rhizobacteria on growth and iron content of maize and canola plants. J. Plant. Nutr. 2017, 40, 736–746. [Google Scholar] [CrossRef]
- Murali, M.; Gowtham, H.G.; Singh, S.B.; Shilpa, N.; Aiyaz, M.; Niranjana, S.R.; Amruthesh, K.N. Bio-prospecting of ACC deaminase producing rhizobacteria towards sustainable agriculture: A special emphasis on abiotic stress in plants. Appl. Soil Ecol. 2021, 168, 104142. [Google Scholar] [CrossRef]
- Bal, H.B.; Nayak, L.; Das, S.; Adhya, T.K. Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant Soil 2013, 366, 93–105. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Hernandez, A.G.; Glick, B.R.; Rossi, M.J. The extreme plant-growth-promoting properties of Pantoea phytobeneficialis MSR2 revealed by functional and genomic analysis. Environ. Microbiol. 2020, 22, 1341–1355. [Google Scholar] [CrossRef] [PubMed]
- Silambarasan, S.; Logeswari, P.; Valentine, A.; Cornejo, P. Role of Curtobacterium herbarum strain CAH5 on aluminum bioaccumulation and enhancement of Lactuca sativa growth under aluminum and drought stresses. Ecotox. Environ. Saf. 2019, 183, 109573. [Google Scholar] [CrossRef]
- Bourles, A.; Guentas, L.; Chalkiadakis, E.; Majorel, C.; Juillot, F.; Cavaloc, Y.; Burtet-Sarramegna, V.; Medevielle, V.; Jourand, P.; Amir, H. New Caledonian ultramafic conditions structure the features of Curtobacterium citreum strains that play a role in plant adaptation. Can. J. Microbiol. 2019, 65, 880–894. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).