Abstract
In the previously published mitochondrial genome sequence of Artemia urmiana (NC_021382 [JQ975176]), the taxonomic status of the examined Artemia had not been determined, due to parthenogenetic populations coexisting with A. urmiana in Urmia Lake. Additionally, NC_021382 [JQ975176] has been obtained with pooled cysts of Artemia (0.25 g cysts consists of 20,000–25,000 cysts), not a single specimen. With regard to coexisting populations in Urmia Lake, and intra- and inter-specific variations in the pooled samples, NC_021382 [JQ975176] cannot be recommended as a valid sequence and any attempt to attribute it to A. urmiana or a parthenogenetic population is unreasonable. With the aid of next-generation sequencing methods, we characterized and assembled a complete mitochondrial genome of A. urmiana with defined taxonomic status. Our results reveal that in the previously published mitogenome (NC_021382 [JQ975176]), tRNA-Phe has been erroneously attributed to the heavy strand but it is encoded in the light strand. There was a major problem in the position of the ND5. It was extended over the tRNA-Phe, which is biologically incorrect. We have also identified a partial nucleotide sequence of 311 bp that was probably erroneously duplicated in the assembly of the control region of NC_021382 [JQ975176], which enlarges the control region length by 16%. This partial sequence could not be recognized in our assembled mitogenome as well as in 48 further examined specimens of A. urmiana. Although, only COX1 and 16S genes have been widely used for phylogenetic studies in Artemia, our findings reveal substantial differences in the nucleotide composition of some other genes (including ATP8, ATP6, ND3, ND6, ND1 and COX3) among Artemia species. It is suggested that these markers should be included in future phylogenetic studies.
1. Introduction
Mitogenomes have been extensively utilized in phylogeny and population genetic studies because mitochondrial genes share some particular characteristics, including maternal origin, rapid evolutionary rates and lack of recombination [1,2]. Sequences of mitochondrial genes have been widely applied as informative molecular markers; therefore, they have been widely used in the molecular phylogenetic studies [3,4,5,6]. However, compared to partial mitochondrial sequences, such as COX1, 16S, 12S, etc., complete mitochondrial genome sequences could provide more of a higher resolution and provide better understanding about evolutionary relationships and structures of animals [4,7].
The number of complete mitochondrial sequences from metazoa has been rising rapidly in the past decade. With the rapid advancement in genomics techniques, such as next-generation sequencing (NGS) technology, many genomes and mitogenomes have been characterized [6,8,9]. The current methodology provides high quality results and has allowed us to accurately determine the gene order and organization in many taxa and to compare the outcomes with conventional sequencing methods [10,11,12,13]. Complete mitochondrial genomes have proved useful in solving evolutionary and biosystematics questions over a broad taxonomic range [6,14].
The genus Artemia Leach, 1819, is an anostracan zooplankter, which lives in more than 600 saline and hypersaline habitats worldwide, except Antarctica [15]. The first scientific report of the brine shrimp Artemia backs to the first half of the 10th century AD from Urmia Lake by Estakhri (d. 951/957 AD, 10th C.), an Iranian geographer, who named it “aquatic dog” [16,17]. The genus Artemia consists of seven bisexual biological species, a large number of parthenogenetic populations and some undescribed and/or unidentified taxa [18,19]. Four bisexual species are native to the Old World namely Artemia salina (Linnaeus, 1758), Artemia urmiana Günther, 1899, Artemia sinica Cai, 1989, and Artemia tibetiana Abatzopoulos et al. (1998). The other three bisexual species are located in the New World consisting of Artemia monica Verrill, 1869, Artemia franciscana Kellogg, 1906, and Artemia persimilis Piccinelli and Prosdocimi, 1968 [18,20,21]. Obligate parthenogenetic Artemia taxa have di-, tri-, tetra- and pentaploid populations [19].
Overall, the phylogenetic relationship within Artemia is presently still ambiguous [22]. Obligate asexuality is one of the critical challenges in biosystematics of the genus Artemia. Accordingly, asexual forms of Artemia do not form defined species but parthenogenetic population(s) [18,23]. A comprehensive phylogenetic study using mitochondrial and nuclear markers provided evidence that the parthenogenetic Artemia taxa form a polyphyletic group. The diploids and triploids are maternally related to A. urmiana, whereas tetrapod and pentaploid lineages share a common ancestor with Artemia sinica [19]. Although the mitochondrial marker Cytochrome Oxidase Subunit I (COX1) has confirmed that parthenogenetic populations were divided into two polyphyletic groups, the nuclear marker ITS1 suggests a common haplotype with the participation of all ploidy degrees [19,24]. Additionally, Eimanifar et al. [24] showed that parthenogenetic populations share a major haplotype with A. urmiana and A. tibetiana based on the ITS1 nuclear maker.
Maniatsi et al. [25] and Eimanifar et al. [24] have proposed different phylogenetic positions for A. salina and A. persimilis, based on nucleotide sequences of COX1. However, results of morphological and genetic investigations were contradictory [22]. To date, the taxonomy and biosystematics of Artemia are still controversial, especially concerning the Asian species [21]. Mitogenomic information could provide a better reconstruction of the maternal evolutionary mechanism and phylogenetic status of Artemia. However, only four mitochondrial genomes of Artemia species have been published to date.
The previously published mitogenome analysis, performed by Zhang et al. [26] for two Artemia species, Artemia urmiana and Artemia tibetiana, detected an unprecedented unusual nucleotide sequence in the mitochondrial genome of A. urmiana from Urmia Lake, Iran. The length of the complete mitochondrial genome of A. urmiana (GenBank accession number NC_021382 [JQ975176]), is 15,945 bp; its control region would be significantly longer than other species. In addition, the taxonomic status of the assembled mitogenome had not been identified. For this examined mitogenome, Zhang et al. have used Artemia cysts stored at the Laboratory of Aquaculture & Artemia Reference Center (ARC, Gent University), which are labeled as ARC 1227 [26]. This sample has been collected in 1991 and it has been only registered as “from Urmia Lake” in ARC documents (Christ Mahieu, personal communication, ARC, Gent University).
Artemia urmiana has coexisted with a low percentage of parthenogenetic populations in the main body of Urmia Lake in western Iran, whereas coastal areas and neighboring lagoons of Urmia Lake hosted mostly parthenogenetic populations. Barigozzi and Baratelli [27] documented that all samples collected from Urmia Lake in 1987 were parthenogenetic and contained di-, tetra-, pentaploid individuals. However, Azari Takami [28] reported that A. urmiana and parthenogenetic populations coexisted in the lake. Asem et al. [29] identified two partly isolated parthenogenetic populations from the main body of Urmia Lake and lagoons neighboring the lake. Generally, some of the adjacent lagoons are connected with Urmia Lake when the lake level raises annually during rainy seasons in spring and autumn [29], which increases the probability of collecting parthenogenetic specimens along the shoreline of the lake (Atashbar, personal communication, Urmia University). On the other hand, genetic variation between parthenogenetic populations and A. urmiana is quite low [19,21]. Additionally, a current study based on barcoding with the mitochondrial COX1 marker found that parthenogenetic populations in some localities share same haplotypes with A. urmiana [30]. Based on this fact, the use of COX1 sequencing alone to distinguish A. urmiana from di- and triploids is questionable. However, these populations need to be further examined with special emphasis on the status of reproductive mode (bisexual or parthenogenesis) [31], morphological traits [29,32,33] and a Single Nucleotide Polymorphism (SNP) in the Na+/K+ ATPase α-1 subunit [34] to distinguish and identify A. urmiana and parthenogenetic populations. The taxonomic status of Artemia from Urmia Lake should be identified before genome sequencing, see [21,35], which has not been considered by Zhang et al. [26]. There is a critical problem in Zhang et al. [26] mitogenome sequence that it has been procured by pooled cysts of Artemia (0.25 g cyst consists of 20,000–25,000 cysts) from Urmia Lake. It is a pivotal puzzle how differences among individuals have been ignored in assembling process. In this condition, the NC_021382 [JQ975176] mitogenome cannot be a valid sequence and any attempt to attribute it to A. urmiana or a parthenogenetic population is unreasonable.
Through the present study, we confirmed the taxonomic status of A. urmiana sampled from Urmia Lake. The purpose of the current research was to re-sequence the complete mitochondrial genome of A. urmiana using Illumina Hi-Seq X next generation sequencing technology with a high-coverage. For that reason, we first aimed to fully re-annotate and characterize the complete mitochondrial genome of A. urmiana and to compare it with previously published assembly by Zhang et al. [26]. Due to lack of consistency regarding the common molecular markers to study Artemia phylogeny, our results will contribute to understanding interspecific variation among different mitochondrial genes, and to identify mitochondrial markers with high variability, and the evolutionary origin of Asian Artemia.
2. Materials and Methods
2.1. Artemia Sample, DNA Extraction and Sequencing
The cysts of Artemia urmiana were collected from Urmia Lake, Iran (Kholman-khaneh Port; 45°29′ E, 37°64′ N) in 2004. The cysts were transferred to the laboratory and stored at the optimum storage condition until analysis. The cysts were processed and incubated at the standard hatching conditions according to procedures suggested by Sorgeloos et al. [36]. Specimens were identified as belonging to the A. urmiana, as confirmed by the reproductive mode (bisexual or parthenogenetic) [19] and a SNP in the Na+/K+ ATPase α-1 subunit [34]. The reared specimen showed the rudimentary furca characterized by oligosetae, which is a main morphological character for A. urmiana [32], while parthenogenetic populations of Urmia Lake have a furca with two lobes and polysetae [29,30,31,32,33]. In order to distinguish it from other bisexual species of Artemia (especially invasive American A. franciscana), the taxonomic status of the chosen specimen was ultimately re-confirmed by applying the standard nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) using COX1 sequences based on Eimanifar et al. [24] datasets.
Total genomic DNA was extracted from a female using a Rapid Animal Genomic DNA Isolation Kit (Sangon Biotech Co., Ltd., Shanghai, China; NO. B518221), following the manufacturer’s instructions and procedures performed by Asem et al. [8]. The quality of extracted DNA was checked on a 1.5% agarose gel and then quantified using a Micro-volume Spectrophotometer (MaestroGen Inc., Hsinchu City, Taiwan). An amount of 600 ng of total DNA was used to construct the genomic library with paired-end (2 × 150 bp) using NEBNext® UltraTM II DNA library preparation kit, followed by next-generation sequencing (10 Gb) approach. The sequencing was performed by a single constructed library, pooled on Illumina HiSeq X-ten sequencing flow cell (Novogene Co., Tianjin, China).
2.2. Sequence Quality Control, Assembly and Annotation
Adapter residues were removed before further analyses. The Illumina sequence reads were qualitatively checked using FastQC [37]. Assembly and coverage metrics are summarized in Figure S1 and Table S1.
The resultant sequence reads were assembled and mapped to the Artemia sinica mitogenome as a reference genome (MK069595 [8]) using Geneious R9.1 [6] and bowtie v2.2.9 programs [38].
2.3. Gene Identification and Annotation
The position and secondary structure of the tRNA genes were determined by ARWEN (http://130.235.46.10/ARWEN/) online software. Protein-coding genes (PCGs) and ribosomal RNA genes (rRNAs) were annotated based on the gene order on the reference mitochondrial map using BLAST analysis (https://blast.ncbi.nlm.nih.gov). The position and orientation of the PCGs and rRNAs were recognized by the analysis of multiple sequence alignments to the reference mitogenome using BioEdit program [39]. In addition, all PCGs were translated into amino acids by the ExPASy online program (https://web.expasy.org/translate/), and sequences were examined to ensure that each could encode a functional protein.
2.4. Bioinformatics and Phylogenetic Analysis
GenomeVx online tool was utilized to draw the circular map of the mitochondrial genome of A. urmiana [40]. The complete mitochondrial sequences of A. sinica (MK069595), A. tibetiana (NC_021383) and A. franciscana (X69067) were downloaded from GenBank. The nucleotide composition and codon usage were calculated with DAMBE 6 [41], and AT- and GC-skew were also calculated using the following formulas: AT-skew = [A-T]/[A+T] and GC-skew = [G-C]/[G+C] [42]. The secondary structure of tRNAs were visualized using ARWEN online software. Sequences were aligned using MEGA X selected with MUSCLE default setting [43]. Pair-wise distances were computed for 13 PCGs and 2 rRNAs using the uncorrected p-distance nucleotide model as implemented in MEGA X [43]. Heat-maps of Euclidean distance among AT- and GC-skew were generated using the Heatmapper online tool [44].
Phylogenetic analyses of the concatenated datasets, including 13 PCGs and two rRNAs, were carried out using two different tree-building methods, Maximum Likelihood (ML) and Bayesian Inference (BI). The ML analysis was performed using MEGA X [43]. The BI was implemented in MrBayes 3.2.2 on XSEDE [45]. For ML and BI the best-fitting nucleotide substitution model was calculated based on the results of the MrModelltest 2.2 [46] test and HKY+G was chosen as the best-fit model (ML: bootstrap replicates: 1000, maximum parsimony analyses were run using TNT (Nearest-Neighbor-Interchange); BI: mcmcp ngen = 10,000,000, samplefreq = 100, nchains = 4, sump burnin = 25,000). The trees were visualized using FIGTREE v1.4.0 [47]. For the ML bootstraps, the values <70 were considered as low, 70–94 as middle, and ≥95 as high [48]. For the BI posterior probabilities, the values <0.94 were regarded as low, and ≥0.95 as high [49].
2.5. Complementary Experiments
To confirm the presence or absence of the 311 bp partial nucleotide in the mitochondrial sequence of A. urmiana, 48 further specimens have been examined following designed primers by Zhang et al. [26] to amplify this partial DNA region.
In order to consider sequence quality of the previously published mitogenome by Zhang et al. [26], the standard nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was performed to check similarity of 16S rRNA sequence of NC_021382 [JQ975176] among A. urmiana and parthenogenetic populations (di- and triploids).
3. Results
3.1. Genomic Characteristics and Gene Organization
The assembled NGS fragments show that the mitochondrial genome of A. urmiana has a typical circular structure including 22 tRNAs, 13 PCGs, 2 ribosomal RNAs and a control region (CR), with a total length of 15,699 bp (GenBank accession no. MN240408). There was no abnormality in the read sequences, which could refer to heteroplasmy and pseudogenes. The complete sequence was composed of 30.84% A, 20.29% C, 17.17% G and 31.70% T. In the complete mitochondrial genome (CmtG) and concatenated sequences of PCGs and rRNAs (PCGs+rRNAs), we found a strong A+T bias (62.54% and 61.89%, respectively). Nine tRNAs (Ile, Gln, Cys, Tyr, Phe, His, Pro, Leu2, and Val), four PCGs (ND5, ND4, ND4L, and ND1) and 16S and 12S ribosomal RNAs were encoded on the light strand (Figure 1). The longest overlaps and gaps were assigned between ATP8/ATP6 (−7 bp), tRNA-Gln/tRNA-Cys (+57 bp) and ND4/ND4L (+49 bp), respectively. The control region had a length of 1672 bp (Table S2). AT-skew and GC-skew were negative for both CmtG (−0.014 vs. −0.083) and PCG+rRNA (−0.165 vs. −0.053) sequences (Figure 2 and Table S3).
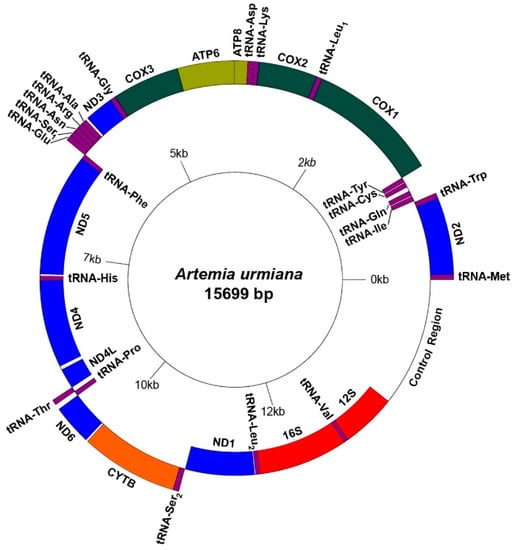
Figure 1.
Complete mitochondrial genome map of Artemia urmiana.
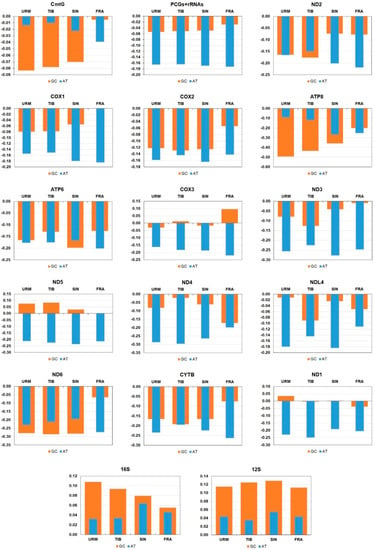
Figure 2.
AT- and GC-skews of the complete mitochondrial genome, protein-coding, ribosomal RNA genes in the mitochondrial genome of A. urmiana and three related species in genus Artemia (URM: this study; TIB: JQ975177; SIN: MK069595; FRA: X69067). URM: A. urmiana, TIB: A. tibetiana, SIN: A. sinica, FRA: A. franciscana.
3.2. Transfer RNAs
As expected, the mitogenome of A. urmiana included all of the 22 tRNAs that are commonly found in metazoan mtDNA. Except for tRNA-Ser1, which shows a D-loop structure, all tRNAs possess the typical cloverleaf structure. The highest and lowest values of AT percentage in tRNAs were detected in tRNA-Glu (80.3%) and tRNA-Ser1 (47.7%), respectively. The shortest and longest tRNAs were tRNA-Ala, tRNA-Ley (59 bp), and tRNA-Ser2 (67 bp), respectively (Figure 3).
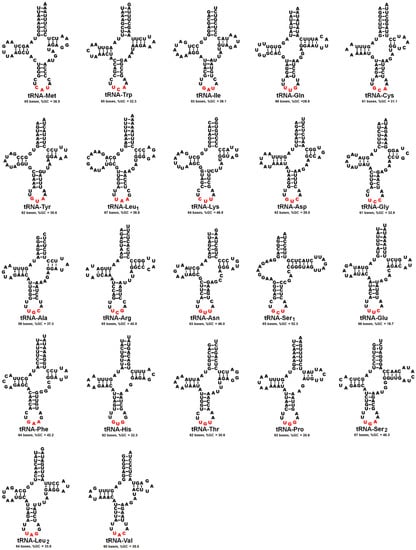
Figure 3.
Putative secondary structures of mitochondrial tRNAs determined in A. urmiana. Nucleotides in red color indicate anticodon sequences.
3.3. Protein-Coding Genes
Seven genes including ND2, COX1, Cox2, COX3, ND5, CYTB and ND1 start with the common ATG start codon. Six genes start with ATC (ATP8 and ND4L), GTG (ATP6 and ND6), or ATT (ND3) and ATA (ND4). Stop codons are TAA (ND2, ATP8, ATP6, COX3, ND4L, CYTB, and ND1), TAG (ND3 and ND6) and incomplete codons T (COX1, COX2 and ND5, and ND4) (Table S2). In A. urmiana, the AT- and GC-skews were negative in all encoded genes, except in ND5 and ND1, showed a positive GC-skew value (Figure 2 and Table S3).
The relative synonymous codon usage (RSCU) results are summarized in Table S4 and Figure 4. All amino acid codons are used in protein coding genes (PCGs) of A. urmiana. Codon usage analysis shows that PCGs totally include 3501 amino acids. Furthermore, AUU (201, 5.74%), UUU (200, 5.71%), UUA (156, 4.46%) were the most frequently used codons that covered almost 16% of all observed codons. Further analysis has indicated that the amino acids leucine (567, 16.2%) and serine (378, 10.80%) had the highest frequency, while cysteine (45, 1.29%), arginine (61, 1.74%), lysine (68, 1.94%) revealed the lowest frequency, respectively. AGG (7, 0.20%), CGG (10, 0.29%), CGC (11, 0.31%) were rarely used codons (Figure 5 and Table S4).
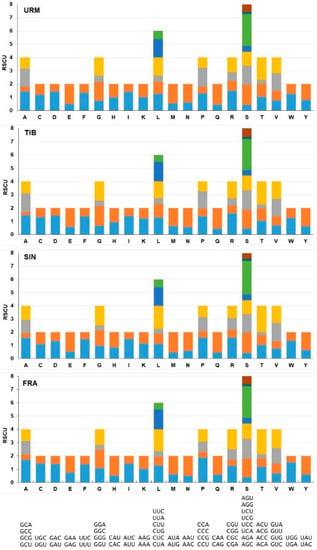
Figure 4.
Relative synonymous codon usage (RSCU) in A. urmiana and three related species in genus Artemia. Codon families were mentioned by the one letter and synonymous codons were listed on the x-axis (URM: A. urmiana, TIB: A. tibetiana, SIN: A. sinica, FRA: A. franciscana).
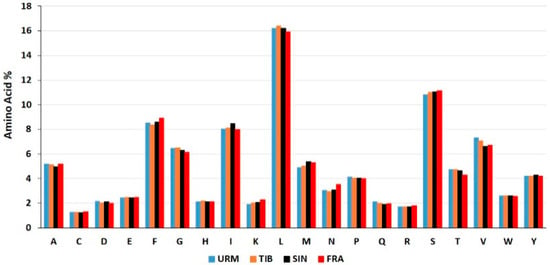
Figure 5.
Amino acid content (%) used in protein-coding genes in the mitochondrial genome of A. urmiana and three related species in genus Artemia (URM: A. urmiana, TIB: A. tibetiana, SIN: A. sinica, FRA: A. franciscana).
3.4. Ribosomal RNAs
The 16S ribosomal RNA and 12S ribosomal RNA are positioned between tRNA-Leu2 and control region, where they are separated by tRNA-Val. Both ribosomal RNA genes are encoded on the light strand from 12,099 to 13,255 (1157 bp) and from 13,317 to 14,027 (711 bp), respectively (Figure 1 and Table S2). The 16S ribosomal gene has a rather higher A+T content than 12S (63.96% vs. 60.90%). Both 16S and 12S ribosomal RNAs show positive values for AT- and GC-skew (Figure 2 and Table S3).
3.5. Correction of the Previously Published A. urmiana Mitogenomic Structure
The current assembled mitochondrial genome of A. urmiana (MN240408) has revealed a shorter length than one that has been deposited in GenBank under NC_021382 [JQ975176] accession number (15,699 bp vs. 15,945 bp). There were high sequence similarities between both mitochondrial genomes (98%) except for a length difference of 311 bp difference. The 16S rRNA sequence of Zhang et al. [26] represented high similarity with parthenogenetic populations (di- and triploids) compared to A. urmiana (96.76–96.21% vs. 95.63–96.07%).
The length difference concerned the sequence of the control region (1672 bp vs. 1932 bp). Our analysis showed that the partial DNA sequence, including 12S ribosomal RNA, and the control region (311 bp length), extending from 13,884 bp to 14,194 bp, consisting of 130 bp from 12S rRNA and 181 bp from the control gene, were duplicated in the control region of the previously published GenBank sequence. The corresponding duplication was from 14,196 bp to 14,506 bp (Figure 6). This part could not be identified in the current study. In addition, the 311 bp duplication (14,196 bp to 14,506 bp) could not be amplified in 48 specimens of A. urmiana using designed primers by Zhang et al. [26].
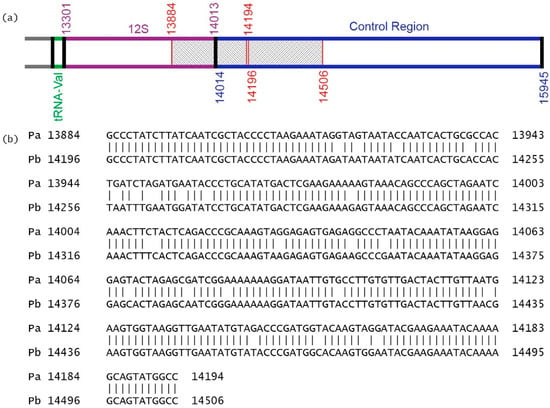
Figure 6.
The partial map (a) and sequence alignment (b) of NC_021382 by Zhang et al. [26] showing duplicated part in control region. (NC_021382 has been corrected by NCBI staff. The reference sequence is identical to JQ975176).
In the GenBank NC_021382 [JQ975176], tRNA-Phe has been mistakenly reported in the heavy strand but it is positioned in the light strand (see Table S2 and Figure 1). This mistake also was observed in annotation of mitogenome of A. franciscana (X69067) and A. tibetiana (JQ975177; NC_021383). Additionally, there was a major problem in position of ND5, which included the tRNA-Phe and would be biologically incorrect. Furthermore, there were several differences in 15 tRNA sequences between NC_021382 [JQ975176] and the current assembly (Figure 7). Our sequence has revealed GTG as a start codon for ATP6 whilst ATG was reported in NC_021382 [JQ975176]. TAA and TAG have been incorrectly reported as stop codons where these were belonging to COX1 and COX2 genes by Zhang et al. [26], while in NC_021382 [JQ975176] and MN240408 assemblies, the incomplete codons T have been revealed in COX1 and COX2 (Table S2).

Figure 7.
Alignment and comparison of determined tRNA sequences in current study (up) and previous mitochondrial assembly NC_021382 by Zhang et al. [26] (down). Nucleotides in red color display differences between two sequences (NC_021382 has been corrected by NCBI staff. The reference sequence is identical to JQ975176).
3.6. Artemia Mitogenome Variation
The complete mitochondrial sequences of four Artemia species had 10,709 conserved and 5047 variable sites, of which 1226 sites were parsimony informative; 3780 sites were singletons; 335 sites were gap. Among the mitogenomes sequenced so far, A. tibetiana (TIB) and A. franciscana (FRA) had the longest (15,826 bp and 15,822 bp) and A. sinica (SIN) and A. urmiana (URM) had the shortest length (15,689 bp and 15,699 bp), respectively. Even so, regardless of the CR region, no significant difference could be observed among the concatenated tRNAs, PCGs and rRNAs sequences of the four recognized species of Artemia (URM: 14,027 bp, TIB: 14,014 bp, SIN: 14,027 bp, FRA: 14,000 bp).
The AT% content of tRNAs among Artemia mitogenomes is summarized in Table 1. Ten tRNAs reveal indicative differences of AT% (>5%), in which the highest dissimilarities were found between tRNA-Ser1 (URM/TIB-FRA: 8.8%) and tRNA-His (URM/TIB-SIN: 8.5%), respectively.

Table 1.
AT content (%) of tRNAs genes in Artemia mitogenomes. “*” shows the tRNA with >5% difference in AT content (URM: A. urmiana, TIB: A. tibetiana, SIN: A. sinica, FRA: A. franciscana).
Nucleotide composition analysis indicates that the AT content of whole mitogenomes ranged from 62.54% (URM) to 64.51% (SIN). The CYTB gene exhibits the lowest AT content in all mitogenomes and varies between 58.18% (TIB) and 60.38% (FRA). The highest AT content was determined in ND3 gene of A. sinica (70.83%). In addition, the ND4L gene displayed a high AT content in other mitogenomes, ranging from 66.28% (TIB) to 69.77% (FRA) (Figure 8 and Table S5).
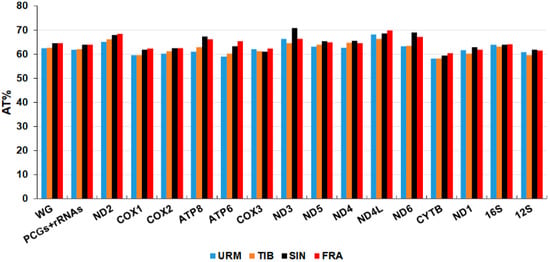
Figure 8.
AT content (%) of the complete mitochondrial genome, protein-coding and ribosomal RNA genes in the mitochondrial genome of A. urmiana and three related species in genus Artemia (URM: A. urmiana, TIB: A. tibetiana, SIN: A. sinica, FRA: A. franciscana).
Detailed information of AT- and GC-skews in each individual protein-condoning gene and the ribosomal ones are provided in Table S3 and Figure 2. It was determined that AT- and GC-skews were positive for both ribosomal RNA genes. Generally, nucleotide skewness was positive with some exceptions. With the exception of FRA, the GC-skews of ND5 were positive for all species, with TIB (0.087) presenting the highest value. In COX1 and ND5, the GC-skew of FRA was reported zero with equal content of G and C. There were two obvious heterogeneity in the value of GC-skew in COX3 and ND1, where TIB (0.013)/FRA (0.096) and URM (0.035)/SIN (0.003) exhibited positive skewness, respectively. The distributional pattern of Artemia species based on AT- and GC-skew values of CmtG and PCG+rRNA sequences were shown on scatter plots (Figure 9). The results have documented the apparent dissimilarity of A. franciscana from other taxa.
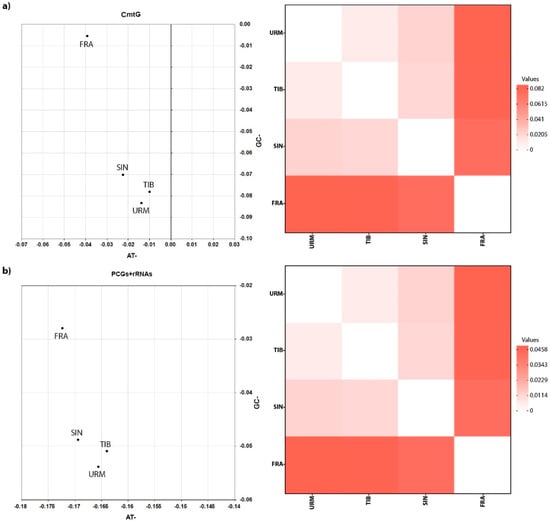
Figure 9.
Distribution of species on scatter plots (left) and heat-map pairwise comparison (right) based on AT- and GC-skew values of CmtG (a) and PCGs+rRNAs (b) (URM: A. urmiana, TIB: A. tibetiana, SIN: A. sinica, FRA: A. franciscana).
3.7. Genetic Distance and Phylogenetic Analysis
The genetic distances among Artemia mitogenomes (PCG+rRNA genes) are summarized in Table 2 in which the highest and lowest distances were determined between A. franciscana–A. tibetiana/A. urmiana (0.214/0.123) and A. tibetiana–A. urmiana (0.090). Phylogenetic analyses (ML and BI) using concatenated PCGs and rRNAs mitogenome sequences have generated a concordant trees topology. According to the phylogenetic analysis, Artemia franciscana was placed as a clade, sister to the Asian spp. (Figure 10).

Table 2.
Pairwise genetic distances using constructed using PCG+rRNA genes (URM: A. urmiana, TIB: A. tibetiana, SIN: A. sinica, FRA: A. franciscana).
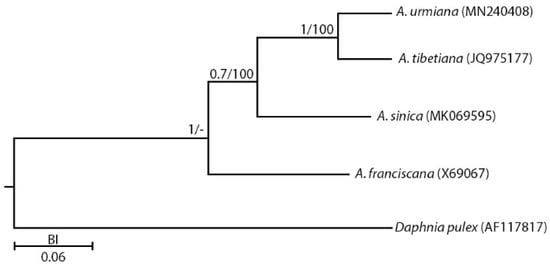
Figure 10.
Phylogeny of Artemia using Daphnia as an outgroup. A phylogenetic tree was constructed using PCG+rRNA genes. The maximum-likelihood bootstrap (left) and Bayesian support values (right) are shown for each major node (URM: A. urmiana, TIB: A. tibetiana, SIN: A. sinica, FRA: A. franciscana).
4. Discussion
In the present study, the complete mitochondrial genome of a taxonomically confirmed A. urmiana specimen from Urmia Lake was re-sequenced and compared with previous mitochondrial assembly (NC_021382 [JQ975176]) published by Zhang et al. [26]. We detected a difference in the sequence length of NC_021382 [JQ975176]: A partial sequence of 311 bp length (consisting of the 130 bp from 12S gene and 181 bp from the CR) is duplicated in the control region. This part covers more than 16% of the whole control region of NC_021382 [JQ975176] and would make it significantly longer than in other Artemia mitogenomes. In addition, we discovered several mistakes in characterization of mitogenome annotation. The absence of 311 bp duplicated partial in 48 specimens documents that this partial cannot be referred to an intraspecific variation in A. urmiana population. Although our results showed that 16S rRNA sequences of NC_021382 [JQ975176] have a similarity with diploid and triploid parthenogenetic populations (96.76–96.21%) than A. urmiana (95.63–95.07%), this value cannot clarify the taxonomic status of Zhang et al.’s [26] mitogenome. Due to coexistence of A. urmiana and parthenogenetic populations in Urmia Lake, and especially intra- and inter-specific variations in the pooled samples, NC_021382 [JQ975176] cannot be a trustworthy sequence to refer to A. urmiana or any parthenogenetic population.
Generally, Artemia species have shorter mitogenomes compared to other anostracans (Phallocryptus tserensodnomi: 16,493 bp and Streptocephalus sirindhornae: 16,887 bp) [50,51]. Our finding shows that the control region of the Artemia mitogenome can differ by 10% between species (A. urmiana: 1672 bp vs. A. franciscana: 1822 bp). Therefore, it is suggested that the control region is less conserved in the genus Artemia. The control region has a particular function in initiation and regulation of DNA transcription and replication [52]. Although mutations in this part may not produce faulty mitogenome products, its variability may change the rate of expression of mitochondrial genes [26,53]
The mitochondrial gene contents of Artemia are uniform with the ancestral Pancrustacea model, exhibiting 22 tRNAs, 13 PCGs and two rRNAs genes (1). Contrary to the dissimilar gene arrangements in mitochondrial genomes among species of decapods [54], as well as the branchiopod Daphnia [55], the gene order recognized for Asian Artemia species was identical to the previously sequenced American A. franciscana. This finding suggests that the gene arrangements in the mitochondrial genome of Artemia from the Old World to New World were highly conserved (see [56]).
As with other branchiopods [55], ATG was found as the start codon in most protein-coding genes in A. urmiana and two other species, A. tibetiana (n = 8) and A. franciscana (n = 5), have displayed the highest and lowest abundances, respectively [26,57]. Most PCGs terminate with a TAA codon in Artemia. We recognize that the COX1, COX2, ND5 and ND4 genes terminate with an incomplete T stop codon (T- or TA-) in Asian species. Additionally, COX3 terminates with an incomplete stop codon in A. sinica. However, no incomplete stop codon was found in the mitogenome of A. franciscana [57].
In this study, the AT content of the A. urmiana mitogenome was calculated, and subsequently compared with the other species. Similar to decapod mitogenomes that are generally AT-rich [54], our results show that the AT content was substantial in Artemia. Though complete mitogenomes of Artemia species had identical AT content (Figure 8 and Table S5), there considerable differences in AT content (>5%) were found in ATP8, ATP6, ND3 and ND6, and almost half of the tRNAs (Table 1) among species. Additionally, the AT- and CG-skew of the mitogenome provide useful information respecting the strand-specific nucleotide frequency bias [42,58,59]. COX3 and ND1 genes exhibited more asymmetries of AT- and GC-skews among the available Artemia mitogenomes.
The mitochondrial markers, COX1 and 16S, have been successfully used in phylogeny of branchiopods [60,61,62,63,64,65]. To date only COX1 has been utilized for phylogenetic studies on Artemia, nevertheless mitogenomic results demonstrated significant difference in the nucleotide composition of ATP8, ATP6, ND3, ND6, ND1 and COX3. Due to the phylogenetic problems in the genus Artemia [21], other mitochondrial markers with high nucleotide variation should be considered in future investigations of intra- and interspecific variation of the genus Artemia. Further studies are required to characterize remaining complete Artemia mitogenomes of bisexual and parthenogenic lineages. This would clarify their phylogenetic relationships and pattern of diversification, as well as suggest new mitochondrial markers for evolutionary and population genetics studies.
Due to phylogenetic problems of the genus Artemia, there is an ongoing project related to sequencing of the complete mitochondrial genomes of all bisexual species and parthenogenetic Artemia with different ploidy levels by Hainan Tropical Ocean University (China) and the result of this study would clarify the phylogenetic relationships among Artemia taxa.
Supplementary Materials
The following are available online at https://www.mdpi.com/1424-2818/13/1/14/s1, Figure S1: The quality of sequencing of A. urmiana, Table S1: Summary of data output quality for A. urmiana, Table S2. The complete mitochondrial genome characteristics of A. urmiana, Table S3. Comparison of skewness in Artemia mitogenomes. (URM: A. urmiana, TIB: A. tibetiana, SIN: A. sinica, FRA: A. franciscana), Table S4. RSCU information in Artemia mitogenomes (URM: A. urmiana, TIB: A. tibetiana, SIN: A. sinica, FRA: A. franciscana). Table S5. AT content (%) of whole mitochondrial genome, protein-coding and ribosomal RNA genes in Artemia mitogenomes (URM: A. urmiana, TIB: A. tibetiana, SIN: A. sinica, FRA: A. franciscana).
Author Contributions
A.A. and A.E. designed the research. Material preparation, data collection and analysis were performed by A.A., A.E., Y.-T.D., Y.Z., H.L., F.M.S., Y.C., P.-Z.W. and W.L. The first draft of the manuscript was written by A.A., A.E., C.-Y.S.; M.W. reviewed the draft. All authors commented on previous versions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by Hainan Province Science and Technology Department Key Research and Development Program (ZDYF2019154) and Key Laboratory of Utilization and Conservation for Tropical Marine Bioresources, Ministry of Education (UCTMB202013).
Acknowledgments
The help of William Shepard (University of California, USA) with the English text and scientific suggestions was highly appreciated.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Miller, W.; Drautz, D.I.; Janecka, J.E.; Lesk, A.M.; Ratan, A.; Tomsho, L.P.; Packard, M.; Zhang, Y.; McClellan, L.R.; Qi, J.; et al. The mitochondrial genome sequence of the Tasmanian tiger (Thylacinus cynocephalus). Genome Res. 2009, 19, 213–220. [Google Scholar] [CrossRef]
- Xia, A.J.; Zhong, L.Q.; Chen, X.H.; Bian, W.J.; Zhang, T.Q.; Shi, Y.B. Complete mitochondrial genome of spined sleeper Eleotris oxycephala (Perciformes, Eleotridae) and phylogenetic consideration. Biochem. Syst. Ecol. 2015, 62, 11–19. [Google Scholar] [CrossRef]
- Zou, Y.; Xie, B.-W.; Qin, C.; Wang, Y.-M.; Yuan, D.-Y.; Li, R.; Wen, Z.-Y. The complete mitochondrial genome of a threatened loach (Sinibotia reevesae) and its phylogeny. Genes Genom. 2017, 39, 767–778. [Google Scholar] [CrossRef]
- Yuan, M.L.; Zhang, Q.L. The complete mitochondrial genome of Gynaephora menyuanensis (Lepidoptera: Lymantriidae) from the Qinghai-Tibetan Plateau. Mitochondrial DNA 2013, 24, 328–330. [Google Scholar] [CrossRef] [PubMed]
- Eimanifar, A.; Kimball, R.T.; Braun, E.L.; Ellis, J.D. Mitochondrial genome diversity and population structure of two western honey bee subspecies in the Republic of South Africa. Sci. Rep. 2018, 8, 1333. [Google Scholar] [CrossRef]
- Powell, A.F.L.A.; Barker, K.F.; Lanyon, S.M. Empirical evaluation of partitioning schemes for phylogenetic analyses of mitogenomic data: An avian case study. Mol. Phylogenet. Evol. 2013, 66, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Asem, A.; Li, W.; Wang, P.-Z.; Eimanifar, A.; Shen, C.-Y.; De Vos, S.; Van Stappen, G. The complete mitochondrial genome of Artemia sinica Cai, 1989 (Crustacea: Anostraca) using next-generation sequencing. Mitochondrial DNA Part B Resour. 2019, 4, 746–747. [Google Scholar] [CrossRef]
- Asem, A.; Lu, H.; Wang, P.-Z.; Li, W. The complete mitochondrial genome of Sinularia ceramensis Verseveldt, 1977 (Octocorallia: Alcyonacea) using next-generation sequencing. Mitochondrial DNA Part B Resour. 2019, 4, 815–816. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Zhang, L.; Yu, D.N.; Storey, K.B.; Zheng, R.Q. Complete mitochondrial genomes of Nanorana taihangnica and N. yunnanensis (Anura: Dicroglossidae) with novel gene arrangements and phylogenetic relationship of Dicroglossidae. BMC Evol. Biol. 2018, 18, 26. [Google Scholar] [CrossRef]
- Mardis, E.R. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008, 24, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Wall, P.K.; Leebens-Mack, J.H.; Chanderbali, A.S.; Barakat, A.; Wolcott, E.; Liang, H.; Landherr, L.L.; Tomsho, L.P.; Hu, Y.; Carlson, J.E.; et al. Comparison of next generation sequencing technologies for transcriptome characterization. BMC Genom. 2009, 10, 347. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, G.; Wen, Z.-Y.; Zou, Y.-C.; Qin, C.-J.; Luo, Y.; Wang, J.; Chen, G.-H. Complete mitochondrial genome of a kind of snakehead fish Channa siamensis and its phylogenetic consideration. Genes Genom. 2019, 41, 147–157. [Google Scholar] [CrossRef]
- Jang, K.H.; Ryu, S.-H.; Hwang, U.W. Mitochondrial Genome of the Eurasian Olter Lutra lutra (Mammalia, Carnivora, Mustelidae). Genes Genom. 2009, 31, 19–27. [Google Scholar] [CrossRef]
- Van Stappen, G. Zoogeography. In Artemia Basic and Applied Biology; Abatzopoulos, T.J., Beardmore, J.A., Clegg, J.S., Sorgeloos, P., Eds.; Kluwer: Dordrecht, The Netherlands, 2002; pp. 171–224. [Google Scholar]
- Asem, A.; Eimanifar, A. Updating historical record on brine shrimp Artemia (Crustacea: Anostraca) from Urmia Lake (Iran) in the first half of the10th century AD. Int. J. Aquatic Sci. 2016, 7, 3–5. [Google Scholar]
- Asem, A. Historical record on brine shrimp Artemia more than one thousand years ago from Urmia Lake. Iran. J. Biol. Res. 2008, 9, 113–114. [Google Scholar]
- Asem, A.; Rastegar-Pouyani, N.; De los Rios, P. The genus Artemia Leach, 1819 (Crustacea: Branchiopoda): True and false taxonomical descriptions. Lat. Am. J. Aquat. Res. 2010, 38, 501–506. [Google Scholar]
- Asem, A.; Eimanifar, A.; Sun, S.C. Genetic variation and evolutionary origins of parthenogenetic Artemia (Crustacea: Anostraca) with different ploidies. Zool. Scr. 2016, 45, 421–436. [Google Scholar] [CrossRef]
- Eimanifar, A.; Asem, A.; Wang, P.-Z.; Li, W.; Wink, M. Using ISSR genomic fingerprinting to study the genetic differentiation of Artemia Leach, 1819 (Crustacea: Anostraca) from Iran and neighbor regions with the focus on the invasive American Artemia franciscana. Diversity 2020, 2, 132. [Google Scholar] [CrossRef]
- Asem, A.; Eimanifar, A.; Rastegar-Pouyani, N.; Hontoria, F.; De Vos, S.; Van Stappen, G.; Sun, S.C. An overview on the nomenclatural and phylogenetic problems of native Asian brine shrimps of the genus Artemia Leach, 1819 (Crustacea: Anostraca). Zookeys 2020, 902, 1–15. [Google Scholar] [CrossRef]
- Asem, A.; Wang, P.; Sun, S.C. Comparative phylogenetic perspectives on the evolutionary relationships in the brine shrimp Artemia Leach, 1819 (Crustacea: Anostraca) based on secondary structure of ITS1 gene. J. Genet. Res. 2018, 4, 72–84. [Google Scholar]
- Baxevanis, A.D.; Kappas, I.; Abatzopoulos, T.J. Molecular phylogenetics and asexuality in the brine shrimp Artemia. Mol. Phylogenet. Evol. 2006, 40, 724–738. [Google Scholar] [CrossRef]
- Eimanifar, A.; Van Stappen, G.; Marden, B.; Wink, M. Artemia biodiversity in Asia with the focus on the phylogeography of the introduced American species Artemia franciscana Kellogg, 1906. Mol. Phylogenet. Evol. 2014, 79, 392–403. [Google Scholar] [CrossRef]
- Maniatsi, S.; Baxevanis, A.D.; Kappas, I.; Deligiannidis, P.; Triantafyllidis, A.; Papakostas, S.; Bougiouklis, D.; Abatzopoulos, T.J. Is polyploidy a persevering accident or an adaptive evolutionary pattern? The case of the brine shrimp Artemia. Mol. Phylogenet. Evol. 2011, 58, 353–364. [Google Scholar] [CrossRef]
- Zhang, H.X.; Luo, Q.B.; Sun, J.; Liu, F.; Wu, G.; Yu, J.; Wang, W.W. Mitochondrial genome sequences of Artemia tibetiana and Artemia urmiana: Assessing molecular changes for high plateau adaptation. Sci. China Life Sci. 2013, 56, 440–452. [Google Scholar] [CrossRef]
- Barigozzi, C.; Baratelli, L. The problem of Artemia urmiana. Artemia Newsl. 1989, 14, 14. [Google Scholar]
- Azari Takami, G. Two strains of Artemia in Urmia Lake (Iran). Artemia Newsl. 1989, 13, 5. [Google Scholar]
- Asem, A.; Atashbar, B.; Rastegar-Pouyani, N.; Agh, N. Biometric comparison of two parthenogenetic populations of Artemia Leach, 1819 from the Urmia Lake basin, Iran (Anostraca: Artemiidae). Zool. Middle East. 2009, 47, 117–120. [Google Scholar] [CrossRef]
- Eimanifar, A.; Van Stappen, G.; Wink, M. Geographical distribution and evolutionary divergence times of Asian populations of the brine shrimp Artemia (Crustacea, Anostraca). Zool. J. Linn. Soc. 2015, 174, 447–458. [Google Scholar] [CrossRef]
- Asem, A.; Sun, S.C. Morphological differentiation of seven parthenogenetic Artemia (Crustacea: Branchiopoda) populations from China, with special emphasis on ploidy degrees. Microsc. Res. Tech. 2016, 79, 258–266. [Google Scholar] [CrossRef]
- Beardmore, J.A.; Abreu-Grobois, F.A. Taxonomy and evolution in the brine shrimp Artemia. In Protein Polymorphism: Adaptive and Taxonomic Significance; Oxford, G.S., Rollinson, D., Eds.; Academic Press: New York, NY, USA, 1983; pp. 153–164. [Google Scholar]
- Asem, A.; Atashbar, B.; Rastegar-Pouyani, N.; Agh, N. Morphological and biometric characterisation of rare male and sexual dimorphism in parthenogenetic Artemia (Crustacea: Anostraca). Zool. Middle East. 2010, 49, 115–117. [Google Scholar] [CrossRef]
- Manaffar, R.; Zare, S.; Agh, N.; Abdolahzadeh, N.; Soltanian, S.; Sorgeloos, P.; Bossier, P.; Van Stappen, G. SNP detection in Na/K ATP-ase gene a1 subunit of bisexual and parthenogenetic Artemia strains by RFLP screening. Mol. Ecol. Resour. 2011, 11, 211–214. [Google Scholar] [CrossRef]
- Abatzopoulos, T.J.; Baxevanis, A.D.; Triantaphyllidis, G.V.; Criel, G.; Pador, E.L.; Van Stappen, G.; Sorgeloos, P. Quality evaluation of Artemia urmiana Günther (Urmia Lake, Iran) with special emphasis on its particular cyst characteristics (International Study on Artemia LXIX). Aquaculture 2006, 254, 442–454. [Google Scholar] [CrossRef]
- Sorgeloos, P.; Lavens, P.; Leger, P.; Tackaert, W.; Versichele, D. Manual for the Culture and Use of Brine Shrimp Artemia in Aquaculture; Ghent University Press: Wachtebeke, Belgium, 1996. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. (04-10-18: Version 0.11.8 Released). 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 11 November 2010).
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Conant, G.C.; Wolfe, K.H. GenomeVx: Simple web-based creation of editable circular chromosome maps. Bioinformatics 2008, 24, 861–862. [Google Scholar] [CrossRef]
- Xia, X. DAMBE6: New tools for microbial genomics, phylogenetics and molecular evolution. J. Hered. 2017, 108, 431–437. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Nylander, J.A.A. MrModeltest v2. Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Rambaut, A. FigTree (Version 1.4.0). 2012. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 11 November 2012).
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Alfaro, M.E.; Zoller, S.; Lutzoni, F. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov Chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol. Biol. Evol. 2003, 20, 255–266. [Google Scholar] [CrossRef]
- Fan, Y.-P.; Lu, B.; Yang, J.-S. The complete mitogenome of the fairy shrimp Phallocryptus tserensodnomi (Crustacea: Anostraca: Thamnocephalidae). Mitochondrial DNA Part A 2016, 27, 3113–3114. [Google Scholar] [CrossRef]
- Liu, X.-C.; Li, H.-W.; Jermnak, U.; Yang, J.-S. The complete mitogenome of the freshwater fairy shrimp Streptocephalus sirindhornae (Crustacea: Anostraca: Streptocephalidae). Mitochondrial DNA Part A 2016, 27, 189–191. [Google Scholar] [CrossRef]
- Geng, X.; Cheng, R.; Xiang, T.; Deng, B.; Wang, Y.; Deng, D.; Zhang, H. The complete mitochondrial genome of the Chinese Daphnia pulex (Cladocera, Daphniidae). Zookeys 2016, 615, 47–60. [Google Scholar]
- Coskun, P.E.; Ruiz-Pesini, E.; Wallace, D.C. Control region mtDNA variants: Longevity, climatic adaptation, and a forensic conundrum. Proc. Natl. Acad. Sci. USA 2003, 100, 2174–2176. [Google Scholar] [CrossRef]
- Tan, M.H.; Gan, H.M.; Lee, Y.P.; Bracken-Grissom, H.; Chan, T.; Miller, A.D.; Austin, C.M. Comparative mitogenomics of the Decapoda reveals evolutionary heterogeneity in architecture and composition. Sci. Rep. 2019, 9, 10756. [Google Scholar] [CrossRef]
- Luchetti, A.; Forni, G.; Skaist, A.M.; Wheelan, S.J.; Mantovani, B. Mitochondrial genome diversity and evolution in Branchiopoda (Crustacea). Zool. Lett. 2019, 5, 15. [Google Scholar] [CrossRef]
- Zhang, S.-M.; Bu, L.; Laidemitt, M.R.; Lu, L.; Mutuku, M.W.; Mkoji, G.M.; Loker, E.S. Complete mitochondrial and rDNA complex sequences of important vector species of Biomphalaria, obligatory hosts of the humaninfecting blood fluke, Schistosoma mansoni. Sci. Rep. 2018, 8, 7341. [Google Scholar] [CrossRef]
- Valverde, J.R.; Batuecas, B.; Moratilla, C.; Marco, R.; Garesse, R. The complete mitochondrial DNA sequence of the crustacean Artemia franciscana. J. Mol. Evol. 1994, 39, 400–408. [Google Scholar] [CrossRef]
- Hassanin, A.; Leger, N.; Deutsch, J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of metazoa, and consequences for phylogenetic inferences. Syst. Biol. 2005, 54, 277–298. [Google Scholar] [CrossRef] [PubMed]
- Fahrein, K.; Talarico, G.; Braband, A.; Podsiadlowski, L. The complete mitochondrial genome of Pseudocellus pearsei (Chelicerata: Ricinulei) and a comparison of mitochondrial gene rearrangements in Arachnida. BMC Genom. 2007, 8, 386. [Google Scholar] [CrossRef] [PubMed]
- Saji, A.; Eimanifar, A.; Soorae, P.S.; Al Dhaheri, S.; Li, W.; Wang, P.-Z.; Asem, A. Phylogenetic analysis of exotic invasive species of Brine Shrimp Artemia Leach, 1819 (Branchiopoda, Anostraca) in Al Wathba Wetland Reserve (UAE; Abu Dhabi). Crustacean 2019, 92, 495–503. [Google Scholar] [CrossRef]
- Asem, A.; Eimanifar, A.; Li, W.; Wang, P.; Brooks, S.A.; Wink, M. Phylogeography and population genetic structure of an exotic invasive brine shrimp Artemia Leach, 1819 (Crustacea: Anostraca) in Australia. Aust. J. Zool. 2018, 66, 307–316. [Google Scholar] [CrossRef]
- Daniels, S.R.; Hamer, M.; Rogers, C. Molecular evidence suggests an ancient radiation for the fairy shrimp genus Streptocephalus (Branchiopoda: Anostraca). Biol. J. Linn. Soc. 2004, 82, 313–327. [Google Scholar] [CrossRef]
- Ketmaier, V.; Pirollo, D.; De Matthaeis, E.; Tiedemann, R.; Mura, G. Large-scale mitochondrial phylogeography in the halophilic fairy shrimp Phallocryptus spinosa (Milne-Edwards, 1840) (Branchiopoda: Anostraca). Aquatic Sci. 2008, 70, 65–76. [Google Scholar] [CrossRef]
- Jeffery, N.W.; Elías-Gutiérrez, M.; Adamowicz, S.J. Species diversity and phylogeographical affinities of the Branchiopoda (Crustacea) of Churchill, Manitoba, Canada. PLoS ONE 2011, 6, e18364. [Google Scholar] [CrossRef]
- Alonso, M.; Ventura, M. A new fairy shrimp Phallocryptus tserensodnomi (Branchiopoda: Anostraca) from Mongolia. Zootaxa 2013, 3670, 349–361. [Google Scholar] [CrossRef][Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).