Abstract
Deep-sea sediments are considered an extreme environment due to high atmospheric pressure and low temperatures, harboring novel microorganisms. To explore marine bacterial diversity in the southern Colombian Caribbean Sea, this study used 16S ribosomal RNA (rRNA) gene sequencing to estimate bacterial composition and diversity of six samples collected at different depths (1681 to 2409 m) in two localities (CCS_A and CCS_B). We found 1842 operational taxonomic units (OTUs) assigned to bacteria. The most abundant phylum was Proteobacteria (54.74%), followed by Bacteroidetes (24.36%) and Firmicutes (9.48%). Actinobacteria and Chloroflexi were also identified, but their dominance varied between samples. At the class-level, Alphaproteobacteria was most abundant (28.4%), followed by Gammaproteobacteria (24.44%) and Flavobacteria (16.97%). The results demonstrated that some bacteria were common to all sample sites, whereas other bacteria were unique to specific samples. The dominant species was Erythrobacter citreus, followed by Gramella sp. Overall, we found that, in deeper marine sediments (e.g., locality CCS_B), the bacterial alpha diversity decreased while the dominance of several genera increased; moreover, for locality CCS_A, our results suggest that the bacterial diversity could be associated with total organic carbon content. We conclude that physicochemical properties (e.g., organic matter content) create a unique environment and play an important role in shaping bacterial communities and their diversity.
1. Introduction
Most microorganisms have specific habitat requirements, and many might be found only in some regions of the planet, particularly areas which have not been widely explored by humans due to limited accessibility. Perhaps one of the most challenging environments to access is the deep sea, which is among the least studied natural habitats and the most extended in the planet. The deep sea combines unique conditions, such as temperatures below 4 °C, lack of light, and high hydrostatic pressure [1]. It has been estimated that marine subsurface habitats harbor majority of the global prokaryotic biomass [2]. These conditions can promote the presence of microbial communities with specific features and requirements, which make them unculturable and difficult to study. It has even been reported that environments with different properties, separated by a few kilometers, can exhibit different microbial communities [3,4]. Currently, metabarcoding techniques allow culturable and nonculturable microorganisms to be identified through 16S ribosomal RNA (rRNA) gene sequencing using high-throughput sequencing (HTS) technology. This includes identifying the taxonomic diversity and community composition of bacteria and archaea domains. The 16S rRNA gene has ubiquitous distribution across all archaeal and bacterial lineages, is evolutionarily conserved, and contains many variable regions that facilitate its use [5]. Besides, it is widely available in public databases (e.g., RDP (http://rdp.cme.msu.edu/), SILVA [6], and Greengenes [7,8]) as a result of the deployment of HTS technology.
Molecular tools have been widely used to conduct crucial global research on metagenomics in marine environments. Large-scale expeditions have been carried out for ocean sampling worldwide, such as the Global Ocean Sampling expedition, which collected ocean water samples from 23 countries and four continents, and the Tara Ocean Foundation expedition in 2003, which obtained nearly 35,000 shallow ocean samples [9,10].There have been several studies evaluating the diversity and distribution of bacteria in deep-sea surface sediments. For example, in the South Atlantic Ocean, 17 samples were analyzed across depths ranging from 5 to 2700 m [11]; in the Pacific Ocean, four samples were studied from Arctic sediments [12]; in the southeastern Atlantic Ocean, five samples were collected from the Cape, Angola, and Guinea Basins at depths ranging from 5032 to 5649 m [3]; in the South Atlantic Polar Front, 15 samples were analyzed across a depth range of 3655 to 4869 m [13]; and in North Sao Paulo Plateau, five samples were collected between 2456 and 2728 m of depth [14]. In Colombia, there have been no studies yet on bacterial communities in deep-sea sediments of the Caribbean Sea. However, some aquatic metagenomic studies have been conducted, for example, research on acidic hot springs in the Colombian Andes [15] and studies analyzing microbiomes associated with corals, algae, sponges, and shallow-water sediment [16,17,18,19]. The southern Colombian Caribbean is a tectonically active, convergent margin with high sediment supply [20] and is probably the main source of nutrients supporting biological productivity in the oligotrophic Caribbean Sea [21]. Therefore, this region could harbor a diversity of microorganisms that have not been studied until now.
In this study, we investigated the bacterial composition and diversity in deep-sea sediments at two sites of the southern Colombian Caribbean Sea as well as the relationship between bacterial communities in marine sediments and the effect of six different depths and physicochemical properties.
2. Materials and Methods
2.1. Sampling
Between October and November of 2018, we collected six deep-sea sediment samples at depths ranging from 1681 to 2409 m in two localities in the southern Colombian Caribbean Sea (Figure 1, Table 1, and Table S1). We obtained three 0.25 m2 samples per locality using a box corer. The sediment samples were stored at −80 °C until they were processed for DNA isolation. Abiotic analyses were performed to measure physicochemical properties, such as granulometry, total organic carbon (TOC), and iron content. The properties at the two sampling sites differed with regard to their bulk parameters. All the samples were dominated by silt and clay (Table 1), except for sample CCS_B1, which was dominated by gravel and sand. The iron concentration increased with depth, while %TOC decreased, except for CCS_B. According to the literature, the salt concentration in the South Caribbean coast at a depth of around of 1000 m is approximately 35 psu, while the temperature is 0 to 5 °C [22,23].
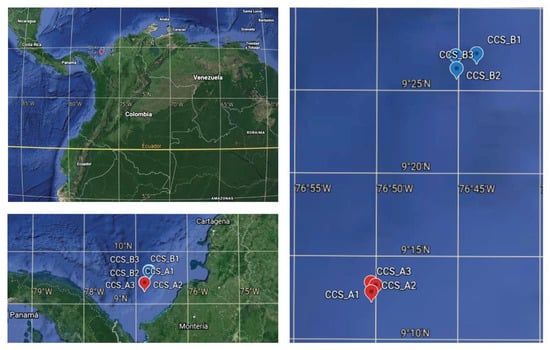
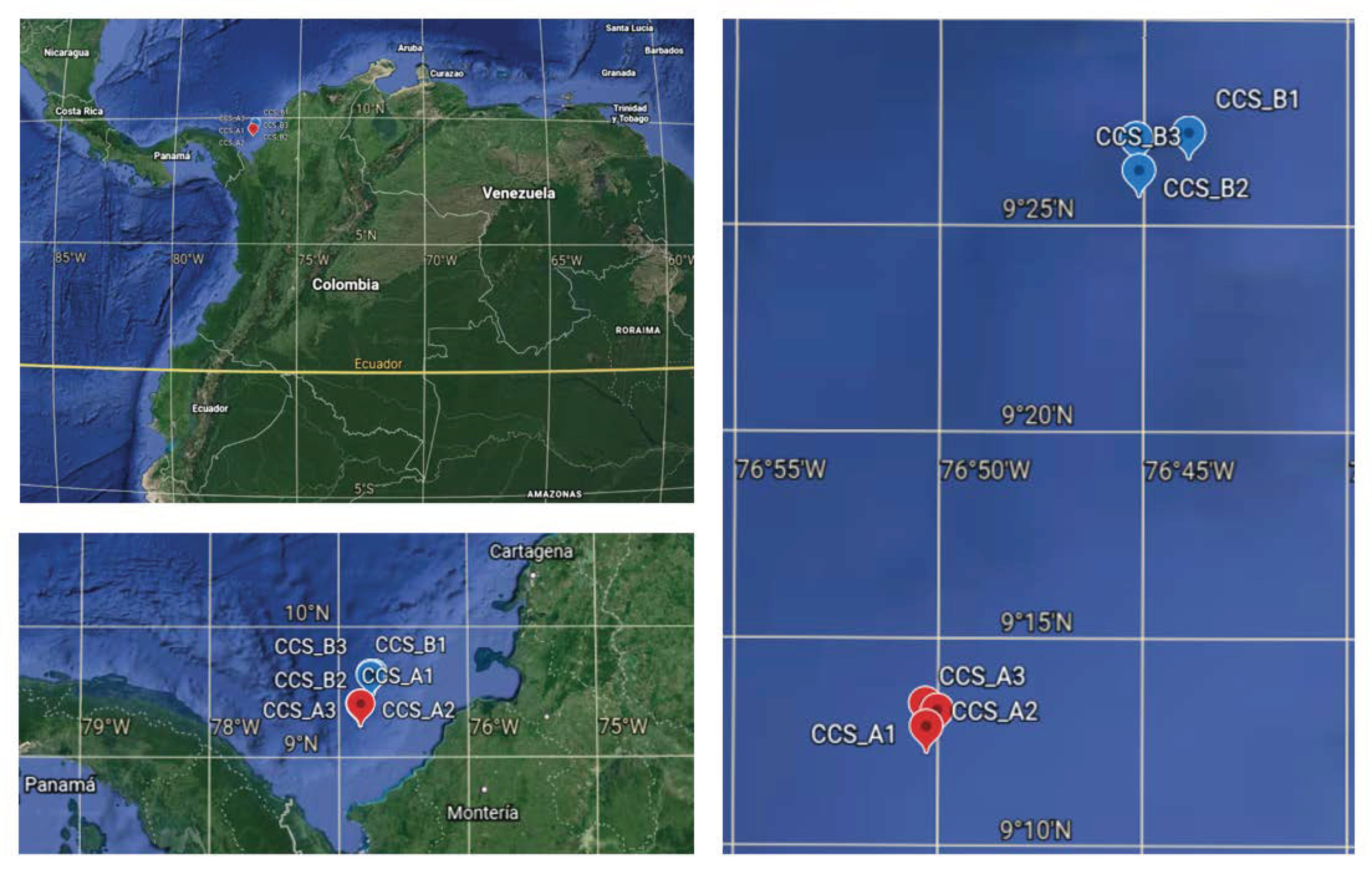
Figure 1.
Geographic location of the two deep-sea sampling localities in the southern Colombian Caribbean Sea: three samples from locality CCS_A (red dots) and three samples from locality CCS_B (blue dots); Table S1.

Table 1.
Metadata of deep-sea sediment samples collected at the southern Colombian Caribbean Sea.
2.2. DNA Isolation and 16S rRNA Gene Sequencing
Total DNA was isolated from 1 g of sediment (homogenized subsample of the top 5 cm collected) for each sample using the DNeasy Powersoil Kit (Qiagen, Hilden, Alemania), according to the manufacturer’s instructions. DNA quality was evaluated by agarose gel electrophoresis and PicoGreen method using a Victor 3 fluorometer. Six barcoding libraries were constructed for the variable V3–V4 region of the 16S rRNA gene using the Nextera XT Index Kit V2 with primers Bakt_341F: CCT ACG GGN GGC WGC AG and Bakt_805R: GAC TAC HVG GGT ATC TAA TCC [24]. All PCR reactions were performed in total reaction volumes of 25 μL containing KAPAHiFi HotStart Ready Mix (2X) (Sigma-Aldrich, St. Louis, MO, USA), 1 μM of forward and reverse primers, and 5 ng/μL template DNA suspended in 10 mM Tris pH 8.5. The thermal cycling profile consisted of initial denaturation at 95 °C for 3 min, followed by 25 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, elongation at 72 °C for 30 s, and a final cycle at 72 °C for 5 min. Libraries were sequenced using the Illumina Miseq platform to obtain ~170,000 PE300bp reads per sample. The 16S rRNA gene sequences obtained in this study have been deposited in the European Nucleotide Archive (ENA) under project number PRJEB40020 and accession numbers ERS4992923 to ERS4992928 (Table S1).
2.3. Bioinformatics Analyses
Taxonomic analyses were performed using QIIME 2 release 2020.2. Quality control of the raw sequences was determined with the DEMUX plugin. The quality variation was inferred using the default error model based on q values using the DADA2 plugin. Chimeric reads were discarded using the consensus method, and amplicon sequence variants (ASVs) were determined. The filtered sequences were aligned and clustered into operational taxonomic units (OTUs) and assigned to clusters using a QIIME feature that applies a Naive Bayes classifier to map each sequence to taxonomy. The classifier was trained on QIIME-compatible Greengenes v13_8, with 99% similarity. Furthermore, the sklearn classifier (http://scikit-learn.org) was trained using 200 to 550 bp reference database sequences generated with primers Bakt_341F and Bakt_805R. Statistical analyses were performed with R software v4.0.2 using the packages qiime2R v.0.99.23, phyloseq v1.30.0 [25], and DESeq2 [26] R/Bioconductor. Abundance plots were computed with ggplot2 [27] to retrieve the nine most abundant classification groups (phylum, class, order, and family), among others. A principal component analysis (PCA) was obtained using the FactorMiner R package to simultaneously analyze the physicochemical variables (e.g., Fe and TOC), 20 bacterial genera, and sediment composition (e.g., sandy, gravel, silt, and clay) to determine the contribution of these variables to each sample. A Venn diagram was built using the Venn R package [28].
2.4. Diversity Analysis
We calculated three aspects of the alpha diversity (richness, evenness, and dominance) for each sample at the two localities. Specifically, we analyzed observed OTUs, Shannon’s index, Pielou’s index, and Berger–Parker’s index using the phyloseq R package.
Beta diversity estimates were calculated based on Bray–Curtis distances between samples. Principal coordinate analysis (PCoA) was computed from the resulting distance matrices to reduce dimensionality to two dimensions (PC1 vs. PC2).
3. Results
3.1. Bacterial Composition Analysis
We sequenced 938,594 raw Illumina reads across the six samples. On average, we obtained 156,432.33 ± 8112.78 (min: 142,822; max: 165,773) raw reads per sample. Furthermore, 18,775.83 ± 5864.36 (min: 12,426; max: 29,108) reads were retained after quality control, denoising, and filtering of chimeric reads (Table S2). We found 1842 OTUs belonging to bacteria (Table S3 and Figure S1)
The reads were taxonomically assigned to 36 phyla (99.3%), 89 classes (98.3%), 93 orders (90%), 104 families (80.2%), 90 genera (74.4%), and 53 species (41.5%) (Table S3 and Figure S1). We found that 12 phyla were shared among the six samples and represented 98.7% of the total reads (Figure S1A). Among these reads, 92.5% were assigned to five phyla: Proteobacteria (56.9%), Bacteroidetes (20.5%), Firmicutes (9.7%), Actinobacteria (3.1%), and Chloroflexi (2.4%) (Figure 2A); additional phyla with minor proportion of reads were assigned to OD1, TM6, Gemmatimonadetes, WS3, Planctomycetes, SBR1093, and Acidobacteria. Overall, in locality CCS_A, phylum Proteobacteria had a relative abundance of 59.04 ± 24.41%, Bacteroidetes of 16.84 ± 27.07%, and Firmicutes of 10.28 + 1.49%. In comparison, locality CCS_B showed a high relative abundance of Proteobacteria (50.43 + 16.79%), followed by Bacteroidetes (31.88 + 17.43%) and Firmicutes (8.68 + 2.56%) (Table S3). We also found seven exclusive phyla, specifically two phyla in sample CCS_A1 (SC4 and CD12), four in sample CCS_A2 (Thermi, TM7, ZB3 and WPS-2), and one in sample CCS_A3 (Spirochaetes) (Figure S1).
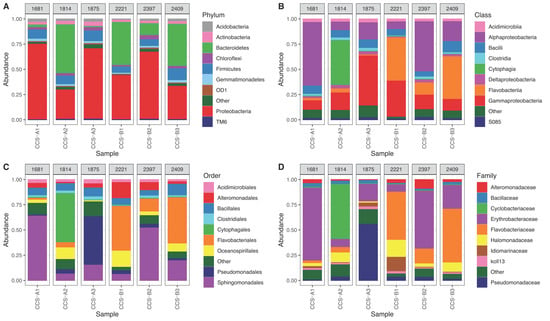
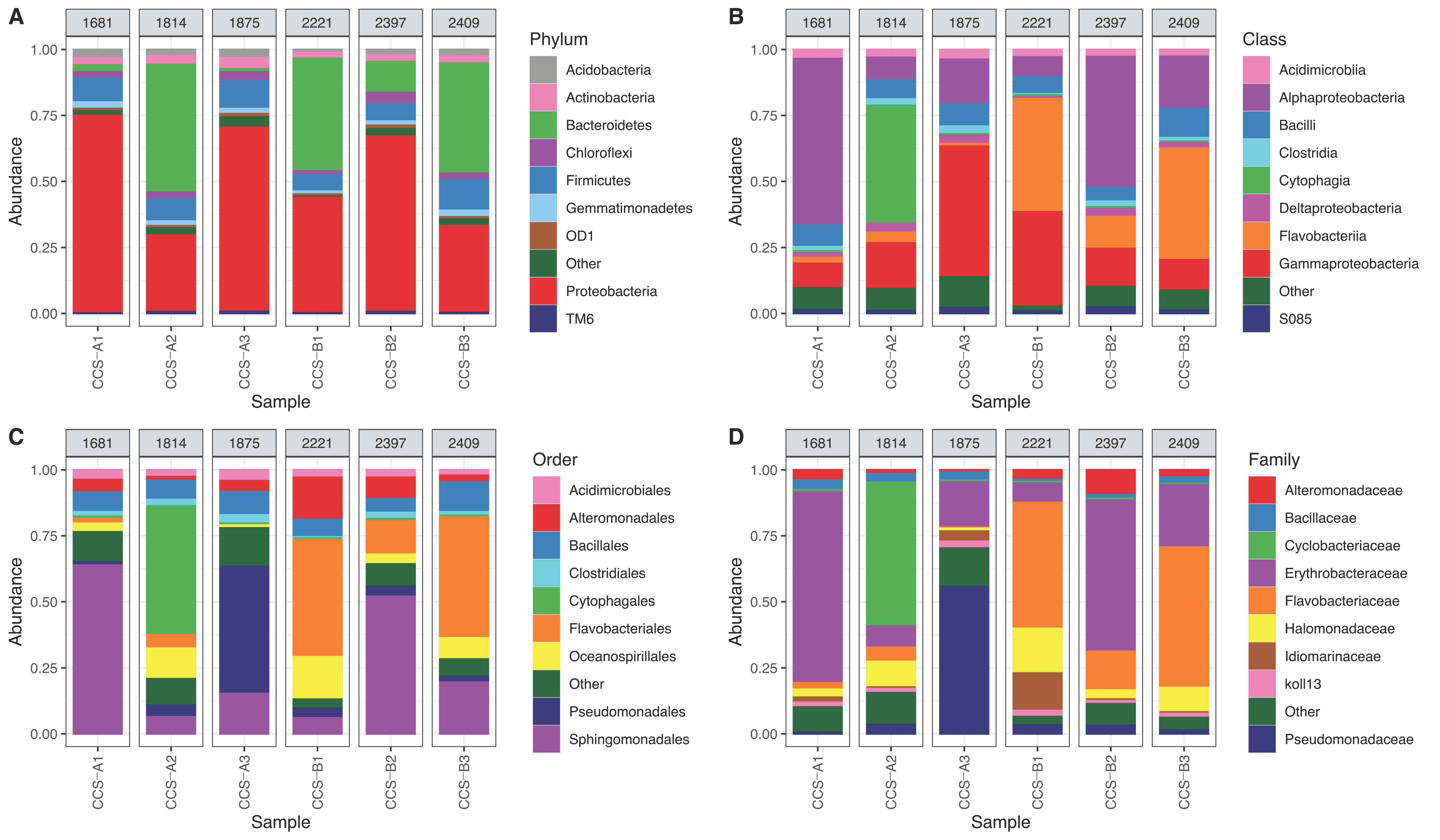
Figure 2.
Relative abundance of bacteria in six samples from the southern Colombian Caribbean Sea at different depths in two localities (CCS-A and CCS-B): (A) phylum, (B) class, (C) order, (D) family. Gray boxes above each sample show depth of the collected sample.
Proteobacteria had the most assigned reads in four of the six samples: CCS_A1 (74.8%), CCS_A3 (71.3%), CCS_B2 (68.6%), and CCS_B1 (47.19%) (Figure 2A). Phylum Bacteroidetes had the most assigned reads in samples CCS_A2 and CCS_B3 (48.1 and 42.5%, respectively) and had the second most number of assigned reads in CCS_B1 (41.4%) and CCS_B2 (11.8%) (Figure 2A). Firmicutes was second in CCS_A3 (11.7%) and CCS_A1 (10.5%) and third in CCS_B3 (11.6%), CCS_A2 (8.7%), CCS_B1 (7.3%), and CCS_B2 (7.2%) (Figure 2A). Moreover, phylum Actinobacteria had the most assigned reads in sample CCS_A3 (4.4%), followed by CCS_A1, CCS_B3, CCS_A2, CCS_B2, and CCS_B1 (3.5, 3.0, 3.0, 1.7, and 1.6%, respectively) (Figure 2A). Chloroflexi was first in CCS_B2 (3.6%), followed by CCS_A3 (3.4%), CCS_A1 (2.2%), CCS_A2 (2.1%), CCS_B3 (1.2%), and CCS_B3 (0.8%) (Figure 2A).
The five classes with the most assigned reads were Alphaproteobacteria (28.2%), Gammaproteobacteria (26%), Flavobacteria (12.7%), Bacilli (8%), and Cytophagia (7.7%). In locality CCS_A, we found a relative abundance of 30.1 + 28.7% of Alphaproteobacteria, 26.4 + 21.7% of Gammaproteobacteria, and 14.9 + 25.68% of Cytophagia. In comparison, locality CCS_B had a relative abundance of 32 + 17.7% of Flavobacteria, 26.7 + 22.3% of Alphaproteobacteria, and 22.5 + 14.7% of Gammaproteobacteria (Figure 2B and Table S2). Gammaproteobacteria showed the highest mean relative abundance in locality CCS_A, and it was higher with higher depths, increasing from 9.8% in sample CCS_A1 to 51% in CCS_A3. Furthermore, this class decreased in relative abundance across samples in locality CCS_B, from 39.3% in sample CCS_B1 to 12.5% in CCS_B3. Alphaproteobacteria did not vary in response to depth, showing the highest relative abundance in CCS_A1 (63%) and CCS_B2 (51%) and least relative abundance in CCS_A2 (9.8%) and CCS_B1 (7.5%). Class Flavobacteria showed higher mean relative abundance in locality CCS_B compared to CCS_A. Lastly, Bacilli did not vary with depth, showing relative abundance between 5.3 and 9.9% (Figure 2B).
The families with the most assigned reads were Erythrobacteraceae (order Sphingomonadales) (25.1%), followed by Flavobacteriaceae (order Flavobacteriales) (12.7%), Pseudomonadaceae (order Pseudomonadales) (12.6%), Cyclobacteriaceae (order Cytophagales) (7.63%), and Halomonadaceae (order Oceanospiralles) (5.05%) (Figure 2C,D). In particular, for Erythrobacteraceae, sample CCS_A1 showed the highest relative abundance with 71.63%, while CCS_B1 showed the lowest with 7.03%. On the other hand, Flavobacteriaceae showed higher mean relative abundance in locality CCS_B (37.72 + 21%) compared to CCS_A (2.26 + 2.12%).
At the genus-level, the genera with the most assigned reads were Erythrobacter (mean relative abundance of 33.6%), Gramella (12.8%), Pseudomonas (12.5%), and Algoriphagus (10%) (Table S3). Gramella was mainly found in five out of six samples and was least abundant in sample CCS_A1 (1.8%) at the lowest depth sampled. Conversely, sample CCS_B3, at the greatest depth sampled, showed a relative abundance of 55.5% for the same genus (Table S3). Five genera (Pseudomonas, Halomonas, Erythrobacter, Bacillus, and Clostridium) were shared among the six samples (Figure S1E). Particularly, for sample CCS_A1, we found seven exclusive genera (Nitrosopumilus, Synechococcus, Pediococcus, Burkholderia, Anaerobacillus, Streptomyces, and Planctomycete); for sample CCS_A2, there were seven exclusive genera (Deinococcus, Pseudoxanthomonas, Muricauda, Rothia, Phaeobacter, Anaerococcus, and Rubritalea), while for sample CCS_A3, there were 20 exclusive genera (Spongiibacter, Psychromonas, Enhydrobacter, Henriciella, Nitratireductor, Alicyclobacillus, Thalassobacillus, Ammoniphilus, Staphylococcus, Pelotomaculum, Desulfosporosinus, Caldanaerocella, Aeromicrobium, Rhodococcus, Gordonia, Candidatus Scalindua, Parachlamydia, Simkania, Nitrospina, and Candidatus Entotheonella) (Figure S1E). For locality CCS_B, we found that only one genus was exclusive to CCS_B1 (Pseudidiomarina), five genera were exclusive to CCS_B2 (Mesoflavibacter, Croceitalea, Alistipes, Ruegeria, and Bdellovibrio), and for CCS_B3, five genera were exclusive (Aquicella, Hyphomonas, Rhodovulum, Rhodoplanes, and Faecalibacterium) (Figure S1E).
At the species-level, Erythrobacter citreus showed the most assigned reads with a percentage similarity of 76–78%, followed by Algoriphagus ornithinivorans (99–100%) and Pseudidiomarina homiensis (78–99%). The OTUs corresponding to Pseudomonas, Pseudomonas stutzeri, and Pseudomonas balearica were identified with 70–100% similarity. Likewise, Chromohalobacter, Chromohalobacter salexigens, and Chromohalobacter thailandesis were identified with 70–99% similarity (Table S3).
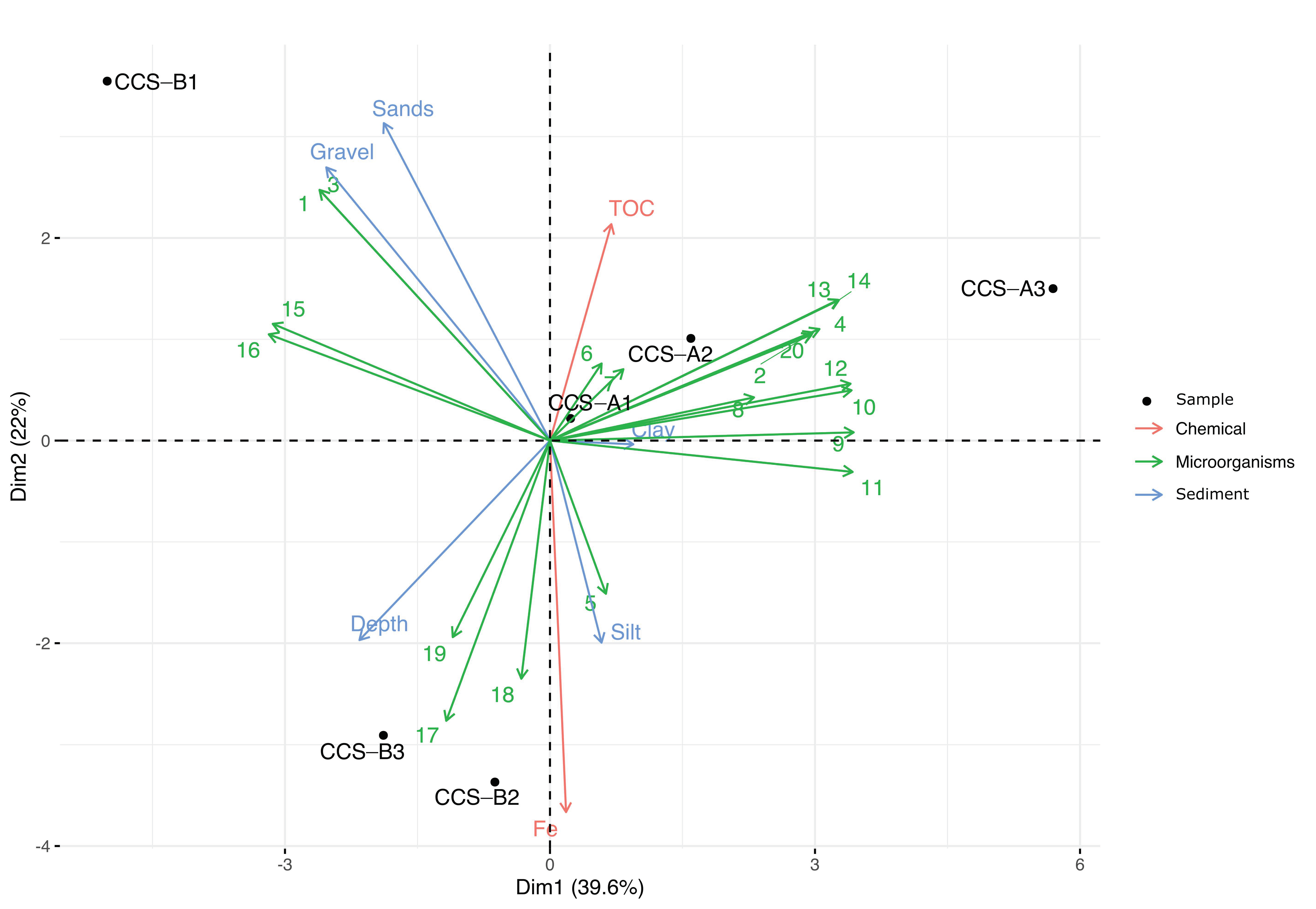
Multifactorial analysis allowed the genera that contributed the most per sample to be identified (Figure 3). Dimension 1 accounted for 39.6% of the data variance and separated the samples from localities CCS_A and CCS_B, while dimension 2 accounted for 22% of the variance and separated sample CCS_B1 from the other samples from locality CCS_B. At location CCS_A, the samples were grouped according to clay sediment type, high TOC content, and 13 bacterial genera (Pseudomonas, Idiomarina, Erythrobacter, Chromohalobacter, Algoriphagus, Hyphomicrobium, Planctomyces, Clostridium, Bacillus, Acinetobacter, Plesiocystis, BD2-6, and Nitratireductor). At locality CCS_B, we observed that sample CCS_B1 was influenced by gravel and sandy sediment types and at least four bacterial genera (Salegentibacter, Pseudidiomarina, Leeuwenhoekiella, and Halomonas). Moreover, samples CCS_B2 and CCS_B3 were defined by depth and three bacterial genera (Gramella, Bacteroides, and Glaciecola). We found eight genera (Planctomyces, Bacillus, Clostridium, Acinetobacter, Plesiocystis, BD2-6, Leeuwenhoekiella, and Halomonas) with significant contribution to dimension 1. Simultaneously, sandy sediment type and iron content were significant contributors to dimension 2 (Table S4).
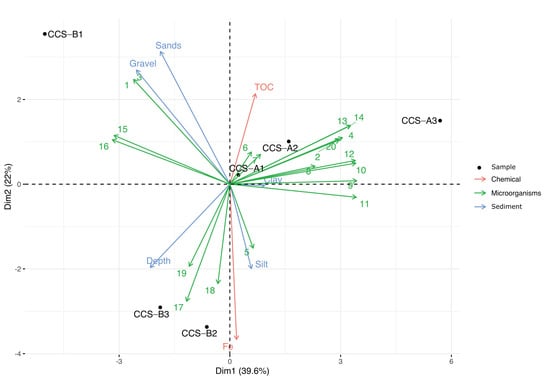
Figure 3.
Principal component analysis (PCA) of the six samples, bacterial genera, and sediment type according to physicochemical variables. The 20 genera analyzed are represented by numbers as follows: 1. Salegentibacter; 2. Pseudomonas; 3. Pseudidiomarina; 4. Idiomarina; 5. Erythrobacter; 6. Chromohalobacter; 7. Algoriphagus; 8. Hyphomicrobium; 9. Planctomyces; 10. Bacillus; 11. Clostridium; 12. Acinetobacter; 13. Plesiocystis; 14. BD2-6; 15. Leeuwenhoekiella; 16. Halomonas; 17. Gramella; 18. Bacteroides; 19. Glaciecola; 20. Nitratireductor.
3.2. Bacterial Diversity Analysis
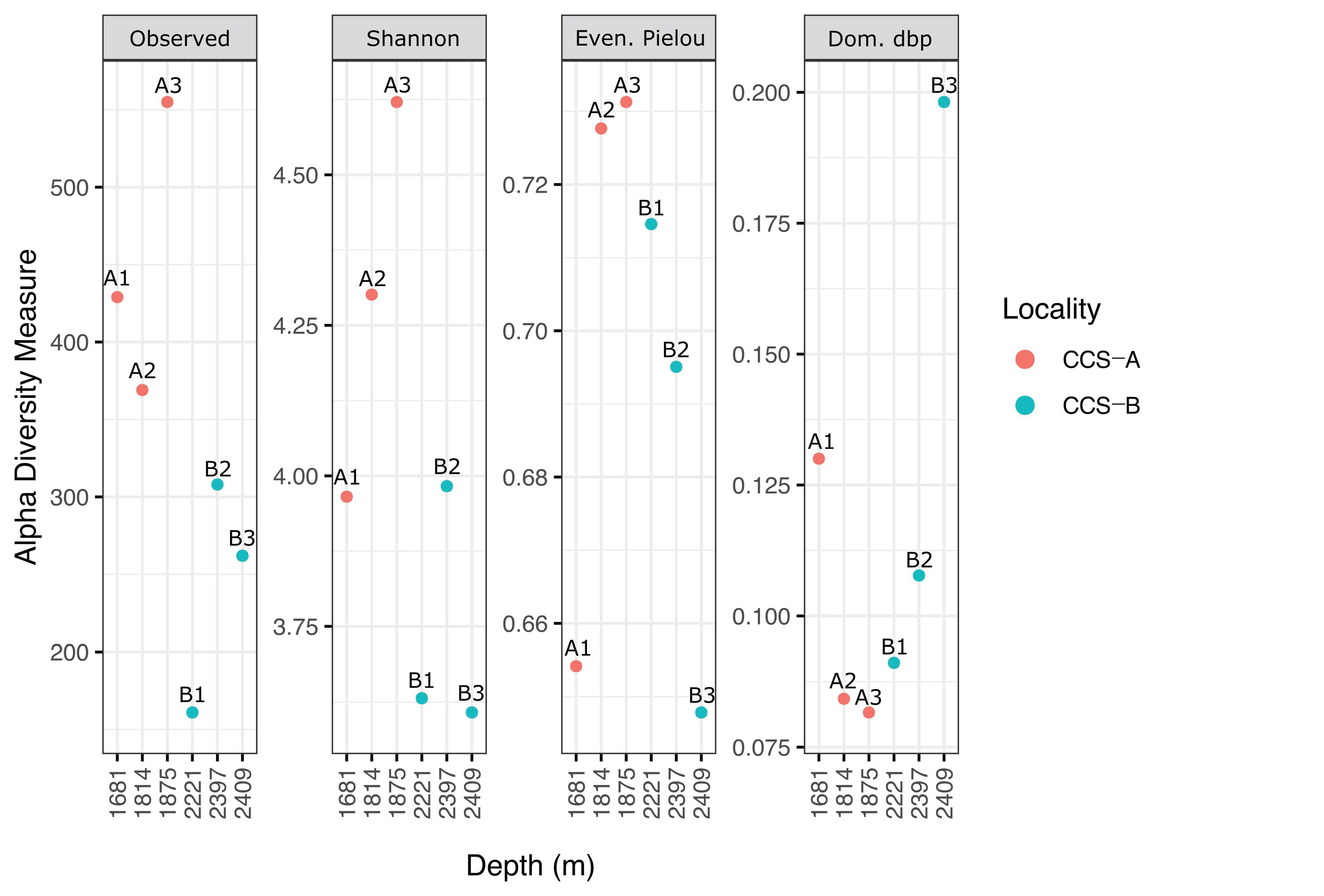
Several diversity indices were used to describe the community composition based on the sequencing data. Accordingly, sample CCS_A3 (555 richness index; 4.6 Shannon’s index) showed the highest species richness, followed by CCS_A1 (429; 4.0), CCS_A2 (369; 4.3), CCS_B2 (308; 4.0), CCS_B3 (262; 3.6), and CCS_B1 (161; 3.6) (Figure 4 and Table S5). The number of genera per sample ranged from 35 to 47 (mean = 39.67 ± 6.43) in locality CCS_A and from 19 to 33 (mean = 26.33 ± 7.02) in CCS_B (Figure S2 and Table S3).
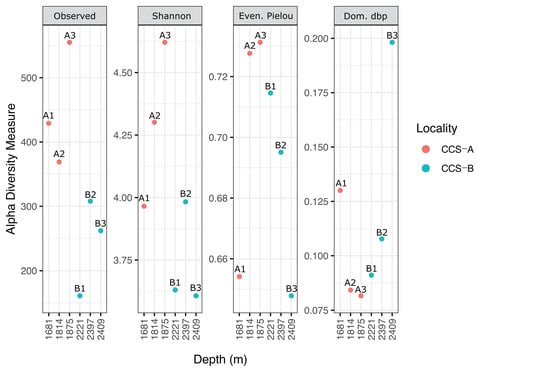
Figure 4.
Depth-related patterns of species richness (observed operational taxonomic units (OTUs)), diversity (Shannon’s index), evenness (Pielou’s index), and dominance (relative) of bacterial communities in the southern Colombian Caribbean Sea for six samples from two localities.
In locality CCS_A, Shannon’s index increased across the three samples in response to depth (Figure 4). Furthermore, Pielou’s evenness index and Berger–Parker’s dominance index showed the expected inverse relationship, with the sample from the lowest depth (CCS_A1) showing the least evenly distributed bacterial communities and dominance of Erythrobacter citreus (Figure 3 and Figure 4 and Table S3). In contrast, locality CCS_B showed lower diversity at greater depths, except for sample CCS_B1. However, Pielou’s evenness index and Berger–Parker’s dominance index showed the expected inverse relationship; for instance, sample CCS_B3, collected at the greatest depth, showed the least evenly distributed bacterial communities, and most reads were assigned to the genus Gramella (Figure 3 and Figure 4 and Table S3).
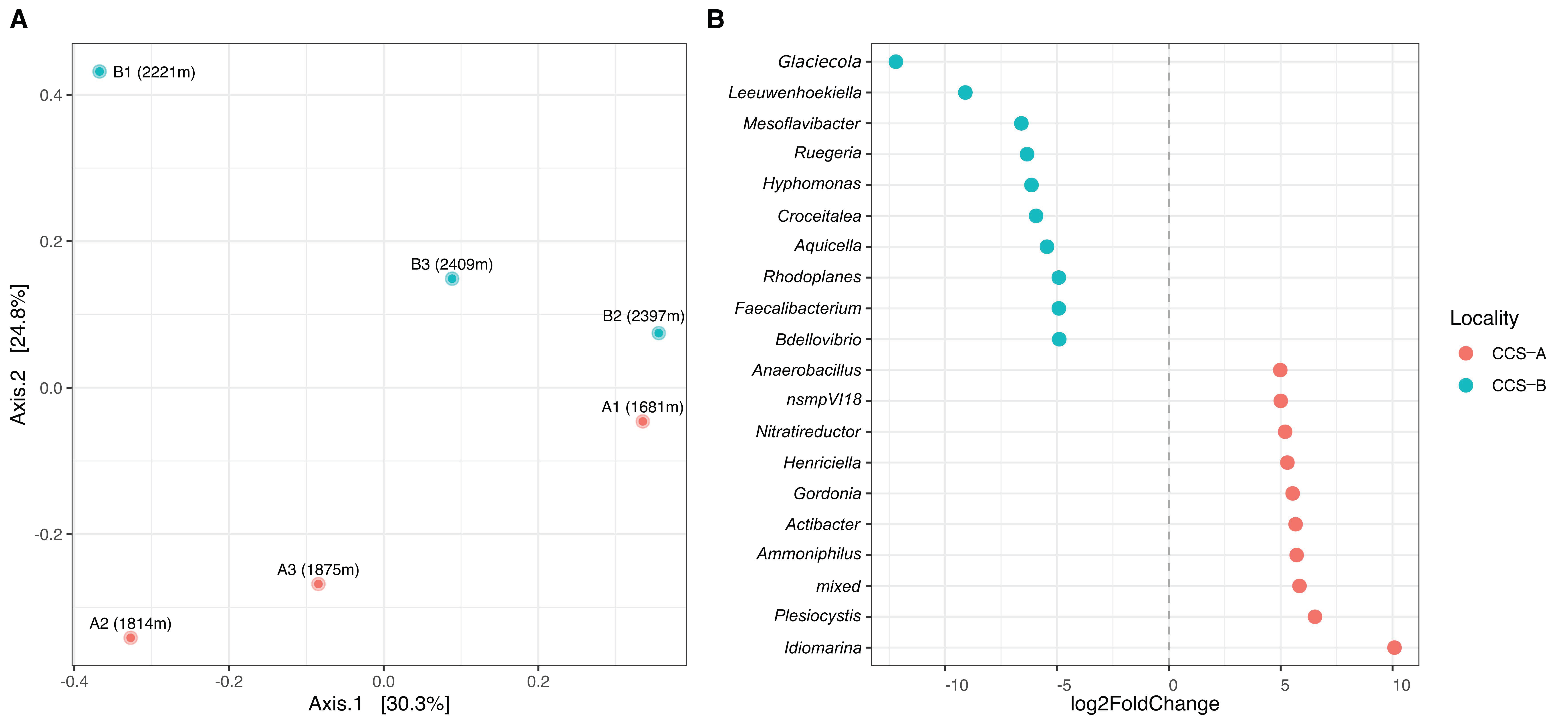
Regarding beta diversity, the PCoA plot with the Bray–Curtis index showed a separation between samples within localities according to bacterial community composition. We found that the first axis of the PCoA accounted for 30.3% of the variance in community composition and did not discriminate between localities (PERMANOVA, F = 1.05; R2 = 0.21; p = 0.5) but possibly separated samples by depth (Figure 5A). The second axis explained 24.8% of the variance and separated the samples by locality (Figure 5A). Bacterial community composition differed between localities in terms of relative abundance (Figure 5A). DESeq2 analysis revealed that two genera (Glaciecola and Leewenhoekiella) had a significantly more assigned reads in CCS_B. In contrast, only one genus (Idiomarina) was significantly more abundant in CCS_A (Figure 5B).
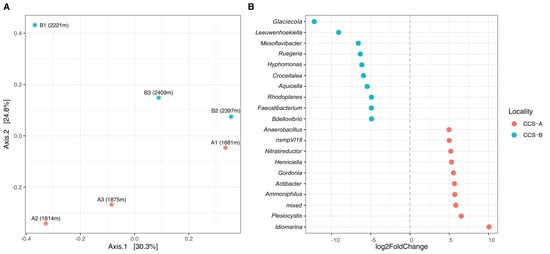
Figure 5.
(A) Principal coordinate analysis (PCoA) using Bray–Curtis on relative abundance for samples and (B) log-transformed ratio of the relative abundance of 20 bacterial genera between localities estimated by DESeq2. CCS_A (red dots) and CCS_B (blue dots).
4. Discussion
Deep-sea sediments constitute one of the most species-rich ecosystems on Earth, yet they are also among the least known [29,30]. This study analyzed deep-sea sediments collected at different depths in two localities in the southern Colombian Caribbean Sea. The sample depths ranged from 1681 to 1875 m at locality CCS_A and from 2221 to 2409 m at locality CCS_B. The samples collected in this study provide a unique opportunity to understand poorly studied microbial ecosystems for which baseline data of microbial diversity is lacking. Considering the hydrostatic pressure of the water column increases by approximately 0.1 MPa every 10 m of depth [11], this pressure change can lead to compositional shifts in microbial diversity.
We found 1842 OTUs belonging to bacteria (Figure S1 and Table S3). Furthermore, we found that bacteria accounted for 99.95% of the sequenced reads for all samples. Our findings agree with observations made in other marine environments, such as the East China Sea in the Pacific Ocean and the Bay of Bengal in the Indian Ocean, where bacteria was the dominant domain [13,31].
We found 36 phyla belonging to bacteria. Among them, the three phyla with the most assigned reads were Proteobacteria, Bacteriodetes, and Firmicutes, which accounted for 88.58% of total reads. Proteobacteria had the most assigned reads in most samples, except for CCS_A2. Previous analyses of shallow-water sediments [12,32,33,34,35] and the aphotic zone [36] have shown that these environments are heavily dominated by classes Alphaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria. We found that Gammaproteobacteria was more abundant (26.38 ± 21.75%) in locality CCS_A compared to the deeper locality CCS_B (22.51 ± 14.67%). The high abundance of this bacterial class is specifically attributed to the orders Pseudomonales and Alteromonadales. A dominance of Gammaproteobacteria is also commonly reported in the South, North, and Mid Atlantic Ocean [11] and the Eastern South Atlantic, mainly in Guinea, Cape, and Angola Basins [3]. Class Alphaproteobacteria had the most abundance (62.8% assigned reads) in sample CCS_A1 collected at the lowest depth. At the same time, Flavobacteria was dominant in the most in-depth sample (e.g., CCS_B3), representing 42.84% of sequences.
Bacteroidetes was the phylum with the second most assigned reads, specifically to classes Flavobacteria (16.97%) and Cytophagia (7.49%). Previous studies have reported that Bacteroidetes are associated with phytoplankton blooms and marine snow particles [37,38,39]. Phylum Firmicutes was the third most abundant, mainly in locality CCS_A. This phylum has been associated with organic matter fall in deep-sea environments [40]. Total organic carbon is lower in marine sediments at greater depths [41]. Our findings agree with Walsh et al. [36], who reported that deep-sea sediment microbial communities also comprise members of Chloroflexi (i.e., the most abundant classes are S085, Anaerolineae, and Dehalococcoidetes) and Actinobacteria.
At the genus level, Erythrobacter had the most assigned reads across the six samples (33.29%); specifically, sample CCS_A1 had the highest abundance (78.3%), followed by CCS_B2 (61.88%). Kai et al. [11] also reported this genus in 17 samples collected at different depths (5–2700 m) in the South Atlantic Ocean. The second most common group of strains identified in this study belonged to the genus Gramella, which was present in five out of six samples. Notably, we found an abundance of 55.47% in the deepest sample collected (CCS_B3), with 6023 assigned reads out of 14,464 total reads, indicating that this genus might be adapted to extreme conditions (Supplementary Table S5 and Figure 4. Gramella has been associated with the hydrolysis of long-chain organic polymers [42] and is relatively common in marine environments, where it has been isolated from sediments, water, and even sea urchins [43]. The next most common genus was Pseudomonas, found in the six samples, showing the most assigned reads in CCS_A3 (62.08%). This genus, along with Halomonas and Psychrobacter, were reported by the authors of [12] for their importance in the production of various types of hydrolytic enzymes, demonstrating the potential of microorganisms found in deep-sea sediments. Regarding species richness, Algoriphagus ornithinivorans was present in samples CCS_A2 (59.55%) and CCS_A1 (0.23%), while Salegentibacter sp. (44.33%) and Pseudidiomarina homiensis (18.58%) were found in CCS_B1. Some species were found at low frequency in all samples, such as Halomonas sp., Chromohalobacter salexigens (except for CCS_A3), and Bacillus sp., whereas other species were unique to a given locality, for example, Glaciecola sp., Leeuwenhoekiella marinoflava, and Pseudomonas balearica in locality CCS_B and Idiomarina sp. and Alteromonas sp. in CCS_A.
We observed that 48 out of 91 genera (Figure S1 and Table S3) were exclusive to specific samples, indicating the presence of nuclei comprising a few microorganisms and high levels of endemism. This phenomenon could be due to sampling or depth effects. According to Petro et al. [44], selection is the predominant force driving the vertical assemblage of microbial communities in the sediment column, and it becomes more relevant at greater depths. Conversely, shallow areas are more influenced by diversification, dispersion, and drift. This selective force is imposed by antagonistic or synergistic abiotic (e.g., soil components, light intensity, and total organic carbon, among others) and biotic pressure (e.g., associated flora and fauna) in the sediment environment [45] as well as complex intrinsic factors at each sampling site that can influence selection [46].
We found a lower number of OTU and species diversity in bacterial communities in deeper sampling sites (Figure 4). Other studies have reported similar findings, such as Petro et al. [44], who found lower Shannon’s diversity and Chao’s 1 richness with depth, which is associated with the presence of many species with equal or low abundance at shallower depths (high index) and few but dominant species at greater depths (low index).
Within each locality, the diversity of the bacterial community varied in response to changes in depth. Differences in diversity, evenness, and dominance of bacterial communities across samples can be explained by the physicochemical composition and texture of the marine sediments. Accordingly, the sediment samples from locality CCS_A showed higher total organic carbon and lower iron content compared to samples CCS_B2 and CCS_B3, where the genera Gramella, Glaciecola, and Bacteroides were highly abundant. The presence of Glaciecola can be associated with phytoplankton bloom as its abundance can increase rapidly after the entry of labile dissolved organic carbon (DOC) derived from phytoplankton [47]. This can be correlated with the high TOC values at location CCS_A. Sample CCS_B1 showed high sandy material and bacterial genera Salegentibacter, Pseudidiomarina, Leeuwenhoekiella, and Halomonas. However, this sample also showed low total organic carbon and iron contents and low abundance of Erythrobacter citreus and Clostridium sp. (Figure 3). Therefore, we found that TOC decreased in response to depth and correlated with the microbial composition of the samples from locality CCS_A (Figure 3). Total organic carbon has been reported to vary according to primary production in surface layers and depends on the speed of descent in the water column [41,48]. Walsh et al. [49] reported the close match between the exponential decline in bacterial richness and the depth distribution of organic degradation rates at open-ocean sites. Only CCS_B1 sample constituted an exception to the correlation described as it had other physical properties (Table 1).
5. Conclusions
Here, we present a pilot study on the taxonomic diversity of bacteria through an exploratory survey of sediments in the southern Colombian Caribbean Sea at different depths. Although the taxonomic composition of the bacterial communities in the six samples was similar, the abundance of most taxa differed between the two locations. Furthermore, our results revealed that Proteobacteria, Bacteroidetes, and Firmicutes dominated these bacterial communities. Their composition depended on ocean depth and physicochemical variables, such as total organic carbon, iron, and deep-sea sediment texture. Future studies will involve analyzing if these marine bacteria can develop under different environmental conditions and thus determine if they are generalists or specialists of marine sediments.
Supplementary Materials
The following are available online at https://www.mdpi.com/1424-2818/13/1/10/s1, Table S1: Sampling sites and their GPS locations in the southern Colombian Caribbean Sea. Table S2: Number of reads retained for the samples after each processing step. Table S3: Taxonomic assignment of the bacterial communities in the southern Colombian Caribbean Sea (from kingdom to species). Table S4: Contribution values for the multifactorial analysis and their significance values. Table S5: Alpha diversity indices of the bacterial communities. Figure S1: Taxonomic distribution comparison of bacterial communities of the six samples from the southern Colombian Caribbean Sea: (A) phylum, (B) class, (C) order, (D) family, (E) genus, (F) OTUs. Figure S2: Rarefaction curves of alpha diversity indices for each sample from the southern Colombian Caribbean Sea subjected to 16S rRNA gene metabarcoding.
Author Contributions
Conceptualization, A.C., L.F.D., and V.P.; methodology, A.C., N.R.F., D.L.-A., and M.Á.G.; software, N.R.F., D.L.-A., and M.Á.G.; validation, A.C., N.R.F., and D.L.-A.; formal analysis, A.C., N.R.F., D.L.-A., and M.Á.G.; investigation, A.C., N.R.F., D.L.-A., M.Á.G., and J.J.G.-F.; resources, V.P. and L.F.D.; data curation, N.R.F. and M.Á.G.; writing—Original draft preparation, A.C., N.R.F., and D.L.-A.; writing—Review and editing A.C., N.R.F., D.L.-A., M.Á.G., J.J.G.-F., V.P., and L.F.D.; visualization, A.C., N.R.F., and D.L.-A.; supervision, A.C.; project administration, A.C.; funding acquisition, A.C., V.P., L.F.D., and D.L.-A. All authors have read and agreed to the published version of the manuscript.
Funding
Anadarko Colombia Company and ECOPETROL funded the sample collection under a monitoring of oil and gas offshore exploratory project. The study has been funded by COLCIENCIAS/Minciencias (Colombia) under ID 80740-069-2020 and the Vice-Rectory of Research Universidad del Valle, Colombia, ID code 71233.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Thanks to Anadarko Colombia Company for allowing us to get the samples for this study. Thanks to other companies such as Aquabiosfera S.A.S. for implementing the offshore monitoring survey in the field and SERPORT S.A. for providing the survey vessel. The authors especially thank Nathalie Abrahams for receiving and transporting the samples from the field station to the laboratory, Andrea Gonzalez for English editing, and Daniel Cuartas for producing the maps.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Malek-Madani, R. Physical Oceanography: A Mathematical Introduction with MATLAB; CRC Press, Inc.: Boca Raton, FL, USA, 2012. [Google Scholar]
- Whitman, W.B.; Coleman, D.C.; Wiebe, W.J. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. USA 1998, 95, 6578–6583. [Google Scholar] [CrossRef] [PubMed]
- Schauer, R.; Bienhold, C.; Ramette, A.; Harder, J. Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J. 2010, 4, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Aravindraja, C.; Viszwapriya, D.; Karutha Pandian, S. Ultradeep 16S rRNA sequencing analysis of geographically similar but diverse unexplored marine samples reveal varied bacterial community composition. PLoS ONE 2013, 8, e76724. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Chun, J. 16S rRNA Gene-based identification of bacteria and archaea using the eztaxon server. In Methods in Microbiology; Academic Press: Cambridge, MA, USA, 2014; Volume 41, pp. 61–74. [Google Scholar]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Salazar, G.; Cornejo-Castillo, F.M.; Benítez-Barrios, V.; Fraile-Nuez, E.; Álvarez-Salgado, X.A.; Duarte, C.M.; Gasol, J.M.; Acinas, S.G. Global diversity and biogeography of deep-sea pelagic prokaryotes. ISME J. 2016, 10, 596–608. [Google Scholar] [CrossRef]
- Pesant, S.; Not, F.; Picheral, M.; Kandels-Lewis, S.; Le Bescot, N.; Gorsky, G.; Iudicone, D.; Karsenti, E.; Speich, S.; Troublé, R.; et al. Open science resources for the discovery and analysis of Tara Oceans data. Sci. Data 2015, 2, 150023. [Google Scholar] [CrossRef]
- Kai, W.; Peisheng, Y.; Rui, M.; Wenwen, J.; Zongze, S. Diversity of culturable bacteria in deep-sea water from the South Atlantic Ocean. Bioengineered 2017, 8, 572–584. [Google Scholar] [CrossRef]
- Li, H.; Yu, Y.; Luo, W.; Zeng, Y.; Chen, B. Bacterial diversity in surface sediments from the Pacific Arctic Ocean. Extremophiles 2009, 13, 233–246. [Google Scholar] [CrossRef]
- Varliero, G.; Bienhold, C.; Schmid, F.; Boetius, A.; Molari, M. Microbial Diversity and Connectivity in Deep-Sea Sediments of the South Atlantic Polar Front. Front. Microbiol. 2019, 10, 665. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, L.L.; Bendia, A.G.; Duarte, R.T.D.; das Graças, D.A.; da Costa da Silva, A.L.; Nakayama, C.R.; Sumida, P.Y.; Lima, A.O.S.; Nagano, Y.; Fujikura, K.; et al. Bacterial diversity in deep-sea sediments under influence of asphalt seep at the São Paulo Plateau. Antonie Van Leeuwenhoek 2020, 113, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, D.J.; Andreote, F.D.; Chaves, D.; Montaña, J.S.; Osorio-Forero, C.; Junca, H.; Zambrano, M.M.; Baena, S. Structural and Functional Insights from the Metagenome of an Acidic Hot Spring Microbial Planktonic Community in the Colombian Andes. PLoS ONE 2012, 7, e52069. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Yela, A.C.; Mosquera-Rendón, J.; Noreña-Puerta, A.; Cristancho, M.M.V.; Lopez-Alvarez, D. Microbial Diversity Exploration of Marine Hosts at Serrana Bank, a Coral Atoll of the Seaflower Biosphere Reserve. Front. Mar. Sci. 2019, 6. [Google Scholar] [CrossRef]
- Gonzalez-Zapata, F.L.; Bongaerts, P.; Ramírez-Portilla, C.; Adu-Oppong, B.; Walljasper, G.; Reyes, A.; Sanchez, J.A. Holobiont Diversity in a Reef-Building Coral over Its Entire Depth Range in the Mesophotic Zone. Front. Mar. Sci. 2018, 5, 29. [Google Scholar] [CrossRef]
- Quintanilla, E.; Ramírez-Portilla, C.; Adu-Oppong, B.; Walljasper, G.; Glaeser, S.P.; Wilke, T.; Muñoz, A.R.; Sánchez, J.A. Local confinement of disease-related microbiome facilitates recovery of gorgonian sea fans from necrotic-patch disease. Sci. Rep. 2018, 8, 14636. [Google Scholar] [CrossRef]
- Villegas-Plazas, M.; Wos-Oxley, M.L.; Sanchez, J.A.; Pieper, D.H.; Thomas, O.P.; Junca, H. Variations in Microbial Diversity and Metabolite Profiles of the Tropical Marine Sponge Xestospongia muta with Season and Depth. Microb. Ecol. 2019, 78, 243–256. [Google Scholar] [CrossRef]
- Naranjo-Vesga, J.; Ortiz-Karpf, A.; Wood, L.; Jobe, Z.; Paniagua-Arroyave, J.F.; Shumaker, L.; Mateus-Tarazona, D.; Galindo, P. Regional controls in the distribution and morphometry of deep-water gravitational deposits along a convergent tectonic margin. Southern Caribbean of Colombia. Mar. Pet. Geol. 2020, 121, 104639. [Google Scholar] [CrossRef]
- Correa-Ramirez, M.; Rodriguez-Santana, Á.; Ricaurte-Villota, C.; Paramo, J. The Southern Caribbean upwelling system off Colombia: Water masses and mixing processes. Deep Sea Res. Part I Oceanogr. Res. Pap. 2020, 155, 103145. [Google Scholar] [CrossRef]
- Rudzin, J.E.; Shay, L.K.; Jaimes, B.; Brewster, J.K. Upper ocean observations in eastern Caribbean Sea reveal barrier layer within a warm core eddy. J. Geophys. Res. Ocean. 2017, 122, 1057–1071. [Google Scholar] [CrossRef]
- Christ, R.D.; Wernli, R.L. (Eds.) Chapter 2—The Ocean Environment. In ROV Manual B.T.-T, 2nd ed.; Butterworth-Heinemann: Oxford, UK, 2014; pp. 21–52. ISBN 978-0-08-098288-5. [Google Scholar]
- Herlemann, D.P.R.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Dusa, A. Package ‘Venn’. Available online: https://cran.r-project.org/web/packages/venn/venn.pdf (accessed on 1 September 2020).
- Sinniger, F.; Pawlowski, J.; Harii, S.; Gooday, A.J.; Yamamoto, H.; Chevaldonné, P.; Cedhagen, T.; Carvalho, G.; Creer, S. Worldwide Analysis of Sedimentary DNA Reveals Major Gaps in Taxonomic Knowledge of Deep-Sea Benthos. Front. Mar. Sci. 2016, 3, 92. [Google Scholar] [CrossRef]
- Guardiola, M.; Wangensteen, O.S.; Taberlet, P.; Coissac, E.; Uriz, M.J.; Turon, X. Spatio-temporal monitoring of deep-sea communities using metabarcoding of sediment DNA and RNA. PeerJ 2016, 4, e2807. [Google Scholar] [CrossRef]
- Da Silva, M.A.C.; Cavalett, A.; Spinner, A.; Rosa, D.C.; Jasper, R.B.; Quecine, M.C.; Bonatelli, M.L.; Pizzirani-Kleiner, A.; Corção, G.; de Lima, A.O.S. Phylogenetic identification of marine bacteria isolated from deep-sea sediments of the eastern South Atlantic Ocean. Springerplus 2013, 2, 127. [Google Scholar] [CrossRef]
- Quaiser, A.; Zivanovic, Y.; Moreira, D.; López-García, P. Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J. 2011, 5, 285–304. [Google Scholar] [CrossRef]
- Zinger, L.; Amaral-Zettler, L.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Huse, S.M.; Welch, D.B.M.; Martiny, J.B.H.; Sogin, M.; Boetius, A.; Ramette, A. Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS ONE 2011, 6, e24570. [Google Scholar] [CrossRef]
- Hamdan, L.J.; Coffin, R.B.; Sikaroodi, M.; Greinert, J.; Treude, T.; Gillevet, P.M. Ocean currents shape the microbiome of Arctic marine sediments. ISME J. 2013, 7, 685–696. [Google Scholar] [CrossRef]
- Dyksma, S.; Bischof, K.; Fuchs, B.M.; Hoffmann, K.; Meier, D.; Meyerdierks, A.; Pjevac, P.; Probandt, D.; Richter, M.; Stepanauskas, R.; et al. Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J. 2016, 10, 1939–1953. [Google Scholar] [CrossRef]
- Walsh, E.A.; Kirkpatrick, J.B.; Rutherford, S.D.; Smith, D.C.; Sogin, M.; D’Hondt, S. Bacterial diversity and community composition from seasurface to subseafloor. ISME J. 2016, 10, 979–989. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.F.; Franks, D.G.; Alldredge, A.L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 1993, 38, 924–934. [Google Scholar] [CrossRef]
- Rath, J.; Wu, K.Y.; Herndl, G.J. High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat. Microb. Ecol. 1998, 14, 261–269. [Google Scholar] [CrossRef]
- Fandino, L.B.; Riemann, L.; Steward, G.F.; Long, R.A. Variations in bacterial community structure during a dinoflagellate bloom analyzed by DGGE and 16S rDNA sequencing. Aquat. Microb. Ecol. 2001, 23, 119–130. [Google Scholar] [CrossRef]
- Goffredi, S.; Orphan, V. Bacterial community shifts in taxa and diversity in response to localized organic loading in the deep sea. Environ. Microbiol. 2009, 12, 344–363. [Google Scholar] [CrossRef]
- Escobar-Briones, E.; García-Villalobos, F.J. Distribución espacial del carbono orgánico total en el sedimento superficial de la planicie abisal del Golfo de México. In Carbono en Ecosistemas Acuáticos de México; Benigno Hernández de la Torre, G.G.C., Ed.; Instituto Nacional de Ecología: Mexico City, Mexico, 2007; p. 508. ISBN 9688178551. [Google Scholar]
- Bauer, M.; Kube, M.; Teeling, H.; Richter, M.; Lombardot, T.; Allers, E.; Würdemann, C.; Quast, C.; Kuhl, H.; Knaust, F.; et al. Whole genome analysis of the marine Bacteroidetes‘Gramella forsetii’ reveals adaptations to degradation of polymeric organic matter. Environ. Microbiol. 2007, 8, 2201–2213. [Google Scholar] [CrossRef]
- Panschin, I.; Becher, M.; Verbarg, S.; Spröer, C.; Rohde, M.; Schüler, M.; Amann, R.; Harder, J.; Tindall, B.; Hahnke, R. Description of Gramella forsetii sp. nov., a marine Flavobacteriaceae isolated from North Sea water, and emended description of Gramella gaetbulicola Cho et al. 2011. Int. J. Syst. Evol. Microbiol. 2016, 67. [Google Scholar] [CrossRef]
- Petro, C.; Starnawski, P.; Schramm, A. Microbial community assembly in marine sediments. Aquat. Microb. Ecol. 2017, 79, 177–195. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Konopka, A.E.; Fredrickson, J.K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef]
- Bienhold, C.; Zinger, L.; Boetius, A.; Ramette, A. Diversity and Biogeography of Bathyal and Abyssal Seafloor Bacteria. PLoS ONE 2016, 11, e0148016. [Google Scholar] [CrossRef]
- Von Scheibner, M.; Sommer, U.; Jürgens, K. Tight Coupling of Glaciecola spp. and Diatoms during Cold-Water Phytoplankton Spring Blooms. Front. Microbiol. 2017, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Tyson, R.V. Sedimentation rate, dilution, preservation and total organic carbon: Some results of a modelling study. Org. Geochem. 2001, 32, 333–339. [Google Scholar] [CrossRef]
- Walsh, E.A.; Kirkpatrick, J.B.; Pockalny, R.; Sauvage, J.; Spivack, A.J.; Murray, R.W.; Sogin, M.L.; D’Hondt, S. Relationship of Bacterial Richness to Organic Degradation Rate and Sediment Age in Subseafloor Sediment. Appl. Environ. Microbiol. 2016, 82, 4994–4999. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).