Abstract
Acquarossa river (Viterbo, Italy) was the site of a prospering Etruscan civilization thanks to metallurgical activity around 625–550 B.C. This caused the spread of heavy metals throughout the area. Rocks along the river probably act as a filter for these elements and they are covered by two different biofilms (epilithons). They differ for both color and bacterial composition. One is red and is enriched with Pseudomonas strains, while the other one is black and Acinetobacter is the most represented genus. Along the river lay the Infernaccio waterfalls, whose surrounding rocks are covered only by the red epilithon. The bacterial composition of this biofilm was analyzed through high throughput sequencing and compared to those ones of red and black epilithons of Acquarossa river. Moreover, cultivable bacteria were isolated and their phenotype (i.e., resistance against antibiotics and heavy metals) was studied. As previously observed in the case of Acquarossa river, characterization of bacterial composition of the Infernaccio red epilithon revealed that the two most represented genera were Acinetobacter and Pseudomonas. Nonetheless, these strains differed from those isolated from Acquarossa, as revealed by RAPD analysis. This work, besides increasing knowledge about the ecological properties of this site, allowed to isolate new bacterial strains, which could potentially be exploited for biotechnological applications, because of their resistance against environmental pollutants.
1. Introduction
The Acquarossa river is located close to the city of Viterbo (Italy) and represents a site of naturalistic and archaeological interest where the Etruscan civilization developed around 625–550 B.C. The metallurgical activity on which the economy of this population was based led to the spread of heavy metals throughout the area [1], mainly iron and arsenic [2]. Along the whole river course, rocks are covered by a red and a black biofilm (epilithon). As previously described [3], these epilithons are characterized by high heavy metal concentrations, suggesting that they might represent natural “filters” for the river. Interestingly, the two kinds of epilithic biofilm are characterized by peculiar bacterial communities, from both genetic and phenotypic viewpoints. These bacterial communities were differently spatially distributed in the two biofilms, depending on the chemical composition of each kind of biofilm. Consequently, the spatial distribution led to the development of specific phenotypic characteristics in the two biofilms (i.e., heavy metal and antibiotic resistance patterns, antagonistic interactions). Along the river course, one encounters the Infernaccio waterfalls (42°31′38.55″ N, 12°07′52.1″ E), where the presence of the red epilithic biofilms persists on the rocks surrounding the bottom of the waterfall and, at the same time, the rocks behind the waterfalls are red too, since iron-enriched water falls continuously from some twenty meters height. Interestingly, contrarily to the rest of the river course, the black epilithic biofilm is almost absent on the rocks around the waterfall.
This site can be considered as an extreme environment in which micro- and macroorganisms might have selected particular traits to survive in the presence of high heavy metal concentrations. From a microbiological viewpoint, this environment is quite interesting since it has been well established by previous studies that environments exhibiting extreme conditions (i.e., low/high temperatures, high heavy metal concentrations, hydrocarbon presence, high salinity level, etc.) can select microorganisms of biotechnological and medical interest able to survive under these conditions. These microorganisms, mainly bacteria, often show a marked antimicrobial activity, even against multidrug resistant human pathogens, such as those belonging to the Burkholderia cepacia complex (Bcc). Many of these antimicrobial-producing extremophilic microorganisms have been isolated in Antarctica [4,5,6,7], in hydrocarbons enriched sites [8], in thermophilic [9,10] and in halophilic environments [11].
Even though some information about the archaeological and historical aspects of the Infernaccio waterfalls and the Acquarossa river are available [12,13,14], from a (micro)biological viewpoint there is an almost complete lack of information, with the only exception represented by the characterization of the red and black epilithic biofilms along the river course [3]. Interestingly, the bacterial community characterization confirmed the presence of strains showing a marked antimicrobial activity against other bacterial strains of the same environments [3]. Moreover, Pseudomonas strains from the Acquarossa river, showed a marked antagonistic activity also against other strains isolated from many other environments [15]. These findings confirm that the Acquarossa river exerts a selective pressure on the microbial diversity, leading to the selection of strains able to antagonize other bacteria, by producing molecules of putative biotechnological interest. The finding that no black epilithon was found in the Infernaccio site suggests that the chemical and physical compositions of red epilithon might differ from those of the red epilithon disclosed in the Acquarossa river, which, in turn, might imply that different bacterial communities might have been selected in Infernaccio red epilithon. Thus, this work was aimed to the characterization of the bacterial communities isolated from the red epilithic biofilm collected on the rocks surrounding the bottom of the Infernaccio waterfalls and to compare data obtained with those previously described [3].
2. Materials and Methods
2.1. Site and Sample Collection
Two samples of waterfall river water (500 mL each) and two samples of red epilithic biofilm (about 5 g each, Figure 1) were collected in July 2016 at the Infernaccio waterfalls near Viterbo (Italy).

Figure 1.
Red epilithic biofilm sampled at Infernaccio waterfalls.
Immediately after sampling, water and biofilm samples were brought to the laboratory for chemico-physical and microbiological analyses. Several parameters were measured both for the water and for the red biofilm, according to the UNI EN 13 657:2004 and UNI EN ISO 11 885:2009 methods: pH, conductivity, concentrations of ammonium (NH4+), nitrites (NO2−), nitrates (NO3−), chemical oxygen demand (COD), sulfates (SO42−), sulfides (S2−), total iron content and heavy metals (i.e., Ni, Cu, Zn, As and Cd). Fe2+ and Fe3+ were measured according to the Standard Methods “3500-Fe B. Phenanthroline Method” (Copyright 1999 by the American Public Health Association, American Water Works Association, Water Environment Federation). The concentration of the following heavy metals was determined within the red biofilm samples: Ni, Cu, Zn, As, Cd, and total iron content. The entire list of parameters analyzed is reported in Table 1.

Table 1.
Chemico-physical characterization of water and red epilithon from Inferaccio waterfalls.
2.2. DNA Extraction, High Throughput Sequencing (HTS) Analysis
About 0.5 g of each of two red epilithic biofilm samples were used for HTS Analysis. DNA extraction from red epilithic biofilm was performed by using a PowerLyzer PowerSoil® DNA Isolation Kit (MO BIO laboratories, Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions. The bacterial V4 region of 16S rRNA gene of each DNA sample was amplified with specific primers (515F, 5′-TGYCAGCMGCCGCGGTAA-3′; 806R 5′-GGACTACNVGGGTWTC TAAT-3′, [16]) using the previously reported protocol [17]. Library preparation and demultiplexing have been carried out as described in [18]. Libraries were sequenced in a single run using Illumina MiSeq technology with pair-end sequencing strategy with a MiSeq Reagent Kit v3.).
2.3. Analysis of 16S rRNA Sequences from Microbial Assemblages
Sequences were clustered into operational taxonomic units (OTUs) using the UPARSE pipeline [19], as described previously [3]. Briefly, raw reads were merged into a single amplicon sequence using the ‘fastq_mergepairs’ command. The identity threshold was set to 80% in order to account for long overlaps (2 × 300 bp, V3–V4). The ‘filter_lowc’ command before quality assessment was used to find and remove low complexity reads (namely those containing repeated sequences and/or regions that are highly enriched for just one nucleotide). Merged reads were then quality checked as described in [20]. An additional quality filtering step and sequences clustering were performed [3]. Representative sequences were taxonomically classified using the ‘sintax’ command along with the RDP training set (version 16). Sequences that were not assigned to Bacteria domain were removed to minimize possible contaminants due to PCR amplification [21]. The OTU table was produced with the ‘otutab’ command. Sequences are part of the previous submission reported in [3] (BioProject accession PRJNA412007). Differences between epilithon types (black vs red) and sampling sites (Infernaccio vs Acquarossa) were tested using permutational multivariate analysis of variance using distance matrices as implemented in the ‘adonis2′ function of the R vegan package (version 2.5.5). The overall distribution of samples was plotted using canonical correspondence analysis (’cca’ function of vegan package). Operational taxonomic units that were resolved at genus level were used to provide a more in-depth insight into the two bacterial communities inhabiting black and red epilithons. LDA Effect Size (LEfSe) [22] was used to find bacterial genera linked to a peculiar type of epilithon, as described in the LEfSe tutorial reported at: https://bitbucket.org/biobakery/biobakery/wiki/lefse.
2.4. Red Epilithic Cultivable Bacterial Community Characterization
1 g of red biofilm of each sample was submitted to serial dilution in saline solution (0.9% w/v NaCl) and spread on Tryptic Soy Agar (TSA) plates. Bacterial growth was checked after 48 h incubation at 21 °C. Isolated bacterial colonies were picked up and streaked on TSA medium for the further molecular and phenotypic characterization. At the same time, the isolated colonies were stored at −80 °C in glycerol (20% v/v final concentration with Tryptic Soy Broth -TSB- medium).
Bacterial cell lysates were prepared by as described in [3]. The 16S rRNA gene of each bacterial isolate according to [3], using the primer set P0 (5′-GAGAGTTTGATCCTGGCTCAG-3′) and P6 (5′-CTACGGCTACCTTGTTACGA-3′) [23] and the amplification conditions described in [3]. Direct sequencing of the amplified 16S rRNA genes was performed with primer P0; each sequence was submitted to GenBank and assigned the accession number shown in Table 2.

Table 2.
List of bacterial strains used in this work. The accession number of 16S rRNA gene sequence of each strain is reported.
Taxonomic affiliation was carried out using the ‘classifier’ tool of the Ribosomal Database Project (RDP) [24]. The alignment of the obtained sequences with those of closely related Type Strains downloaded from the RDP database was carried out using the BioEdit Software [25]; the resulting alignment was then used to build the relative phylogenetic tree using the MEGA5 Software [26] and the parameters reported in [3].
2.5. Random Amplified Polymorphic DNA (RAPD) Analysis
Random amplification of DNA fragments [27] was performed on Acinetobacter and Pseudomonas bacterial isolates [3]. Briefly, a 25 μL-total volume reaction was performed for each strain using primer 1253 (5′-GTTTCCGCCC-3′) [28] and amplicons generated were analyzed by 2% w/v agarose gel. Isolates sharing the same RAPD profile were grouped together into the same cluster (haplotype).
Alpha diversity indices (Shannon, Evenness and Chao1) were calculated using PAST3 software [29] on the results obtained from haplotype attribution, considering the number of strains attributed to each haplotype.
2.6. Phenotypic Characterization of Acinetobacter and Pseudomonas spp. Bacterial Strains: Resistance to Antibiotics and Heavy Metals, and Statistical Analysis
The resistance patterns to six antibiotics and to six heavy metals were obtained through the broth microdilution method in Muller Hinton Broth (MHB), according to [30] as previously described [3]. The following antibiotics and heavy metals were tested at different concentrations: kanamycin, streptomycin and ciprofloxacin (0.50–1.00–2.50–5.00–10.00–50.00 μg/mL), tetracycline (0.50–1.25–2.50–5.00–12.50–25.00 μg/mL), chloramphenicol (1.00–2.50–5.00–10.00–25.00–50.00 μg/mL), rifampicin (5.00–10.00–25.00–50.00–100.00 μg/mL, As(V) [KH2AsO4, 0.5–1.0–2.5–5.0–7.5–10.0–12.5–25.0 mM], Zn [ZnSO4, 5.0–10.0–15.0–25.0 mM], Cu [CuCl2, 1.0–2.5–5.0–10.0 mM], Cd [Cd(NO3)2, 5.0–10.0–15.0–25.0 mM], Ni [NiCl2, 5.0–10.0–15.0–25.0 mM], and As(III) [NaAsO2 0.5–1.0–2.5–5.0–10.0–15.0 mM]. Data obtained from broth microdilution methods were validated by using TECAN microplate reader (Tecan, Durham, NC, USA) at 600 nm wavelength, after 48 h incubation at 21 °C.
For both heavy metal and antibiotic resistance pattern experiments, a positive control and a negative control were also set up in triplicate in each microplate The OD600 value for the positive control was considered as 100%. All the other measured values were reported as percentage of growth in proportion to the positive control. The data matrix containing the performances of growth expressed as percentage on the positive control has been used for statistical analysis (UPGMA with Bray-Curtis distance measure, and PCA) with PAST3 software [29], as described in [31].
3. Results
3.1. Chemico-Physical Characterization of Water and Biofilm
The chemico-physical analysis of the water and the red epilithon of the Infernaccio waterfalls is reported in Table 1, together with the same parameters measured in the water, in the red and black epilithons collected in the Acquarossa river. The comparison of the two water samples collected in the Acquarossa river [3] and in the Infernaccio waterfalls did not reveal differences between them. On the other site, the red epilithon of the Infernaccio waterfalls exhibited higher concentrations of P, Al, Fe, Mn, Ba, Cu, V, Co, Be, Ni, and Zn in comparison to the red epilithon of the Acquarossa site.
3.2. High Throughput Sequencing (HTS) Analysis
Results of HTS of the two epilithon samples were merged together and shown in Figure 2. The overall bacterial composition was mainly influenced by the different epilithon types (permutational multivariate analysis of variance using distance matrices, reported in Table S1). Even if the two sampling sites seemed to influence bacterial distribution only marginally (R squared values from Table S1 < 0.14), red epilithon samples from Infernaccio showed a distinct distribution over the CCA2 axis of the canonical correspondence analysis reported in Figure 2 and Figure 3, suggesting that the different heavy metal concentrations disclosed in the two sites might have driven the structuring of the bacterial communities. At genus level some differences were detected as reported in Figure S1.
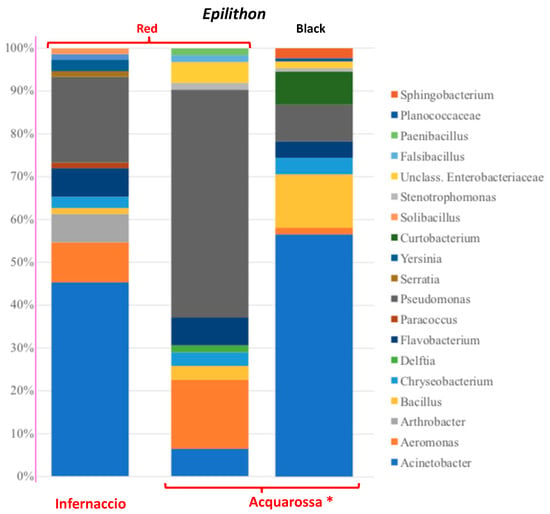
Figure 2.
Composition of bacterial communities isolated from red epilithons of Infernaccio and Acquarossa river and black epilithon from Acquarossa river. * Data from [3].
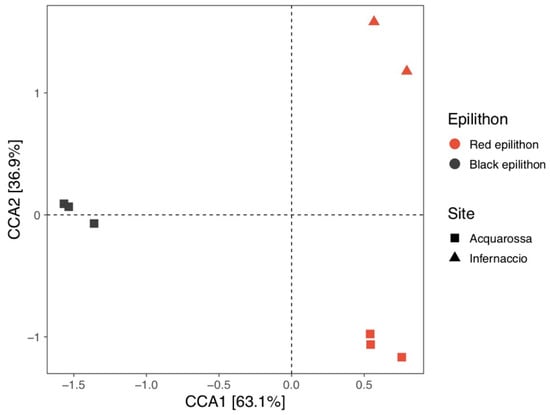
Figure 3.
Canonical correspondence analysis of bacterial communities from different epilithon types and different sampling sites. Epilithon types were reported using different colors whereas sampling sites were reported using different point shapes. Values between squared brackets represent the value of inertia explained by the given axis.
3.3. Composition of Culturable Bacterial Community Isolated from Red Epilithon
The titer of heterotrophic cultivable bacteria in the Infernaccio red epilithon was about 5.0 × 105 CFU/g, similar to those obtained in the Acquarossa red epilithon [3]. To determine the composition of the cultivable fraction of the bacterial community collected from the red epilithon, the 16S rRNA gene sequences from 75 randomly chosen isolates were amplified and sequenced.
Each of the 75 sequences obtained was compared with those available in databases through the BLASTn option of the BLAST software. Data obtained revealed that the 75 bacterial isolates were affiliated to 12 genera with a dominance of Acinetobacter and Pseudomonas (Figure 4).
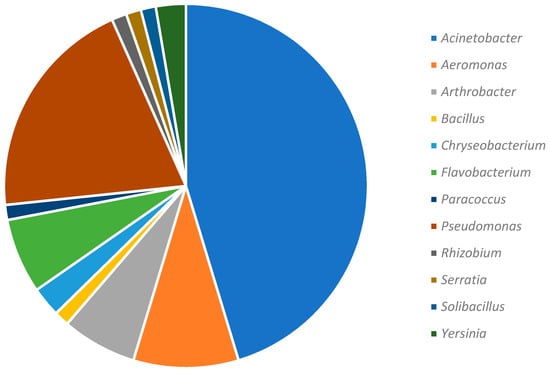
Figure 4.
Taxonomic distribution of 75 bacterial strains isolated from the Infernaccio red epilithon.
3.4. Phenotypic and Molecular Characterization of Pseudomonas and Acinetobacter Isolates
3.4.1. Structure of the Pseudomonas and Acinetobacter Communities
Since Acinetobacter and Pseudomonas isolates represented a high percentage (65%) of the total isolates (45% and 20%, respectively), a deeper analysis on these isolates was performed. First, a RAPD analysis was carried out as described in Materials and Methods for each of the 34 Acinetobacter and 15 Pseudomonas isolates. By assuming that isolates sharing the same RAPD profile correspond to the same strain, the comparative analysis of the RAPD profiles obtained allowed to split the Acinetobacter sp. community in 19 haplotypes and the Pseudomonas sp. community in 9 haplotypes, corresponding to 19 and 9 strains, respectively (Table 3 and Table 4), revealing a high degree of biodiversity at the strain level for both genera. The comparison of Acinetobacter and Pseudomonas RAPD profiles with those obtained from the Acquarossa river [3] revealed the absence of common profiles between the two sites, strongly suggesting that the different composition/concentration of heavy metals in the two red epilithons might have selected different strains belonging to the same genus.

Table 3.
Distribution of Acinetobacter RAPD haplotypes.

Table 4.
Distribution of Pseudomonas RAPD haplotypes.
The alpha diversity indices (Shannon Evenness and Chao1) calculated on the RAPD distribution pattern for both Acinetobacter and Pseudomonas strains (Table 5) showed a higher Shannon and Chao-1 index values for Acinetobacter, and a higher Evenness value for Pseudomonas (0.9244 vs 0.848 of Acinetobacter).

Table 5.
Alpha diversity indices calculated on RAPD profiles for Acinetobacter and Pseudomonas strains.
3.4.2. Phylogenetic Analysis
Phylogenetic analysis of Acinetobacter and Pseudomonas bacterial strains was performed as described in Materials and Methods and data obtained are shown in Figure 3, Figure 4, Figures S2 and S3. The Acinetobacter strains can be split into three main taxonomic groups: the first one including most of the strains, was taxonomically related to A. johnsonii; a second group related to A. pakistanensis (strains 14.13, 14.24, 11.40), and a third group related to A. guillouiae (strains 14.2, 14.3, 14.21 and 11.39); all the other isolates joined a single cluster (Figure 5).
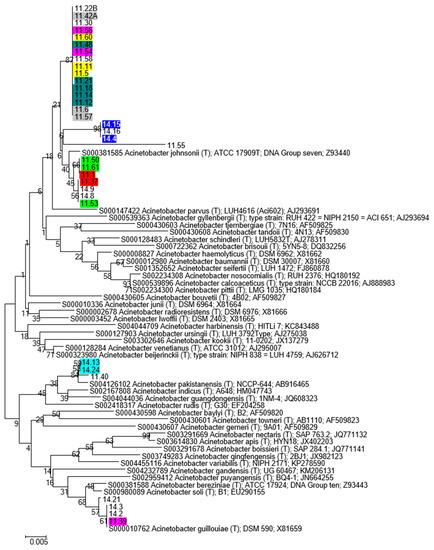
Figure 5.
Phylogenetic tree showing relationships between Acinetobacter sp. isolates, and Acinetobacter sp. Type Strains retrieved from RDP database. The tree was constructed based on 16S rRNA gene sequences, with Neighbor-Joining algorithm and 100 bootstrap values. Isolates exhibiting the same RAPD profile are indicated with the same color.
As it may be expected, the Acinetobacter strains belonging to the same RAPD haplotype exhibit the same taxonomic affiliation (with the only exception being represented by strain 11.39 belonging to haplotype 26).
Also, in the Pseudomonas 16S rRNA gene phylogenetic tree three main groups can be recognized (Figure 6): the first one embedding isolates 11.26, 11.46, 11.32 and 11.2 and taxonomically related to the group of P. reinekei/P. granadensis; a second group of strains related to P. protegens (most of the isolated bacteria) and strain 14.6 taxonomically related to P. kireensis. Pseudomonas strains belonging to the same RAPD haplotype are taxonomically related to the same species.
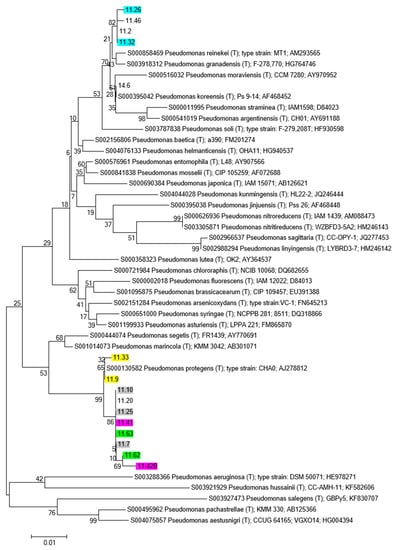
Figure 6.
Phylogenetic tree showing relationships between Pseudomonas sp. isolates, and Pseudomons sp. Type Strains retrieved from RDP database. The tree was constructed based on 16S rRNA gene sequences, with Neighbor-Joining algorithm and 100 bootstrap values. Isolates exhibiting the same RAPD profile are indicated with the same color.
3.4.3. Phenotypic Characterization of Bacterial Strains: Antibiotic and Heavy Metal Resistance Patterns
As previously shown [3], bacteria isolated from black and red epilithons from the Acquarossa river exhibited different patterns of resistance to both antibiotics and heavy metals, suggesting that the presence of such compounds might drive the structuring of the two epilhitic bacterial communities. In order to have some insights into the phenotypic traits of Acinetobacter and Pseudomonas strains isolated from the Infernaccio red epilithon, the resistance profile toward heavy metals and antibiotics of each strain was tested, as described in Materials and Methods. Data concerning the resistance patterns to a panel of six antibiotics and six heavy metals are shown in in Figure 7 whose analysis revealed that Acinetobacter strains were in general more sensitive to antibiotics, especially to streptomycin, tetracycline, and chloramphenicol, whereas Pseudomonas strains were more sensitive to arsenite and arsenate.
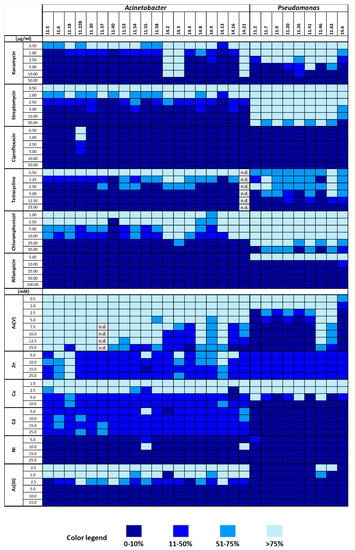
Figure 7.
Resistance patterns towards antibiotics and heavy metals of the Acinetobacter and Pseudomonas strains isolated from the Infernaccio waterfalls. Colors: navy 0–10% growth; blue 11–50% growth; deep sky blue 51–75% growth; light blue > 75% growth. Abbreviations: n.d., not determined.
Accordingly, the UPGMA performed on the resistance patterns data matrix confirmed the clear separation between strains belonging to the two genera (Figure 8).
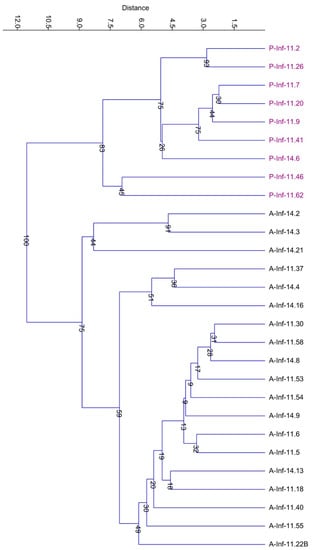
Figure 8.
UPGMA analysis (Bray-Curtis distance) performed on the whole antibiotic and heavy metal resistance pattern data matrix. Abbreviations: A and P indicate Acinetobacter and Pseudomonas strains, respectively.
4. Discussion
The aim of this work was the analysis of the composition of the total bacterial community and the molecular and phenotypic analysis of the heterotrophic bacterial communities isolated from the Infernaccio waterfalls, where rocks were covered by a red epilithon; differently from the Acquarossa river, no black epilithon was found in Infernaccio. This suggested a different chemico-physical composition either of water and/or red epilithons, which, in turn, might reflect into different bacterial communities inhabiting on the rock surfaces.
According to this idea, the analysis of the chemical and physical parameters of the two red epilithons revealed the existence of strong difference between them, whereas the water samples in the two sites showed very similar profiles. The different accumulation over time of the compounds analyzed might have driven a different composition of total and cultivable bacterial community of two red epilithic biofilms (i.e., Infernaccio and Acquarossa). Indeed, both the HTS and cultivable fraction analysis revealed that the composition of the bacterial communities from Infernaccio was different in respect to that of the red epilithic biofilm in the Acquarossa river [3]. Red epilithon of Infernaccio shares similar features with black epilithic biofilm of Acquarossa, as for example the dominance of Acinetobacter genus. Indeed, since in Acquarossa red epilithon the Acinetobacter sp. abundance was 6.45% of the total, in the Infernaccio red epilithon it was 45.33% of the total, that is more similar to the black epilithon (56.58%). At the same time, in Acquarossa red epilithon the Pseudomonas sp. abundance was 53.22% of the total, while in the Infernaccio red epilithon it is 20% of the total, analogously to the black epilithon (8.52%). Moreover, in Infernaccio waterfalls the less represented bacterial genera such as Arthrobacter sp. (6.6%), Solibacillus sp., Serratia sp., Rhizobium sp., Paracoccus sp. (each one 1.33%), and Yersinia sp. (2.6%), have not been detected in the Acquarossa red epilithon. On the other side, Acquarossa red epilithon Stenotrophomonas sp. is absent in the Infernaccio waterfall samples.
These findings strongly suggest that the different chemico-physical composition of Infernaccio and Acquarossa red epilithons might have played an important role in selecting bacteria at the genus level. Deeper molecular and phenotypic analyses were carried out on isolates of the two most represented genera (i.e., Acinetobacter and Pseudomonas). The 16S rRNA gene phylogenetic analysis performed on the Infernaccio Acinetobacter and Peudomonas isolates revealed that they were located on different branches of the respective phylogenetic tree, strongly suggesting that both Acinetobacter and Pseudomonas isolates belong to different species. Composite phylogenetic trees embedding Acinetobacter or Pseudomonas isolates from both Infernaccio and Acquarossa were constructed to check the existence of differences/similarities between the Acinetobacter or Psedudomonas isolates from the two sites. The two phylogenetic trees are shown in the Supplementary Material. The analysis of the two trees revealed that:
- (i)
- The major fraction of Acinetobacter isolates was located on the same branch of the phylogenetic tree (very close to A. johnsonii and A. parvus.). In this branch the isolates coming from Infernaccio and Acquarossa were intermixed.
- (ii)
- Some Acinetobacter isolates coming from Infernaccio clustered together joining different branches of the tree and were separated from the Acquarossa isolates.
- (iii)
- A similar scenario was disclosed for the Pseudomonas tree.
The whole body of phylogenetic data strongly suggested that the Acinetobacter as well as Pseudomonas communities from the two sites (Infernaccio and Acquarossa) showed some differences at the species level, even though isolates from the two sites can share the same taxonomical affiliation. In order to check the structure of the Acinetobacter and Pseudomonas communities, a RAPD analysis was performed on Acinetobacter and Pseudomonas isolates. The comparison of RAPD profiles splits the Acinetobacter and Pseudomonas communities into 19 and nine groups, respectively, corresponding to at least 19 and nine strains. This result suggests a high degree of diversity at strain level in the Acinetobacter community. The high degree of diversity is also confirmed by the Chao-1 index, which is higher in Acinetobacter (32.75) respect to Pseudomonas (10.2). The higher Evenness index value of the Acinetobacter community (0.9244 vs 0.848 of Pseudomonas) reflects a more homogeneous distribution of strains among the different RAPD groups. This result is in agreement with previous data on the Acquarossa site [3]. On the other side, the Shannon index of the Acquarossa site communities, suggested a more diverse Pseudomonas community respect of Acinetobacter, while in the present work, the result was the opposite.
The taxonomic distribution of both Acinetobacter and Pseudomonas strains mirrored the RAPD haplotypes distribution, with isolates showing the same RAPD haplotype (very likely corresponding to the same strain) joining the same clade in the phylogenetic tree.
The comparative analysis of the Infernaccio and Acquarossa Acinetobacter RAPD profiles obtained with the same primer under the same experimental conditions did not reveal the existence of any profile sharing between isolates from the two sites (data not shown). This was also true for Pseudomonas isolates, strongly suggesting that the force(s) driving the structuring of the bacterial communities in the two red epilithic biofilms (Infernaccio and Acquarossa) might strongly act at the strains level and at a lesser extent at the species and genus level.
This suggests that the different chemical composition of the two red epilithons might have played a key role in shaping both the composition and the structure of the bacterial communities.
The statistics of the phenotypic traits related to the resistance patterns towards heavy metals and antibiotics (Figure 7) revealed the differences between Acinetobacter and Pseudomonas strains. Indeed, the resistance patterns towards heavy metals and antibiotics are different for the two bacterial genera, and this is confirmed by statistic (100 bootstrap value supporting the separation between Acinetobacter and Pseudomonas in UPGMA analysis). Overall, these results are in agreement with those obtained for the bacteria isolated in the Acquarossa river [3] with a few exceptions. Indeed, on one hand, among Pseudomonas strains isolated from Infernaccio, some were more sensitive to either tetracycline or As in comparison to their counterpart isolated from Acquarossa. On the other hand, some of the Acinetobacter strains isolated from Infernaccio were more resistant against Cu and Cd if compared to other Acinetobacter strains from Acquarossa.
Taxonomic distribution of both Acinetobacter and Pseudomonas bacterial strains does not mirror the clustering of the UPGMA tree (Figure 8). This might be expected, since the genes responsible for the resistance to antibiotics and/or heavy metals might be related more to the genomic traits of microorganisms rather than taxonomy and to the conditions of the micro-environments inside the biofilm. Indeed, it is well known from literature that a bacterial biofilm is a complex structure in which different micro-environment can be recognized [32,33]. Probably, the different behaviors of bacterial strains belonging to the same taxonomic group are due to the different distribution of chemical species in the biofilm. Moreover, it cannot be a priori excluded the possibility that in the epilithons mobile genetic elements may transfer between bacteria belonging to different species the genetic traits allowing the surviving in the extreme conditions.
5. Conclusions
In this work, the taxonomic and phenotypic characterization of bacterial strains isolated from a red epilithic biofilm has been performed, together with the chemico-physical characterization of both waterfall samples, and red epilithon samples. Data have been correlated and compared with previous data on samples collected in the same environment, but from a different sampling site [3]. This work expands our knowledge of the microbiological aspects of the site, which is one of the most interesting in Italy from an historical, ecological, and naturalistic point of view. Moreover, it helps elucidate new aspects of the microscale interactions and structuring of this red epilithic biofilm, that is peculiar of such environment.
Supplementary Materials
The following are available online at https://www.mdpi.com/1424-2818/11/10/175/s1: Figure S1: LDA scores of bacterial taxa significantly influenced by epilithon type. Only taxa with a p-value lower than 0.05 and a LDA score higher than 2 were reported. The full lineage of each taxon was reported on the y-axis. Figure S2: Phylogenetic tree showing relationships between Acinetobacter spp. isolated from Infernaccio (in blue) and Acquarossa (in red). Figure S3: Phylogenetic tree showing relationships between Pseudomonas spp. isolated from Infernaccio (in blue) and Acquarossa (in red). Table S1: Permutational multivariate analysis of variance using distance matrices. Results were reported for each factor inspected (epilithon type and sampling sites). Df, degree of freedom; Sum of Sq., sum of squares; R2, R squared value; F, value of F statistic; P-value, p-value corresponding to the given F statistic.
Author Contributions
Conceptualization, G.B., F.C., and R.F.; Data curation, S.C., A.V. and G.B.; Formal analysis, G.B.; Investigation, C.C., S.M., E.M., C.F., E.C., and D.F.; Methodology, G.B. and F.C.; Resources, C.C., E.M., C.F., G.B., F.C. and R.F.; Software, C.C.; Supervision, S.C., A.V., G.B., F.C. and R.F.; Validation, S.M.; Writing—original draft, C.C. and R.F.; Writing—review & editing, C.C., S.C., A.V. and R.F.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Harrison, A.P.; Cattani, I.; Turfa, J.M. Metallurgy, environmental pollution and the decline of Etruscan civilisation. Environ. Sci. Pollut. Res. Int. 2010, 17, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Hook, D. The composition and technology of selected Bronze Age and Early Iron Age Copper alloy artefacts from Italy. In Prehistoric Metal Artefacts from Italy (3500–720 BC) in the British Museum; Bietti Sestieri, A.M., Macnamara, E., Eds.; The British Museum: London, UK, 2007; pp. 308–323. [Google Scholar]
- Chiellini, C.; Miceli, E.; Bacci, G.; Fagorzi, C.; Coppini, E.; Fibbi, D.; Bianconi, G.; Mengoni, A.; Canganella, F.; Fani, R. Spatial structuring of bacterial communities in epilithic biofilms in the Acquarossa river (Italy). FEMS Microbiol. Ecol. 2018, 94, fiy181. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, M.C.; Fondi, M.; Maida, I.; Perrin, E.; Lo Giudice, A.; Michaud, L.; Mangano, S.; Bartolucci, G.; Romoli, R.; Fani, R. Sponge-associated microbial Antarctic communities exhibiting antimicrobial activity against Burkholderia cepacia complex bacteria. Biotechnol. Adv. 2012, 30, 272–293. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, M.C.; Romoli, R.; Bartolucci, G.; Maida, I.; Perrin, E.; Fondi, M.; Orlandini, V.; Mengoni, A.; Emiliani, G.; Tutino, M.L.; et al. Bioactive volatile organic compounds from Antarctic (sponges) bacteria. N. Biotechnol. 2013, 30, 824–838. [Google Scholar] [CrossRef] [PubMed]
- Orlandini, V.; Maida, I.; Fondi, M.; Perrin, E.; Papaleo, M.C.; Bosi, E.; de Pascale, D.; Tutino, M.L.; Michaud, L.; Lo Giudice, A.; et al. Genomic analysis of three sponge-associated Arthrobacter Antarctic strains, inhibiting the growth of Burkholderia cepacia complex bacteria by synthesizing volatile organic compounds. Microbiol. Res. 2014, 169, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Maida, I.; Fondi, M.; Papaleo, M.C.; Perrin, E.; Orlandini, V.; Emiliani, G.; de Pascale, D.; Parrilli, E.; Tutino, M.L.; Michaud, L.; et al. Phenotypic and genomic characterization of the Antarctic bacterium Gillisia sp. CAL575, a producer of antimicrobial compounds. Extremophiles 2014, 18, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Fondi, M.; Orlandini, V.; Maida, I.; Perrin, E.; Papaleo, M.C.; Emiliani, G.; de Pascale, D.; Parrilli, E.; Tutino, M.L.; Michaud, L.; et al. Draft genome sequence of the volatile organic compound-producing Antarctic bacterium Arthrobacter sp. strain TB23, able to inhibit cystic fibrosis pathogens belonging to the Burkholderia cepacia complex. J. Bacteriol. 2012, 194, 6334–6335. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Muhammad, S.A.; Ahmad, S.; Hameed, A. Report: Antibiotic production by thermophilic Bacillus specie SAT-4. Pak. J. Pharm. Sci. 2009, 22, 339–345. [Google Scholar]
- Dobretsov, S.; Abed, R.M.M.; Al Maskari, S.M.S.; Al Sabahi, J.N.; Victor, R. Cyanobacterial mats from hot springs produce antimicrobial compounds and quorum-sensing inhibitors under natural conditions. J. Appl. Phycol. 2011, 23, 983–993. [Google Scholar] [CrossRef]
- Ballav, S.; Kerkar, S.; Thomas, S.; Augustine, N. Halophilic and halotolerant actinomycetes from a marine saltern of Goa, India producing anti-bacterial metabolites. J. Biosci. Bioeng. 2015, 119, 323–330. [Google Scholar] [CrossRef]
- Staccioli, R.A. Considerazioni sui complessi monumentali di Murlo e di Acquarossa. In L’Italie Préromaine Et La Rome Républicaine; École Française de Rome: Rome, Italy, 1976; Volume 27, pp. 961–972. [Google Scholar]
- Meyers, G.E. Etrusco-Italic Monumental Architectural Space from the Iron Age to the Archaic Period: An Examination of Approach and Access. Ph.D. Thesis, Faculty of the Graduate School of the University of Texas at Austin, The University of Texas at Austin, Austin, TX, USA, December 2003. [Google Scholar]
- Meyers, G.E. Approaching monumental architecture: Mechanics and movement in Archaic Etruscan palaces. Pap. Br. Sch. Rome 2013, 81, 39–66. [Google Scholar] [CrossRef]
- Chiellini, C.; Lombardo, K.; Mocali, S.; Miceli, E.; Fani, R. Pseudomonas strains isolated from different environmental niches exhibit different antagonistic ability. Ethol. Ecol. Evol. 2019. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Illumina Support Center. Available online: https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 29 July 2019).
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Bacci, G.; Bazzicalupo, M.; Benedetti, A.; Mengoni, A. StreamingTrim 1.0: A Java software for dynamic trimming of 16S rRNA sequence data from metagenetic studies. Mol. Ecol. Resour. 2014, 14, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Beckers, B.; Op De Beeck, M.; Thijs, S.; Truyens, S.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Performance of 16s rDNA primer pairs in the study of rhizosphere and endosphere bacterial microbiomes in metabarcoding studies. Front. Microbiol. 2016, 7, 650. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Di Cello, F.; Fani, R. A molecular strategy for the study of natural bacterial communities by PCR-based techniques. Minerva Biotecnol. 1996, 8, 126–134. [Google Scholar]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.G.K.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef] [PubMed]
- Mori, E.; Liò, P.; Daly, S.; Damiani, G.; Perito, B.; Fani, R. Molecular nature of RAPD markers from Haemophilus influenzae Rd genome. Res. Microbiol. 1999, 150, 83–93. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Jorgensen, J.H. Antimicrobial susceptibility testing of bacteria that grow aerobically. Infect. Dis. Clin. N. Am. 1993, 7, 393–409. [Google Scholar]
- Mengoni, A.; Maida, I.; Chiellini, C.; Emiliani, G.; Mocali, S.; Fabiani, A.; Fondi, M.; Firenzuoli, F.; Fani, R. Antibiotic resistance differentiates Echinacea purpurea endophytic bacterial communities with respect to plant organs. Res. Microbiol. 2014, 165, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Stoodley, P. Developmental regulation of microbial biofilms. Curr. Opin. Biotechnol. 2002, 13, 228–233. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).