Abstract
We herein report on the synthesis of a pentacyclic triterpene functionalized through derivation of betulinic acid with hydroxybenzotriazole. The compound was fully characterized by proton (1H-NMR), carbon-13 (13C-NMR), heteronuclear single quantum coherence (HSQC) and distortionless enhancement by polarization transfer (DEPT-135 and DEPT-90) nuclear magnetic resonance. Ultraviolet (UV), and Fourier-transform infrared (FTIR) spectroscopies as well as and high-resolution mass spectrometry (HRMS) were also adopted. Computational studies were conducted to foresee the interactions between compound 3 and phosphodiesterase 9, a relevant target in the field of neurodegenerative diseases. Additionally, preliminary calculation of physico-chemical descriptors was performed to evaluate the drug-likeness of compound 3.
1. Introduction
Dementia is a syndrome that has become a public health problem, affecting approximately 47 million people [1], and it is the result of a combination of diseases, such as Alzheimer’s disease (AD) and vascular dementia (VaD) [1,2]. The symptoms that characterize this condition are losses of memory, attention, speech ability, personal identity, and a progressive cognitive decline [3].
Given the high incidence of AD, many studies have been carried out over the years, and they point out abnormal amyloid β neuritic plaques, neurofibrillary tangles containing tau protein, and degeneration of cholinergic neurons at the cerebral level [3,4]. According to the described evidence, different drugs (e.g., galantamine, rivastigmine, and donepezil) have been designed to address several targets, such as acetylcholinesterase, which hydrolyzes acetylcholine (ACh) and contributes to preventing the loss of cholinergic neurons [5]. VaD, the second most common form of senile dementia [2], is closely associated with ischemic strokes. Indeed, 25–30% of survivors develop immediate or delayed vascular dementia, which nowadays does not have a specific real therapy [6]. This effect is also because pathophysiologic mechanisms of VaD are knotty and involve alteration of many homeostatic processes coordinated by many signaling pathways. One of the most important is the cyclic nucleotide signal system constituted by cyclic adenosine 3′,5′-monophosphate (cAMP) and cyclic guanosine 3′,5′-monophosphate (cGMP) [1,7,8]. This system is involved in cell proliferation and differentiation, gene expression, and inflammation processes [9].
cAMP and cGMP are synthesized by adenylyl cyclase or guanylyl cyclase, and transduce the signal to several cellular effectors, such as neurotransmitters and cyclic nucleotide phosphodiesterases (PDEs) families [6]. PDEs hydrolyze these second messengers with different degrees of selectivity according to the family in which they belong [10] to obtain 5′-AMP and 5′-GMP, contributing to cellular regulation [8,11]. There are eleven PDEs (PDE1 to 11); among them PDE5, PDE6, and PDE9 are cGMP-selective, while PDE4, PDE7, and PDE8 are cAMP-selective. Further, some PDE families have double selectivities (PDE1, PDE2, PDE3, PDE10, and PDE11) [12]. From a structural point of view, these eleven families share a conserved catalytic domain (C domain) and a sequence identity of 75% [13]; however, the amino acid sequence can differentiate enzymes belonging to different families in the outer zone of the protein. These structural differences are also related to the functionality and affinity of one substrate over another [8,11]. Given their relevance in modulating the cellular signal transduction systems, PDEs have become the target for designing small molecules capable of inhibiting their action and leading to neuroprotective effects.
In this work, we focused on PDE9, a studied target to counteract diabetes mellitus, obesity, cardiovascular diseases, and dementia, since this enzyme is highly expressed in the brain of patients affected by AD [10,14,15]. As with other PDEs, PDE9 is constituted by a catalytic domain (C domain) formed by 16 helices (residue 181-506) and associated as dimers. Of note, two important residues (Gln453 and Phe456) for ligand-target interaction are present and conserved among PDEs families [13,16]. Moreover, metal ions, such as zinc and magnesium, are present in PDE9 and have a role in activating the enzyme. The magnesium ion is reported to be coordinated by Asp293 and five molecules of water, while the zinc ion is noted to be coordinated by His256, His292, Asp293, Asp402, and two molecules of water [13,17].
Many studies have reported how treatment with inhibitors of PDE9 could lead to neuroprotective effects [13,18] through treatments with small molecules that can be of natural or synthetic origin [6,10]. Moreover, the designed synthetic small molecule PF-04447943, entered Phase II trials as a drug for treating AD since it was found to cross the blood-brain barrier (BBB) and reach the central nervous system (CNS) [19,20]. This way, through a preclinical evaluation of the cerebrospinal fluid, it was reported that PF-04447943 could improve cGMP concentration and lead to beneficial effects [21].
Pentacyclic triterpenoids are a class of pharmacologically active and structurally rich natural products with privileged motifs for further modifications [22]. The structure of these compounds contains substituents, such as isopropenyl moiety and hydroxyl and/or carboxyl groups, that can be easily functionalized [23]. Betulinic acid (BA, Figure 1), a lupane-type pentacyclic triterpenoid, is widespread in many plants, including Betula cordifolia [24]. BA and its derivatives demonstrate multiple bioactivities, particularly an antitumor effect and antimicrobial, anti-inflammatory, anti-HIV, antimalarial, and anthelmintic activities (Figure 1) [25,26,27]. More specifically, in the context of identifying novel anti-dementia agents, BA has neuroprotective properties and was reported to reduce toxic amyloid β oligomers [6,26,28].
In this work, we report the preparation of compound 3 (Figure 1) since, based on previous reports demonstrating the activity of BA on PDEs [6,29], this derivatization of BA could represent a good starting point for obtaining a molecule aimed at PDE9 inhibition.
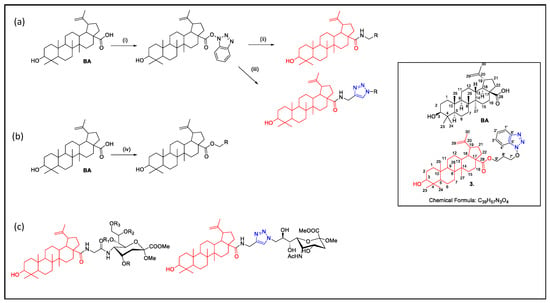
Figure 1.
Synthesis of betulinic acid (BA) derivatives: (a) (i) TBTU, DIPEA, THF (ii) amine, K2CO3, DMF, 1 h; (iii) CuSO4, Na ascorbate, THF- water and (b) (iv) halogen derivatives, DMF, K2CO3; (c) Example of betulinic acid derivatives [30,31]); Square: structure of BA and compound 3 reported in this paper.
2. Results and Discussion
The synthetic route of the BA derivative, 3-[(1H-benzo[d][1,2,3]triazol-1-yl)oxy]propyl 9-hydroxy-5a,5b,8,8,11a-pentamethyl-1-(prop-1-en-2-yl)icosahydro-3aH-cyclopenta[a]chrysene-3a-carboxylate (3), started from 1H-benzo[d][1,2,3]triazol-1-ol (HOBt, 1) (Scheme 1).
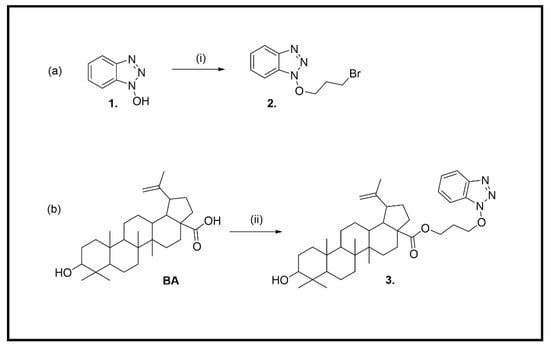
Scheme 1.
(a) Synthesis of 1-(3-bromopropoxy)-1H-benzo[d][1,2,3]triazole (2) (i) BrCH2CH2CH2Br, K2CO3, DMF (b) Synthesis of 3-[(1H-benzo[d][1,2,3]triazol-1-yl)oxy]propyl 9-hydroxy-5a,5b,8,8,11a-pentamethyl-1-(prop-1-en-2-yl)icosahydro-3aH-cyclopenta[a]chrysene-3a-carboxylate (3); (ii) 2, K2CO3, DMF.
1-(3-Bromopropoxy)-1H-benzo[d][1,2,3]triazole (2) was prepared by the reaction of 1 with 1,3-dibromopropane (2 eq) in anhydrous DMF at 65 °C for 4 h (Scheme 1a) [32]. The crude product was purified by column chromatography to afford 2 in 70% yield. The structure of 2 was confirmed by 1H and 13C NMR spectra (Supplementary material, Figures S3 and S4).
Finally, the hybrid compound 3 was obtained using a protocol reported recently by Zhang et al. [33]. A mixture of BA (1 g, 0.002 mol, 1 eq) and potassium carbonate (0.33 g, 0.002 mol, 1.2 eq) in DMF (20 mL) was stirred at 85 °C. Next, 2 was added to this mixture at 85 °C, and the reaction was stirred for 4 h and then left overnight at room temperature. The mixture was washed twice with water. Combined organic layers were dried over Na2SO4 and concentrated under reduced pressure to obtain the crude product. The residue was purified by column chromatography (dichloromethane) in a high yield (68%).
The structure of 3 was determined by 1H and 13C-NMR spectroscopy (Supplementary material, Figures S5 and S6). The 1H-NMR spectrum of 3 showed peaks above 7 ppm corresponding to aromatic protons of the benzotriazole ring and methylene groups (7′–9′) corresponding to the three chain groups at 4.66, 4.33, and 2.26 ppm.
For the 13C-NMR signals, the shift of peaks of the acid group (179.19 ppm) to an ester (175.96 ppm), the appearance of six C-13 aromatic signals at 108–144 ppm, and the C-7′–9′ methylene signal at 70.5, 59.8 and 27.8 ppm were considered as relevant to assess product formation.
Heteronuclear single quantum coherence spectroscopy (HSQC) and distortionless enhancement by polarization transfer (DEPT-135 and DEPT-90) were also used to assign 13C signals of compound 3, as shown in Table S1 (see Supplementary material for 2D spectra, Figures S7–S9). The 13C-NMR spectrum of 3 exhibited 39 carbon signals, classified by DEPT experiments as six methyl groups, thirteen methylenes, six methines, one olefinic methylene, four aromatic, and nine quaternary carbons.
HRMS of 3 was also obtained for further characterization, validating the proposed structure determined by NMR spectra (Supplementary material, Figure S10).
The IR spectrum of compound 3 showed characteristic OH stretching at 3549 cm−1, C-H stretching at 2939 and 2870 cm−1, and C=O stretching at 1689 cm−1 (Supplementary material, Figure S11). Other vibrational peaks at 1450 and 1373 cm−1 corresponded to the methyl group’s C-H bending.
The UV spectrum of compound 3 showed an absorption peak at 257 nm and another lower absorption peak at 285 nm (n→π transition) (Supplementary material, Figure S12).
The interaction of compound 3 with PDE9 was evaluated through computational investigation. Firstly, molecular modeling studies were done. X-ray structure, composed of crystallized PDE9 enzyme and co-crystallized PF-04447943 ligand, were retrieved from RCSB Protein Data Bank (PDB ID: 4E90) [21]. Once that site-specific molecular docking was performed, the obtained PF-04447943 docked pose (−9.7 kcal/mol) was compared to the co-crystallized one to confirm the method’s reliability. Root-mean-square deviation (RMSD) parameter was then calculated (2.271 Å).
Then, the same molecular docking protocol was used to obtain the best-docked pose of compound 3 (−9.1 kcal/mol, Figure 2). According to the predicted model, the bicyclic aromatic systems of compound 3 and PF-04447943 colocalized within the binding pocket of PDE9 (Figures S13 and S14 in the Supplementary material).
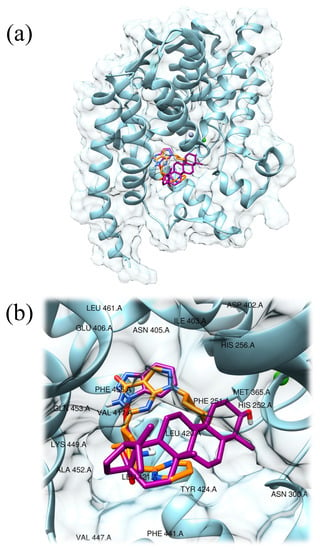
Figure 2.
Predicted interaction motif for compound 3 (magenta) with PDE9 and best-docked pose of PF-04447943 (−9.7 kcal/mol; orange) within PDE9 (light blue, PDB ID: 4E90) (a); detailed view of compound 3 (magenta) in the active site of PDE9 and comparison with PF-04447943 (orange), the residues present in a zone of 5 Å radius from the center of the ligand have been labeled (b).
Once both docked poses of PF-04447943 and compound 3 ligands were obtained, protein-compound interaction profiler tool (PLIP) [34] was used to evaluate and compare established interactions between PDE9 and ligands.
Since, in literature, it is reported that Gln453 and Phe456 residues are important in the catalytic site for achieving inhibitory effects of the PDE9 enzyme, interactions with these residues were evaluated [13,21].
Compound 3 displayed hydrophobic interactions, π–π stacking interactions, and hydrogen bonds. Interestingly the ligand established twelve hydrophobic interactions with PDE9 at the level of Asn300 (3.42 Å), Met365 (3.66 Å), Ile403 (3.63 Å), Tyr424 (3.76 Å and 3.98 Å), Phe441 (3.65, 3.72, and 3.83 Å), Phe456 (3.12, 3.69, and 3.88 Å), and Val460 (3.66 Å).
One further π–π stacking interaction was found at the level of Phe251 (center distance = 5.24 Å). Again, one hydrogen bond was established with residue Gln453 (2.91 Å) (Table S2 in the Supplementary material). Together these interactions suggest that compound 3 could be a considerable ligand candidate for PDE9 enzymes since relevant interactions with residues Gln453 and Phe456 are established (Figure S15 in the Supplementary material).
Then, absorption, distribution, metabolism, and excretion characteristics were checked for compound 3 (see Materials and Methods section). Firstly, compound 3 was evaluated for its drug-likeness properties through SwissADME [34]. Under the Lipinski’s rule, the molecule can make at most one violation to be considered as drug-like. The parameters are the following: (1) an octanol-water partition coefficient logP ≤ 5, (2) molecular weight ≤ 500 g/mol, (3) hydrogen bond donors ≤ 5, and (4) hydrogen bond acceptors ≤ 10 [35]. According to SwissADME data, compound 3 has two violations since it has (1) an octanol-water partition coefficient logP = 5.83, (2) molecular weight = 631.89 g/mol, (3) hydrogen bond donors = 1, and (4) hydrogen bond acceptors = 6 (Table S3 in Supplementary material). As expected, according to these criteria, prediction of pharmacokinetic properties reports that compound 3 can be passively absorbed in a limited manner through the gastrointestinal tract and the BBB [36]. Pajouhesh and Lenz [37] efficiently determined the chemical and structural features of a successful CNS drug candidate. The conditions that should be respected are: (1) the molecular weight ≤ 400 g/mol, (2) the number of hydrogen bond donors ≤ 7, (3) the number of hydrogen bond acceptors ≤ 3, and (4) the topological polar surface area should be ≤ 140 or ideally ≤ 60 Å2. Consistently, compound 3 satisfies these requirements except for the molecular weight.
Thus, since the localization of the target is found at the level of CNS and requires the crossing of the BBB, optimization of this small molecule is needed to reach the target and obtain beneficial effects.
3. Materials and Methods
3.1. Chemistry
Silica gel (FCP 230–400 mesh) was used for column chromatography. Thin-layer chromatography was carried out on E. Merck pre-coated silica gel 60 F254 plates and visualized with phosphomolybdic acid, iodine, or a UV-visible lamp.
All chemicals were purchased from Bide Pharmatech., Ltd. (Shanghai, China) and J & K scientific (Hong Kong, China). 1H-NMR and 13C-NMR spectra were collected in CDCl3 at 25 °C on a Bruker Ascend®-600 NMR spectrometer (600 MHz for 1H and 150 MHz for 13C). All chemical shifts were reported in the standard δ notation of parts per million using the peak of residual proton signals of CDCl3 or DMSO-d6 as an internal reference (CDCl3, δC 77.2, δH 7.26; DMSO-d6, δC 39.5, δH 2.50). High-resolution mass spectra (HRMS) were measured using electrospray ionization (ESI). The measurements were done in a positive ion mode (interface capillary voltage 4500 V); the mass ratio was from m/z 50 to 3000 Da; external/internal calibration was done with Electrospray Calibration Solution.
HRMS analyses were performed by an Agilent 6230 electrospray ionization (ESI) time-of-flight (TOF) mass spectrometer with an Agilent C18 column (4.6 mm × 150 mm, 3.5 μm). The mobile phase was isocratic (water + 0.01% TFA; CH3CN) at a flow rate of 0.35 mL/min. The peaks were determined at 254 nm under UV.
UV analysis was performed by a Shimadzu UV—2600 with 1 cm quartz cell and a slit width of 2.0 nm. The analysis was carried out using wavelengths in the range of 200–400 nm.
3.1.1. Synthesis of 1-(3-Bromopropoxy)-1H-benzo[d][1,2,3]triazole (2)
1H-Benzo[d][1,2,3]triazol-1-ol (1.8 g, 13 mmol) was dissolved in 10 mL of anhydrous DMF. Anhydrous K2CO3 (2.7 g, 19.5 mmol) was then added to the solution, and the mixture was stirred at 30 °C for 30 min. 1,3 dibromo propane (2.2 mL, 19.5 mmol) was added slowly to the reaction mixture and subsequently stirred at 30 °C for 6 h, upon which TLC indicated completion of the reaction. The reaction mixture was then diluted with 50 mL of water and extracted with ethyl acetate (3 × 50 mL). These extracts were combined, washed with water (2 × 50 mL), dried over anhydrous Na2SO4, and evaporated in a vaccuum to yield the product residue that was then recrystallized from CH2Cl2/hexane 1:1 mixture to yield the compound 2 in 80% yield (2.6 g). δH (600 MHz, CDCl3) 2.43–2.44 (2H, m, CH2), 3.73–3.74 (2H, m, CH2-Br), 4.72–4.73 (2H, m, CH2-O), 7.42 (1H, t, J = 8.04 Hz, HAr), 7.52 (1H, t, J = 7.3 Hz, HAr), 7.62 (1H, d, J = 7.3 Hz, HAr), 8.02 (1H, d, J = 7.3 Hz, HAr); δC (150 MHz, CDCl3) 28.7, 30.8, 77.8, 108.5, 120.2, 124.8, 128.7, 143.7.
3.1.2. Synthesis of 3-[(1H-Benzo[d][1,2,3]triazol-1-yl)oxy]propyl 9-hydroxy-5a,5b,8,8,11a-pentamethyl-1-(prop-1-en-2-yl)icosahydro-3aH-cyclopenta[a]chrysene-3a-carboxylate (3)
Betulinic acid (BA) in DMF (0.76 g, 1.6 × 10−3 mol, 5 mL) was treated with K2CO3 (0.33 g, 0.002 mol, 1.2 eq) and 2 eq of 2 (0.82 g, 3.2 × 10−3 mol). After stirring for 24 h at room temperature, deionized water (15 mL) was added, and the mixture was extracted with CH2Cl2. The combined organic layers were washed with brine, dried using anhydrous Na2SO4, filtered, and evaporated in a vacuum until dry. The residue was further purified by flash column chromatography on silica gel (CH2Cl2 and MeOH) to produce an oil sample Yield 75% (0.94 g). δH (600 MHz, CDCl3) 0.66–0.67 (1H, m, H-5), 0.75 (3H, s, Me), 0.79 (3H, s, Me), 0.86 (3H, s, Me), 0.96 (6H, s, Me), 1.03 (1H, dd, H-12), 1.14–1.16 (1H, m, H-15), 1.18–1.21 (1H, m), 1.25–1.27 (1H, m, H-9), 1.30–1.33 (2H, m, H-7), 1.35–1.37 (6H, m), 1.49–1.65 (11H, m), 1.84–1.91 (2H, m, H-21), 2.20 (1H, dd, H-13), 2.22–2.26 (2H, m, H-8′), 3.02–3.04 (1H, m, H-19), 3.18 (H, dd, J = 11.5 and 4.65 Hz, H-3), 4.35–4.43 (1H, m, H-7′), 4.60 (1H, s, H-30a), 4.66 (1H, t, H-9′), 4.72 (1H, br, H-30b), 7.39 (1H, t, J = 7.7 Hz, Har), 7.51 (1H, t, J = 7.1 Hz, Har), 7.59 (1H, d, J = 8.3 Hz, Har), 8.02 (1H, d, J = 8.4 Hz, Har); δC (150 MHz, CDCl3) 14.6 (C-27), 15.3 (C-24), 15.9 (C-25), 16.1 (C-26), 18.2 (C-6), 19.4 (C-29), 20.8 (C-11), 25.5, 27.4, 27.8, 27.9, 29.7, 30.6, 32.1, 34.3, 37, 37.1, 38.3, 38.7, 38.8, 40.7, 42.4, 46.9, 49.4, 50.5 (C-9), 55.3 (C-5), 56.6 (C-17), 59.8 (C-7′), 77.4 (C-9′), 78.9 (C-3), 108.5 (C-4′), 109.7 (C-30), 120.3 (C-1′), 124.7 (C-3′), 127.2 (C-5′), 128.1 (C-2′), 143.5 (C-6′), 150.3 (C-20), 175.9 (CO); [α]D20 = −50 (c = 0.1, MeOH); HRMS-ESI m/z 632.4408 [M + H]+ (calcd. for C39H57N3O4, m/z 632.4422), 654.4244 [M + Na]+ (calcd. for C39H57N3O4, m/z 654.4241), UV (CH2Cl2) peaks 257 (logε = 1.68) and 285 nm (logε = 1.20), IR (KBr) 3549, 2939, 2870, 1689, 1450, 1373 cm−1.
3.2. Computational Studies
The 3D X-ray crystal structure of human PDE9 complexed with PF-04447943 (7RG) was retrieved from the RCSB Protein Data Bank (www.rcsb.org; accessed on 29 June 2022; PDB ID 4E90, resolution 2.50 Å) [21].
Before site-specific docking studies, PDE9 was prepared, and chains with zinc and magnesium residues were isolated and considered. Further, the obtained structure was processed through the DockPrep tool of Chimera to fix potentially missing residues in the structure [38,39]. A zone within 5 Å centered on PF-04447943 ligand was selected to identify the binding site region.
To perform a site-specific molecular docking study, Chimera software based on AutoDock Vina was used [38,40]. From this point, a grid-enclosing box was centered on the identified site and a receptor search volume space was created with the following coordinates and dimensions: x = 80.4359, y = 57.1372, z = 42.2024; size: 37.0000 × 34.0000 × 44.0000 Å. The number of generated docking poses was set to 10, and the docking energy conformation value was expressed in −kcal/mol. The best scoring pose was selected for further analysis. Residue numbering used in the PDB file was adopted.
Further, compound 3 was docked into the previously defined binding site of the PDE9 enzyme, and the best pose was selected for further studies. To validate the adopted site-specific molecular docking protocol, the co-crystallized PF-04447943 compound was redocked, and the root-mean-square deviation (RMSD) value was calculated using the Chimera rmsd command [38]. UCSF Chimera molecular viewer [38] and PyMol software (The PyMOL Molecular Graphics System, Version 2.3.5, Schrödinger, LLC.) were used to produce the artworks. ADME properties for compound 3 were retrieved using the SwissADME tool [36]. To evaluate interaction pattern data PLIP tool was used [34].
4. Conclusions
Nowadays, the identification of small molecules able to treat neurodegeneration is one of the most challenging issues faced by drug discovery. Our work attempted to generate a small molecule able to bind PDE9 enzyme with the aim of inhibiting it. This was performed through site-specific molecular docking studies that displayed a good binding energy score. Then, ligand-target interactions were evaluated and demonstrated that crucial residues were involved. Further, physical-chemical properties were checked, and results suggested that, since PDE9 enzyme is located at the level of the CNS, optimization for compound 3 should be considered as a potential anti-neurodegeneration drug.
Supplementary Materials
The following are available online, NMR spectrometry, Mass spectrometry, IR spectrometry, UV-VIS spectrometry, Computational studies of the compounds mentioned in the article are shown in the supplementary materials [41,42].
Author Contributions
Conceptualization, P.C.; methodology, G.R. and P.C.; investigation, K.L., M.A., A.J. and J.L.; data curation, A.J.; writing—original draft preparation, K.L., M.A. and A.J.; writing—review and editing, A.G., G.R. and P.C.; supervision, G.R. and P.C.; project administration, G.R. and P.C.; funding acquisition, G.R. and P.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by FDCT grants from Macao Science and Technology University to PC (Project Code: FRG-22-077-SP) and by University of Brescia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Materials.
Acknowledgments
We would like thank Yuhan Xie for support during UV studies.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Arvanitakis, Z.; Shah, R.C.; Bennett, D.A. Diagnosis and Management of Dementia: A Review. J. Am. Med. Assoc. 2019, 322, 1589–1599. [Google Scholar] [CrossRef]
- Kalaria, R.N. Neuropathological Diagnosis of Vascular Cognitive Impairment and Vascular Dementia with Implications for Alzheimer’s Disease. Acta Neuropathol. 2016, 131, 659–685. [Google Scholar] [CrossRef]
- Dooley, M.; Lamb, H.M. Donepezil: A Review of Its Use in Alzheimer??S Disease. Drugs Aging 2000, 16, 199–226. [Google Scholar] [CrossRef]
- Hane, F.T.; Robinson, M.; Lee, B.Y.; Bai, O.; Leonenko, Z.; Albert, M.S. Recent Progress in Alzheimer’s Disease Research, Part 3: Diagnosis and Treatment. J. Alzheimer’s Dis. 2017, 57, 645–665. [Google Scholar] [CrossRef] [PubMed]
- Silman, I.; Sussman, J.L. Acetylcholinesterase: ‘Classical’ and ‘Non-Classical’ Functions and Pharmacology. Curr. Opin. Pharmacol. 2005, 5, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Kaundal, M.; Zameer, S.; Najmi, A.K.; Parvez, S.; Akhtar, M. Betulinic Acid, a Natural PDE Inhibitor Restores Hippocampal CAMP/CGMP and BDNF, Improve Cerebral Blood Flow and Recover Memory Deficits in Permanent BCCAO Induced Vascular Dementia in Rats. Eur. J. Pharmacol. 2018, 832, 56–66. [Google Scholar] [CrossRef]
- De La Monte, S.M.; Sohn, Y.K.; Etienne, D.; Kraft, J.; Wands, J.R. Role of Aberrant Nitric Oxide Synthase-3 Expression in Cerebrovascular Degeneration and Vascular-Mediated Injury in Alzheimer’s Disease. Ann. N. Y. Acad. Sci. 2000, 903, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Maurice, D.H.; Ke, H.; Ahmad, F.; Wang, Y.; Chung, J.; Manganiello, V.C. Advances in Targeting Cyclic Nucleotide Phosphodiesterases. Nat. Rev. Drug Discov. 2014, 13, 290–314. [Google Scholar] [CrossRef]
- Conti, M.; Beavo, J. Biochemistry and Physiology of Cyclic Nucleotide Phosphodiesterases: Essential Components in Cyclic Nucleotide Signaling. Annu. Rev. Biochem. 2007, 76, 481–511. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, D.; Mudedla, S.; Jang, S.; Kim, H.; Park, H.; Choi, Y.; Oh, J.; Wu, S. Computational Study on Selective PDE9 Inhibitors on PDE9-Mg/Mg, PDE9-Zn/Mg, and PDE9-Zn/Zn Systems. Biomolecules 2021, 11, 709. [Google Scholar] [CrossRef]
- Francis, S.H.; Blount, M.A.; Corbin, J.D. Mammalian Cyclic Nucleotide Phosphodiesterases: Molecular Mechanisms and Physiological Functions. Physiol. Rev. 2011, 91, 651–690. [Google Scholar] [CrossRef]
- Bender, A.T.; Beavo, J.A. Cyclic Nucleotide Phosphodiesterases: Molecular Regulation to Clinical Use. Pharmacol. Rev. 2006, 58, 488–520. [Google Scholar] [CrossRef] [PubMed]
- Ribaudo, G.; Memo, M.; Gianoncelli, A. A Perspective on Natural and Nature-Inspired Small Molecules Targeting Phosphodiesterase 9 (PDE9): Chances and Challenges against Neurodegeneration. Pharmaceuticals 2021, 14, 58. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sharma, V.; Singh, V.P.; Kaundal, M.; Gupta, M.K.; Bariwal, J.; Deshmukh, R. Herbs to Curb Cyclic Nucleotide Phosphodiesterase and Their Potential Role in Alzheimer’s Disease. Mech. Ageing Dev. 2015, 149, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Ribaudo, G.; Ongaro, A.; Zagotto, G.; Memo, M.; Gianoncelli, A. Therapeutic Potential of Phosphodiesterase Inhibitors against Neurodegeneration: The Perspective of the Medicinal Chemist. ACS Chem. Neurosci. 2020, 11, 1726–1739. [Google Scholar] [CrossRef] [PubMed]
- Cameron, R.T.; Coleman, R.G.; Day, J.P.; Yalla, K.C.; Houslay, M.D.; Adams, D.R.; Shoichet, B.K.; Baillie, G.S. Chemical Informatics Uncovers a New Role for Moexipril as a Novel Inhibitor of CAMP Phosphodiesterase-4 (PDE4). Biochem. Pharmacol. 2013, 85, 1297–1305. [Google Scholar] [CrossRef][Green Version]
- Huai, Q.; Wang, H.; Zhang, W.; Colman, R.W.; Robinson, H.; Ke, H. Crystal Structure of Phosphodiesterase 9 Shows Orientation Variation of Inhibitor 3-Isobutyl-1-Methylxanthine Binding. Proc. Natl. Acad. Sci. USA 2004, 101, 9624–9629. [Google Scholar] [CrossRef]
- Richards, D.A.; Aronovitz, M.J.; Liu, P.; Martin, G.L.; Tam, K.; Pande, S.; Karas, R.H.; Bloomfield, D.M.; Mendelsohn, M.E.; Blanton, R.M. CRD-733, a Novel PDE9 (Phosphodiesterase 9) Inhibitor, Reverses Pressure Overload–Induced Heart Failure. Circ. Heart Fail. 2021, 14, e007300. [Google Scholar] [CrossRef]
- Schwam, E.; Nicholas, T.; Chew, R.; Billing, C.; Davidson, W.; Ambrose, D.; Altstiel, L. A Multicenter, Double-Blind, Placebo-Controlled Trial of the PDE9A Inhibitor, PF-04447943, in Alzheimer’s Disease. Curr. Alzheimer Res. 2014, 11, 413–421. [Google Scholar] [CrossRef]
- Vardigan, J.D.; Converso, A.; Hutson, P.H.; Uslaner, J.M. The Selective Phosphodiesterase 9 (PDE9) Inhibitor PF-04447943 Attenuates a Scopolamine-Induced Deficit in a Novel Rodent Attention Task. J. Neurogenet. 2011, 25, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Verhoest, P.R.; Fonseca, K.R.; Hou, X.; Proulx-LaFrance, C.; Corman, M.; Helal, C.J.; Claffey, M.M.; Tuttle, J.B.; Coffman, K.J.; Liu, S.; et al. Design and Discovery of 6-[(3S,4S)-4-Methyl-1-(Pyrimidin-2-Ylmethyl)Pyrrolidin-3-Yl]-1-(Tetrahydro-2H-Pyran-4-Yl)-1,5-Dihydro-4H-Pyrazolo [3,4-d]Pyrimidin-4-One (PF-04447943), a Selective Brain Penetrant PDE9A Inhibitor for the Treatment of Cognitive Disorders. J. Med. Chem. 2012, 55, 9045–9054. [Google Scholar] [CrossRef] [PubMed]
- Kvasnica, M.; Urban, M.; Dickinson, N.J.; Sarek, J. Pentacyclic Triterpenoids with Nitrogen- and Sulfur-Containing Heterocycles: Synthesis and Medicinal Significance. Nat. Prod. Rep. 2015, 32, 1303–1330. [Google Scholar] [CrossRef] [PubMed]
- Chrobak, E.; Jastrzębska, M.; Bębenek, E.; Kadela-Tomanek, M.; Marciniec, K.; Latocha, M.; Wrzalik, R.; Kusz, J.; Boryczka, S. Molecular Structure, In Vitro Anticancer Study and Molecular Docking of New Phosphate Derivatives of Betulin. Molecules 2021, 26, 737. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.M.; Bentley, M.D.; Campbell, C.S.; Cole, B.J.W. Betulin and Lupeol in Bark from Four White-Barked Birches. Phytochemistry 1988, 27, 2175–2176. [Google Scholar] [CrossRef]
- Zhang, D.-M.; Xu, H.-G.; Wang, L.; Li, Y.-J.; Sun, P.-H.; Wu, X.-M.; Wang, G.-J.; Chen, W.-M.; Ye, W.-C. Betulinic Acid and Its Derivatives as Potential Antitumor Agents: Betulinic Acid and Its Derivatives with Antitumor Activities. Med. Res. Rev. 2015, 35, 1127–1155. [Google Scholar] [CrossRef] [PubMed]
- Yogeeswari, P.; Sriram, D. Betulinic Acid and Its Derivatives: A Review on Their Biological Properties. Curr. Med. Chem. 2005, 12, 657–666. [Google Scholar] [CrossRef]
- Mukherjee, R.; Kumar, V.; Srivastava, S.K.; Agarwal, S.K.; Burman, A.C. Betulinic Acid Derivatives as Anticancer Agents: Structure Activity Relationship. Anti-Cancer Agents Med. Chem. 2006, 6, 271–279. [Google Scholar] [CrossRef]
- Planchard, M.S.; Samel, M.A.; Kumar, A.; Rangachari, V. The Natural Product Betulinic Acid Rapidly Promotes Amyloid-β Fibril Formation at the Expense of Soluble Oligomers. ACS Chem. Neurosci. 2012, 3, 900–908. [Google Scholar] [CrossRef]
- Kim, J.; Lee, Y.S.; Kim, C.-S.; Kim, J.S. Betulinic Acid Has an Inhibitory Effect on Pancreatic Lipase and Induces Adipocyte Lipolysis: Antiobesity Effect of Betulinic Acid. Phytother. Res. 2012, 26, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Si, L.-L.; Shi, Y.-Y.; Fan, Z.-B.; Wang, S.-X.; Tian, Z.-Y.; Li, M.; Sun, J.-Q.; Jiao, P.-X.; Ran, F.-X.; et al. Synthesis and In Vitro Anti-Influenza Virus Evaluation of Novel Sialic Acid (C-5 and C-9)-Pentacyclic Triterpene Derivatives. Molecules 2017, 22, 1018. [Google Scholar] [CrossRef]
- Han, X.; Shi, Y.; Si, L.; Fan, Z.; Wang, H.; Xu, R.; Jiao, P.; Meng, K.; Tian, Z.; Zhou, X.; et al. Design, Synthesis and Biological Activity Evaluation of Novel Conjugated Sialic Acid and Pentacyclic Triterpene Derivatives as Anti-Influenza Entry Inhibitors. MedChemComm 2016, 7, 1932–1945. [Google Scholar] [CrossRef]
- Huang, L.; Shi, A.; He, F.; Li, X. Synthesis, Biological Evaluation, and Molecular Modeling of Berberine Derivatives as Potent Acetylcholinesterase Inhibitors. Bioorg. Med. Chem. 2010, 18, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ye, S.; Wang, Y.; Wang, C.; Zhu, Y.; Wu, Y.; Zhang, Y.; Zhang, H.; Miao, Z. Discovery and Optimization of Betulinic Acid Derivatives as Novel Potent CD73 Inhibitors. Bioorg. Med. Chem. 2022, 59, 116672. [Google Scholar] [CrossRef] [PubMed]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the Scope of the Protein–Ligand Interaction Profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Pajouhesh, H.; Lenz, G.R. Medicinal Chemical Properties of Successful Central Nervous System Drugs. NeuroRx 2005, 2, 541–553. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera? A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Fai, L.K.; Anyanwu, M.; Ai, J.; Xie, Y.; Gianoncelli, A.; Ribaudo, G.; Coghi, P. 4-(4-(((1H-Benzo[d][1,2,3]triazol-1-Yl)oxy)methyl)-1H-1,2,3-triazol-1-Yl)-7-chloroquinoline. Molbank 2022, 2022, M1404. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Chirchir, K.D.; Cheplogoi, K.P.; Omolo, O.J.; Langat, K.M. Chemical Constituents of Solanum mauense (Solanaceae) and Dovyalis abyssinica (Salicaceae). Int. J. Biol. Chem. Sci 2018, 12, 999. [Google Scholar] [CrossRef]
- Noviany; Osman, H. Structure Elucidation of Betulinic Acid from Sesbania Grandiflora Root. J. Phys. Conf. Ser. 2021, 1751, 012090. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).