3,5-Bis((E)-4-methoxybenzylidene)-1-(2-morpholinoethyl)piperidin-4-one
Abstract
1. Introduction
2. Results
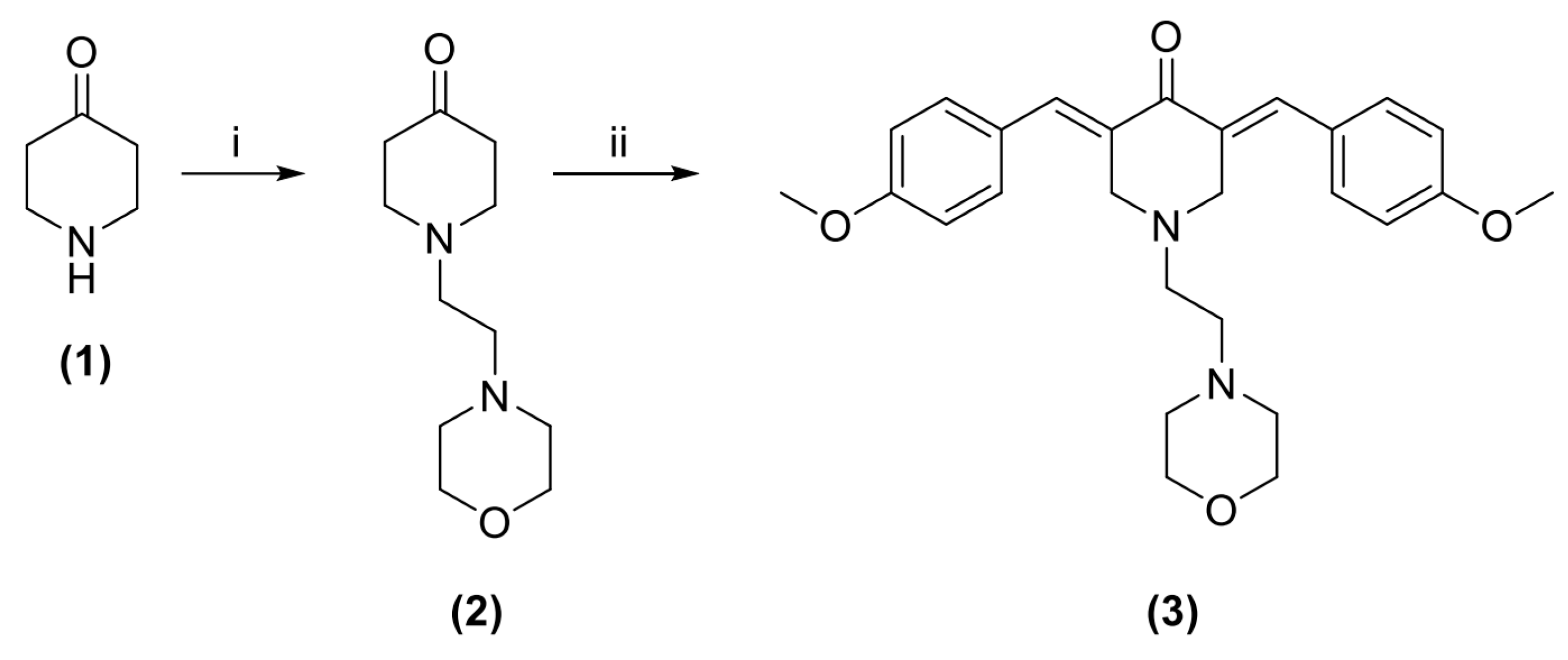
2.1. Chemistry
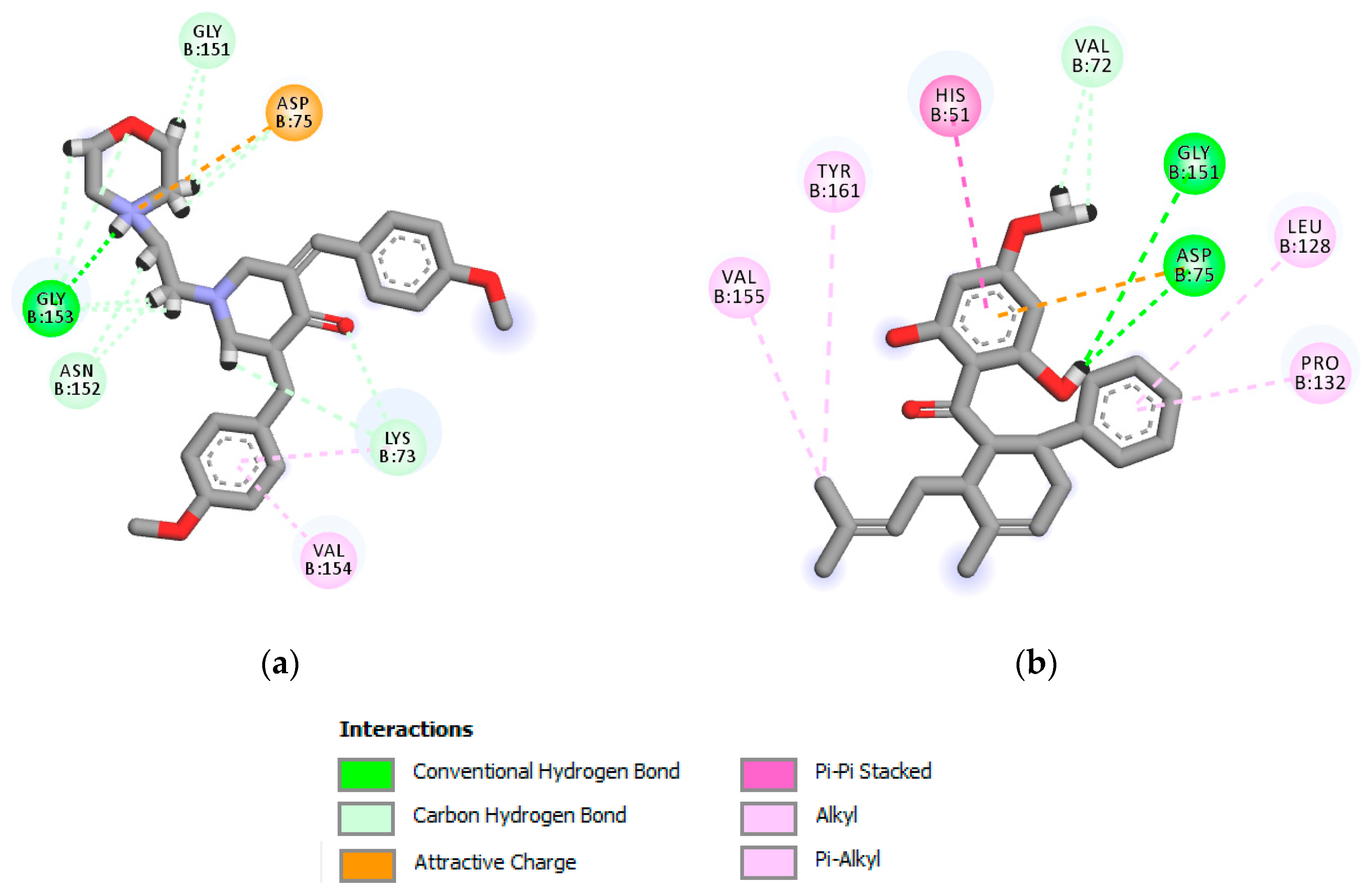
2.2. Anti-Dengue Activity Evaluation
3. Materials and Methods
3.1. Synthesis of 1-(2-Morpholinoethyl)piperidin-4-one (2)
3.2. Synthesis of 3,5-Bis((E)-4-methoxybenzylidene)-1-(2-morpholinoethyl)piperidin-4-one (3)
3.3. DENV2 NS2B-NS3 Protease Inhibition Assay
3.4. Molecular Docking Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. Dengue Vaccine: WHO Position Paper—September 2018. Available online: https://www.who.int/newsroom/factsheets/detail/dengueand-severe-dengue/ (accessed on 20 March 2020).
- Osman, H.; Idris, N.H.; Kamarulzaman, E.E.; Wahab, H.A.; Hassan, M.Z. 3,5-Bis(arylidene)-4-piperidones as potential dengue protease inhibitors. Acta Pharm. Sin. B 2017, 7, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Galula, J.U.; Shen, W.; Chuang, S.; Chang, G.J.; Chao, D. Virus-like particle secretion and genotype-dependent immunogenicity of dengue virus serotype 2 DNA vaccine. J. Virol. 2014, 88, 10813–10830. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Frimayanti, N.; Zain, S.M.; Lee, V.S.; Wahab, H.A.; Yusof, R.; Rahman, N.A. Fragment-based molecular design of new competitive dengue DEN2 NS2B/NS3 inhibitors from the components of fingerroot (Boesenbergia rotunda). In Silico Biol. 2012, 11, 29–37. [Google Scholar]
- Yin, Z.; Patel, S.J.; Wang, W.; Chan, W.; Rao, K.R.R.; Wang, G.; Ngew, X.; Patel, V.; Beer, D.; Knox, J.E.; et al. Peptide inhibitors of dengue virus NS3 protease. Part 2: SAR study of tetrapeptide aldehyde inhibitors. Bioorg. Med. Chem. Lett. 2006, 16, 40–43. [Google Scholar] [CrossRef] [PubMed]
- Behnam, M.A.; Nitsche, C.; Boldescu, V.; Klein, C.D. The medicinal chemistry of dengue virus. J. Med. Chem. 2016, 59, 5622–5649. [Google Scholar] [CrossRef] [PubMed]
- Sumathi, C.S.; Rajesh, P.; Kannan, V.R. The biological potentials of indian traditional medicine, curcumin for treating human diseases. Cardiovasc. Hematol. Agents Med. Chem. 2017, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Marbawati, D.; Umniyati, S.R. The effects of curcumin against dengue-2 virus based on immunocytochemistry technique. Open Trop. Med. J. 2013, 3, 95–102. [Google Scholar]
- Zhang, Y.; Zhao, L.; Wu, J.; Jiang, X.; Dong, L.; Xu, F.; Zou, P.; Dai, Y.; Shan, X.; Yang, S.; et al. Synthesis and evaluation of a series of novel asymmetrical curcumin analogs for the treatment of inflammation. Molecules 2014, 19, 7287–7307. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, A.; Pilankatta, R.; Teramoto, T.; Sajith, A.M.; Nwulia, E.; Kulkarni, A.; Padmanabhan, R. Inhibition of dengue virus by curcuminoids. Antiviral Res. 2019, 162, 71–78. [Google Scholar] [CrossRef] [PubMed]
- El-Subbagh, H.I.; Abu-Zaid, S.M.; Mahran, M.A.; Badria, F.A.; Al-Obaid, A.M. Synthesis and biological evaluation of certain α,β-unsaturated ketones and their corresponding fused pyridines as antiviral and cytotoxic agents. J. Med. Chem. 2000, 43, 2915–2921. [Google Scholar] [CrossRef] [PubMed]
- Zamri, A.; Teruna, H.Y.; Rahmawati, E.N.; Frimayanti, N.; Ikhtiarudin, I. Synthesis and in silico studies of a benzenesulfonyl curcumin analogue as a new anti-dengue virus type 2 (DEN2) NS2B/NS3. Indones. J. Pharm. 2019, 30, 84–90. [Google Scholar]
- Habibi, R.; Herfindo, N.; Hendra, R.; Teruna, H.Y.; Zamri, A. Synthesis and molecular docking study of 1-(3-chloropropyl)-3,5-bis((E)-4-methoxybenzylidene)piperidin-4-one as dengue virus type 2 (DEN2) NS2B/NS3 protease inhibitor candidate. Pharmacol. Clin. Pharm. Res. 2020, 5, 14–22. [Google Scholar] [CrossRef]
- Eryanti, Y.; Herlina, T.; Zamri, A.; Halim, S.N.A.; Shiono, Y.; Syah, Y.M.; Awang, K.; Supratman, U. 3,5-Bis(2-hydroxybenzylidene)piperidin-4-one. Molbank 2014, 2014, M825. [Google Scholar] [CrossRef]
- Valdez, C.A.; Leif, R.N.; Mayer, B.P. An Efficient, Optimized Synthesis of Fentanyl and Related Analogs. PLoS ONE 2014, 9, e108250. [Google Scholar] [CrossRef] [PubMed]
- Noble, C.G.; Seh, C.C.; Chao, A.T.; Shi, P.Y. Ligand-Bound Structures of the Dengue Virus Protease Reveal the Active Conformation. J. Virol. 2011, 86, 438–446. [Google Scholar] [CrossRef]
- Frimayanti, N.; Chee, C.F.; Zain, S.M.; Rahman, N.A. Design of new competitive dengue NS2B/NS3 protease inhibitors-a computational approach. Int. J. Mol. Sci. 2011, 12, 1089–1100. [Google Scholar] [CrossRef]
- Rothan, H.A.; Abdulrahman, A.Y.; Sasikumer, P.G.; Othman, S.; Rahman, N.A.; Yusof, R. Protegrin-1 inhibits dengue NS2B-NS3 serine protease and viral replication in MK2 cells. J. Biomed. Biotechnol. 2012, 2012, 251482. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuswardani, T.; Herfindo, N.; Frimayanti, N.; Hendra, R.; Zamri, A. 3,5-Bis((E)-4-methoxybenzylidene)-1-(2-morpholinoethyl)piperidin-4-one. Molbank 2020, 2020, M1159. https://doi.org/10.3390/M1159
Kuswardani T, Herfindo N, Frimayanti N, Hendra R, Zamri A. 3,5-Bis((E)-4-methoxybenzylidene)-1-(2-morpholinoethyl)piperidin-4-one. Molbank. 2020; 2020(4):M1159. https://doi.org/10.3390/M1159
Chicago/Turabian StyleKuswardani, Tyas, Noval Herfindo, Neni Frimayanti, Rudi Hendra, and Adel Zamri. 2020. "3,5-Bis((E)-4-methoxybenzylidene)-1-(2-morpholinoethyl)piperidin-4-one" Molbank 2020, no. 4: M1159. https://doi.org/10.3390/M1159
APA StyleKuswardani, T., Herfindo, N., Frimayanti, N., Hendra, R., & Zamri, A. (2020). 3,5-Bis((E)-4-methoxybenzylidene)-1-(2-morpholinoethyl)piperidin-4-one. Molbank, 2020(4), M1159. https://doi.org/10.3390/M1159





