Probing the Feasibility of Single-Cell Fixed RNA Sequencing from FFPE Tissue
Abstract
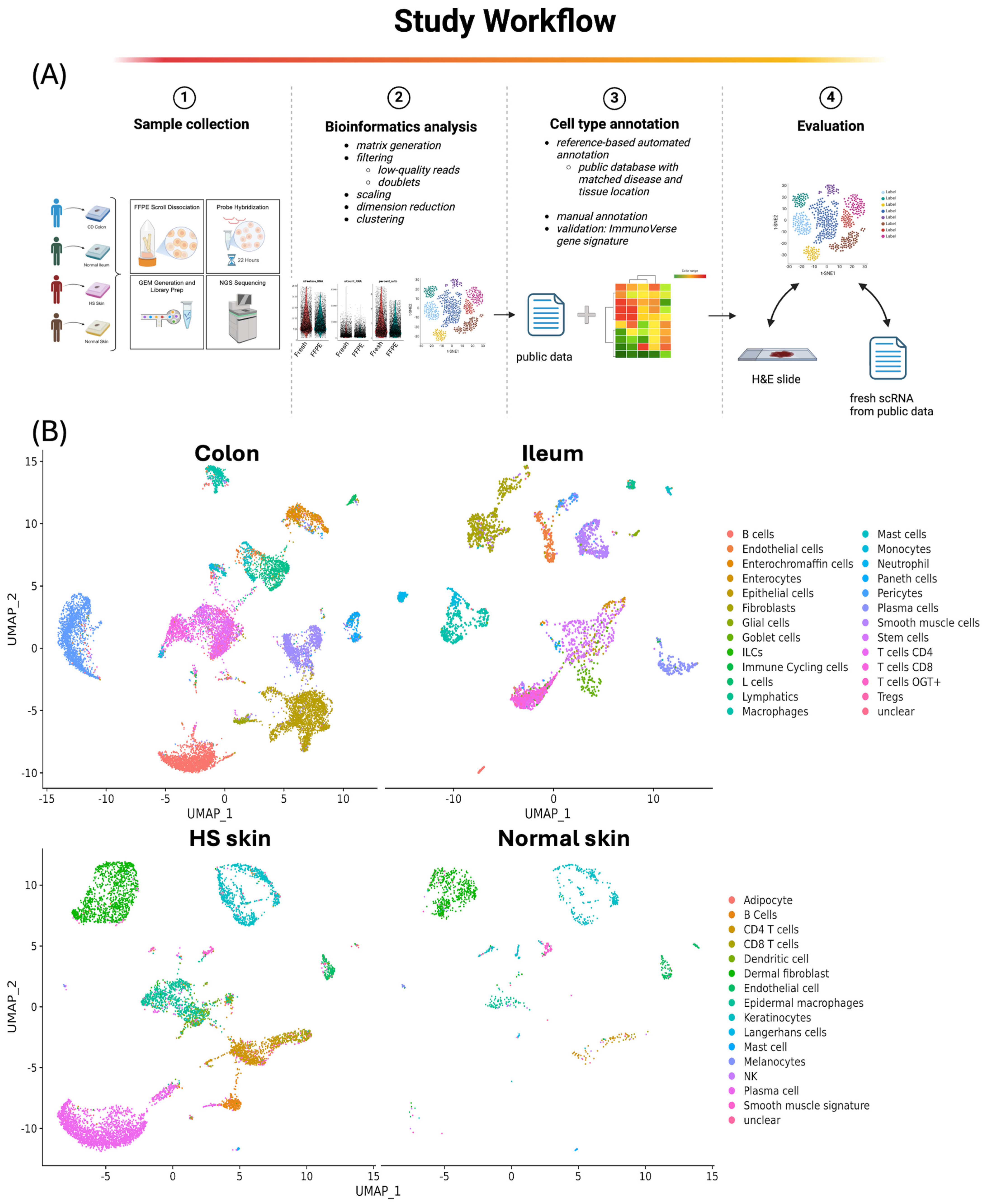
1. Introduction
2. Results
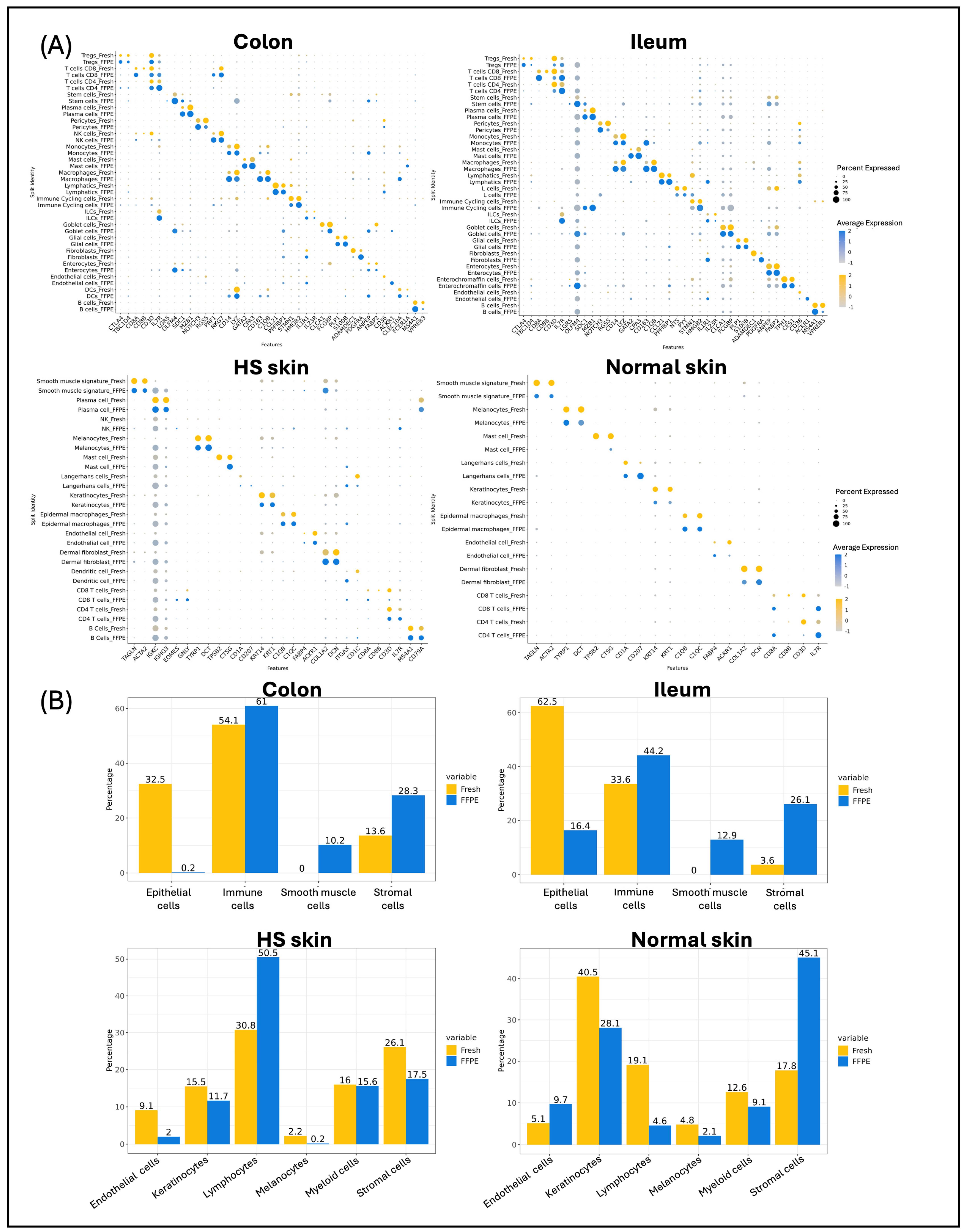
2.1. Single-Cell Quality Matrices in FFPE Samples
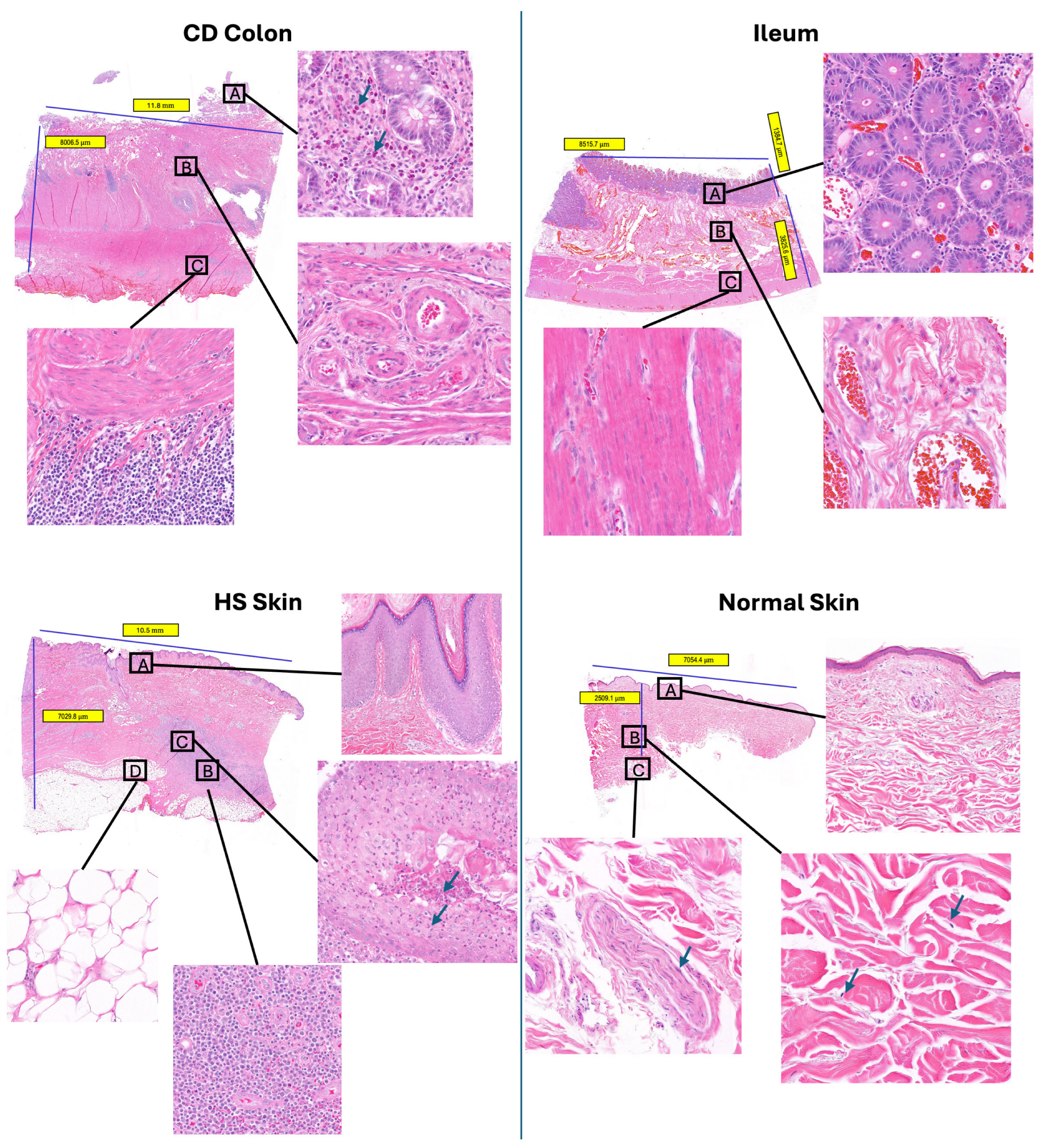
2.2. Cell Abundance Evaluation in scFFPE-Seq via H&E Imaging
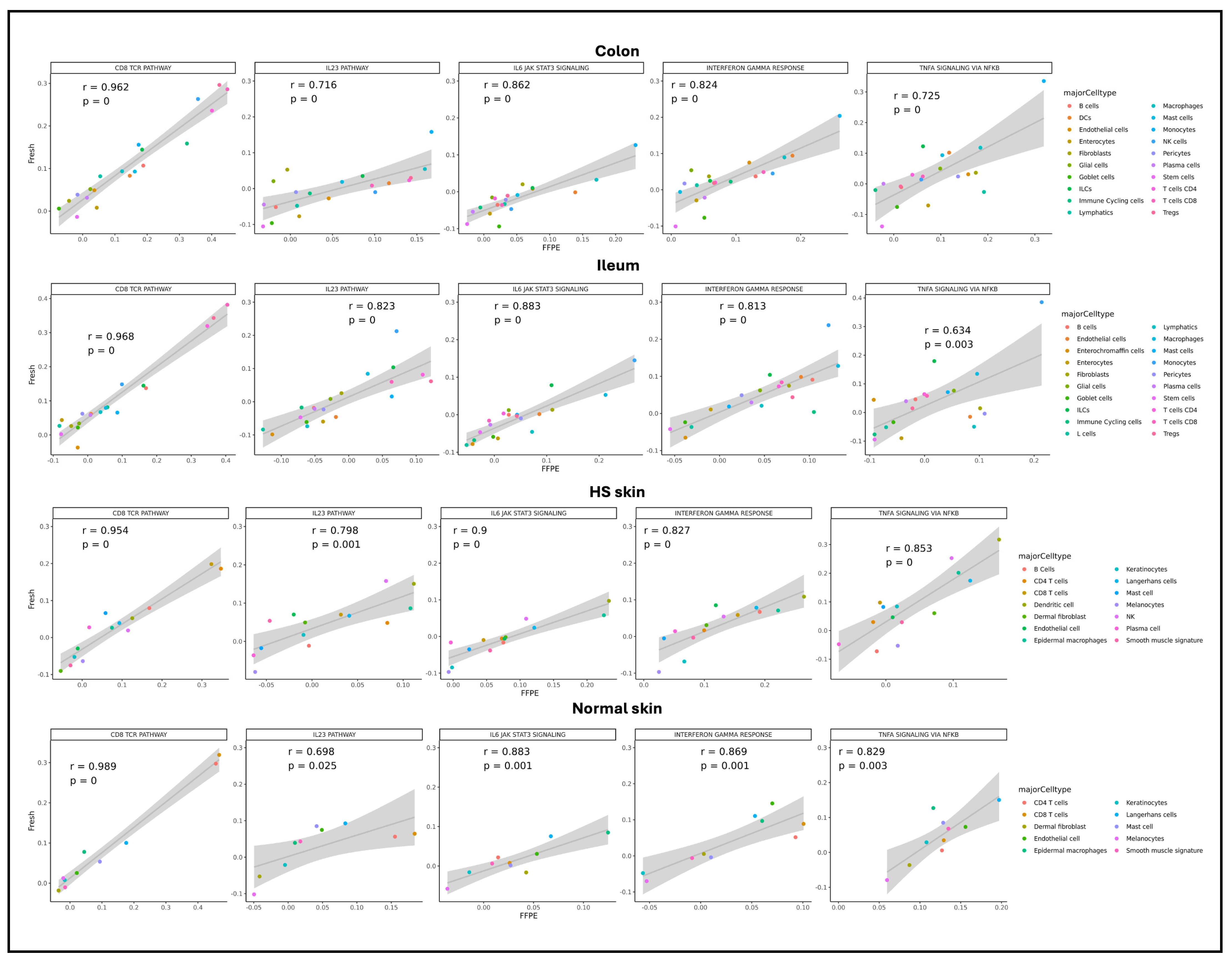
2.3. Gene Expression Comparison Between scFFPE-Seq and Fresh scRNA
2.4. Cell Abundance in scFFPE-Seq Versus Fresh scRNA
2.5. Clinical Conditions in scFFPE vs. Fresh scRNA
3. Discussion
4. Materials and Methods
4.1. Sample Collection, scFFPE-Seq Library Preparation, and Sequencing
4.2. H&E Staining and Image Analysis
4.3. Selection of Public scRNA Data
4.4. Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kulkarni, A.; Anderson, A.G.; Merullo, D.P.; Konopka, G. Beyond Bulk: A Review of Single Cell Transcriptomics Methodologies and Applications. Curr. Opin. Biotechnol. 2019, 58, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Ning, B.; Shi, T. Single-Cell RNA-Seq Technologies and Related Computational Data Analysis. Front. Genet. 2019, 10, 317. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.X.Y.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively Parallel Digital Transcriptional Profiling of Single Cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, W.; Ye, K.; He, L.; Ge, Q.; Huang, Y.; Zhao, X. Single-Nucleus RNA-Seq: Open the Era of Great Navigation for FFPE Tissue. Int. J. Mol. Sci. 2023, 24, 13744. [Google Scholar] [CrossRef]
- Vallejo, A.F.; Harvey, K.; Wang, T.; Wise, K.; Butler, L.M.; Polo, J.; Plummer, J.; Swarbrick, A.; Martelotto, L.G. snPATHO-Seq: Unlocking the FFPE Archives for Single Nucleus RNA Profiling. bioRxiv 2022. [Google Scholar] [CrossRef]
- Trinks, A.; Milek, M.; Beule, D.; Kluge, J.; Florian, S.; Sers, C.; Horst, D.; Morkel, M.; Bischoff, P. Robust Detection of Clinically Relevant Features in Single-Cell RNA Profiles of Patient-Matched Fresh and Formalin-Fixed Paraffin-Embedded (FFPE) Lung Cancer Tissue. Cell. Oncol. 2024, 47, 1221–1231. [Google Scholar] [CrossRef]
- Wang, K.; Kumar, T.; Wang, J.; Minussi, D.C.; Sei, E.; Li, J.; Tran, T.M.; Thennavan, A.; Hu, M.; Casasent, A.K.; et al. Archival Single-Cell Genomics Reveals Persistent Subclones during DCIS Progression. Cell 2023, 186, 3968–3982.e15. [Google Scholar] [CrossRef]
- Janesick, A.; Shelansky, R.; Gottscho, A.D.; Wagner, F.; Rouault, M.; Beliakoff, G.; de Oliveira, M.F.; Kohlway, A.; Abousoud, J.; Morrison, C.A.; et al. High Resolution Mapping of the Breast Cancer Tumor Microenvironment Using Integrated Single Cell, Spatial and in Situ Analysis of FFPE Tissue. bioRxiv 2022. [Google Scholar] [CrossRef]
- Chung, H.; Melnikov, A.; McCabe, C.; Drokhlyansky, E.; Wittenberghe, N.V.; Magee, E.M.; Waldman, J.; Spira, A.; Chen, F.; Mazzilli, S.; et al. SnFFPE-Seq: Towards Scalable Single Nucleus RNA-Seq of Formalin-Fixed Paraffin-Embedded (FFPE) Tissue. bioRxiv 2022. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, T.; Chen, H.; Zhu, Y.; Lv, Y.; Zhang, S.; Chen, J.; Chen, H.; Yang, L.; Jiang, W.; et al. High-Throughput Single Nucleus Total RNA Sequencing of Formalin-Fixed Paraffin-Embedded Tissues by snRandom-Seq. Nat. Commun. 2023, 14, 2734. [Google Scholar] [CrossRef]
- Wang, T.; Roach, M.J.; Harvey, K.; Morlanes, J.E.; Kiedik, B.; Al-Eryani, G.; Greenwald, A.; Kalavros, N.; Dezem, F.S.; Ma, Y.; et al. snPATHO-Seq, a Versatile FFPE Single-Nucleus RNA Sequencing Method to Unlock Pathology Archives. Commun. Biol. 2024, 7, 1340. [Google Scholar] [CrossRef]
- de Sande, B.V.; Lee, J.S.; Mutasa-Gottgens, E.; Naughton, B.; Bacon, W.; Manning, J.; Wang, Y.; Pollard, J.; Mendez, M.; Hill, J.; et al. Applications of Single-Cell RNA Sequencing in Drug Discovery and Development. Nat. Rev. Drug Discov. 2023, 22, 496–520. [Google Scholar] [CrossRef] [PubMed]
- González-Martínez, S.; Palacios, J.; Carretero-Barrio, I.; Lanza, V.F.; Piqueras, M.G.-C.; Caniego-Casas, T.; Hardisson, D.; Esteban-Rodríguez, I.; Cortés, J.; Pérez-Mies, B. Single-Cell RNA Sequencing on Formalin-Fixed and Paraffin-Embedded (FFPE) Tissue Identified Multi-Ciliary Cells in Breast Cancer. Cells 2025, 14, 197. [Google Scholar] [CrossRef]
- Villacampa, E.G.; Larsson, L.; Mirzazadeh, R.; Kvastad, L.; Andersson, A.; Mollbrink, A.; Kokaraki, G.; Monteil, V.; Schultz, N.; Appelberg, K.S.; et al. Genome-Wide Spatial Expression Profiling in Formalin-Fixed Tissues. Cell Genom. 2021, 1, 100065. [Google Scholar] [CrossRef]
- Ben-Moshe, N.B.; Gilad, S.; Perry, G.; Benjamin, S.; Balint-Lahat, N.; Pavlovsky, A.; Halperin, S.; Markus, B.; Yosepovich, A.; Barshack, I.; et al. mRNA-Seq Whole Transcriptome Profiling of Fresh Frozen versus Archived Fixed Tissues. BMC Genom. 2018, 19, 419. [Google Scholar] [CrossRef]
- Avram, V.; Yadav, S.; Sahasrabudhe, P.; Chang, D.; Wang, J. IBDTransDB: A Manually Curated Transcriptomic Database for Inflammatory Bowel Disease. Database 2024, 2024, baae026. [Google Scholar] [CrossRef]
- Buxton, E.J.; Luesley, D.M.; Shafi, M.I.; Rollason, M. Colposcopically Directed Punch Biopsy: A Potentially Misleading Investigation. BJOG Int. J. Obstet. Gynaecol. 1991, 98, 1273–1276. [Google Scholar] [CrossRef]
- Elston, D.M.; Stratman, E.J.; Miller, S.J. Skin Biopsy Biopsy Issues in Specific Diseases. J. Am. Acad. Dermatol. 2016, 74, 1–16. [Google Scholar] [CrossRef]
- Curry, J.L.; Davies, M.A.; Calderone, T.L.; Nathanson, K.; Prieto, V.G.; Gershenwald, J.E. Molecular Diagnostics for Melanoma, Methods and Protocols. Methods Mol. Biol. 2013, 1102, 679–695. [Google Scholar] [CrossRef]
- Petousis, S.; Christidis, P.; Margioula-Siarkou, C.; Sparangis, N.; Athanasiadis, A.; Kalogiannidis, I. Discrepancy between Colposcopy, Punch Biopsy and Final Histology of Cone Specimen: A Prospective Study. Arch. Gynecol. Obstet. 2018, 297, 1271–1275. [Google Scholar] [CrossRef] [PubMed]
- Grieshaber-Bouyer, R.; Radtke, F.A.; Cunin, P.; Stifano, G.; Levescot, A.; Vijaykumar, B.; Nelson-Maney, N.; Blaustein, R.B.; Monach, P.A.; Nigrovic, P.A.; et al. The Neutrotime Transcriptional Signature Defines a Single Continuum of Neutrophils across Biological Compartments. Nat. Commun. 2021, 12, 2856. [Google Scholar] [CrossRef]
- Gudjonsson, J.E.; Tsoi, L.C.; Ma, F.; Billi, A.C.; van Straalen, K.R.; Vossen, A.R.J.V.; Zee, H.H.; Harms, P.W.; Wasikowski, R.; Yee, C.M.; et al. Contribution of Plasma Cells and B-Cells to Hidradenitis Suppurativa Pathogenesis. JCI Insight 2020, 5, e139930. [Google Scholar] [CrossRef]
- Moran, B.; Smith, C.M.; Zaborowski, A.; Ryan, M.; Karman, J.; Dunstan, R.W.; Smith, K.M.; Hambly, R.; Musilova, J.; Petrasca, A.; et al. Targeting the NLRP3 Inflammasome Reduces Inflammation in Hidradenitis Suppurativa Skin. Br. J. Dermatol. 2023, 189, 447–458. [Google Scholar] [CrossRef]
- Lowe, M.M.; Naik, H.B.; Clancy, S.; Pauli, M.; Smith, K.M.; Bi, Y.; Dunstan, R.; Gudjonsson, J.E.; Paul, M.; Harris, H.; et al. Immunopathogenesis of Hidradenitis Suppurativa and Response to Anti-TNF-A Therapy. JCI Insight 2020, 5, 19, Correction in JCI Insight 2022, 7, 20. https://doi.org/10.1172/jci.insight.165502. [Google Scholar] [CrossRef] [PubMed]
- Rojahn, T.B.; Vorstandlechner, V.; Krausgruber, T.; Bauer, W.M.; Alkon, N.; Bangert, C.; Thaler, F.M.; Sadeghyar, F.; Fortelny, N.; Gernedl, V.; et al. Single-Cell Transcriptomics Combined with Interstitial Fluid Proteomics Defines Cell Type–Specific Immune Regulation in Atopic Dermatitis. J. Allergy Clin. Immunol. 2020, 146, 1056–1069. [Google Scholar] [CrossRef]
- Kong, L.; Pokatayev, V.; Lefkovith, A.; Carter, G.T.; Creasey, E.A.; Krishna, C.; Subramanian, S.; Kochar, B.; Ashenberg, O.; Lau, H.; et al. The Landscape of Immune Dysregulation in Crohn’s Disease Revealed through Single-Cell Transcriptomic Profiling in the Ileum and Colon. Immunity 2023, 56, 444–458.e5. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xiang, Q.; Xu, F.; Huang, J.; Yu, N.; Zhang, Q.; Long, X.; Zhou, Z. Single-Cell RNA-Seq of Cultured Human Adipose-Derived Mesenchymal Stem Cells. Sci. Data 2019, 6, 190031. [Google Scholar] [CrossRef]
- McGinnis, C.S.; Murrow, L.M.; Gartner, Z.J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019, 8, 329–337.e4. [Google Scholar] [CrossRef] [PubMed]
- Clarke, Z.A.; Andrews, T.S.; Atif, J.; Pouyabahar, D.; Innes, B.T.; MacParland, S.A.; Bader, G.D. Tutorial: Guidelines for Annotating Single-Cell Transcriptomic Maps Using Automated and Manual Methods. Nat. Protoc. 2021, 16, 2749–2764. [Google Scholar] [CrossRef]
- Tran, H.T.N.; Ang, K.S.; Chevrier, M.; Zhang, X.; Lee, N.Y.S.; Goh, M.; Chen, J. A Benchmark of Batch-Effect Correction Methods for Single-Cell RNA Sequencing Data. Genome Biol. 2020, 21, 12. [Google Scholar] [CrossRef]
- Aran, D.; Looney, A.P.; Liu, L.; Wu, E.; Fong, V.; Hsu, A.; Chak, S.; Naikawadi, R.P.; Wolters, P.J.; Abate, A.R.; et al. Reference-Based Analysis of Lung Single-Cell Sequencing Reveals a Transitional Profibrotic Macrophage. Nat. Immunol. 2019, 20, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Li, T.; Xu, Y.; Zhang, X.; Li, F.; Bai, J.; Chen, J.; Jiang, W.; Yang, K.; Ou, Q.; et al. CellMarker 2.0: An Updated Database of Manually Curated Cell Markers in Human/Mouse and Web Tools Based on scRNA-Seq Data. Nucleic Acids Res. 2022, 51, D870–D876. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M.; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated Analysis of Multimodal Single-Cell Data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; II, M.H.W.; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the Multicellular Ecosystem of Metastatic Melanoma by Single-Cell RNA-Seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
| CD Colon | Normal Ileum | HS Skin | Normal Skin | |
|---|---|---|---|---|
| Cells sequenced | 16,130 | 4387 | 9091 | 1723 |
| Cells passed QC | 13,778 | 3720 | 7705 | 1406 |
| Median reads per cell | 7156 | 11,994 | 6208 | 25,061 |
| Median UMI counts per cell | 1753 | 2264 | 1474 | 1577 |
| Median genes per cell | 1150 | 1416 | 818 | 1044 |
| Total genes detected | 16,950 | 16,478 | 16,989 | 15,659 |
| Confidently mapped reads in cells | 94.90% | 95.00% | 90.83% | 82.98% |
| Sequencing saturation | 73.43% | 79.68% | 73.17% | 93.11% |
| Epithelial Cells | Neutrophils | Keratinocyte | ||||
|---|---|---|---|---|---|---|
| Colon | Ileum | Colon | Ileum | HS Skin | Normal Skin | |
| H&E estimation | 4738 (0.9%) | 26,226 (17.2%) | 1282 (0.2%) | 357 (0.2%) | 19,551 (9.3%) | 1515 (16.1%) |
| scFFPE-seq | 21 (0.2%) | 609 (16.37%) | 51 (0.2%) | 106 (0.7%) | 900 (10.8%) | 395 (28.1%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Liu, X.; Naughton, K.; Karsen, S.D.; Bentley, P.; Duggan, L.; Chaudhary, N.; Smith, K.M.; Phillips, L.; Chang, D.; Mahi, N.A. Probing the Feasibility of Single-Cell Fixed RNA Sequencing from FFPE Tissue. Int. J. Mol. Sci. 2026, 27, 1605. https://doi.org/10.3390/ijms27031605
Liu X, Naughton K, Karsen SD, Bentley P, Duggan L, Chaudhary N, Smith KM, Phillips L, Chang D, Mahi NA. Probing the Feasibility of Single-Cell Fixed RNA Sequencing from FFPE Tissue. International Journal of Molecular Sciences. 2026; 27(3):1605. https://doi.org/10.3390/ijms27031605
Chicago/Turabian StyleLiu, Xiaochen, Katherine Naughton, Samuel D. Karsen, Patricia Bentley, Lori Duggan, Neha Chaudhary, Kathleen M. Smith, Lucy Phillips, Dan Chang, and Naim A. Mahi. 2026. "Probing the Feasibility of Single-Cell Fixed RNA Sequencing from FFPE Tissue" International Journal of Molecular Sciences 27, no. 3: 1605. https://doi.org/10.3390/ijms27031605
APA StyleLiu, X., Naughton, K., Karsen, S. D., Bentley, P., Duggan, L., Chaudhary, N., Smith, K. M., Phillips, L., Chang, D., & Mahi, N. A. (2026). Probing the Feasibility of Single-Cell Fixed RNA Sequencing from FFPE Tissue. International Journal of Molecular Sciences, 27(3), 1605. https://doi.org/10.3390/ijms27031605






