Transduction of Lentiviral Vectors and ADORA3 in HEK293T Cells Modulated in Gene Expression and Alternative Splicing
Abstract
1. Introduction
2. Results
2.1. Overexpression of GFP and ADORA3
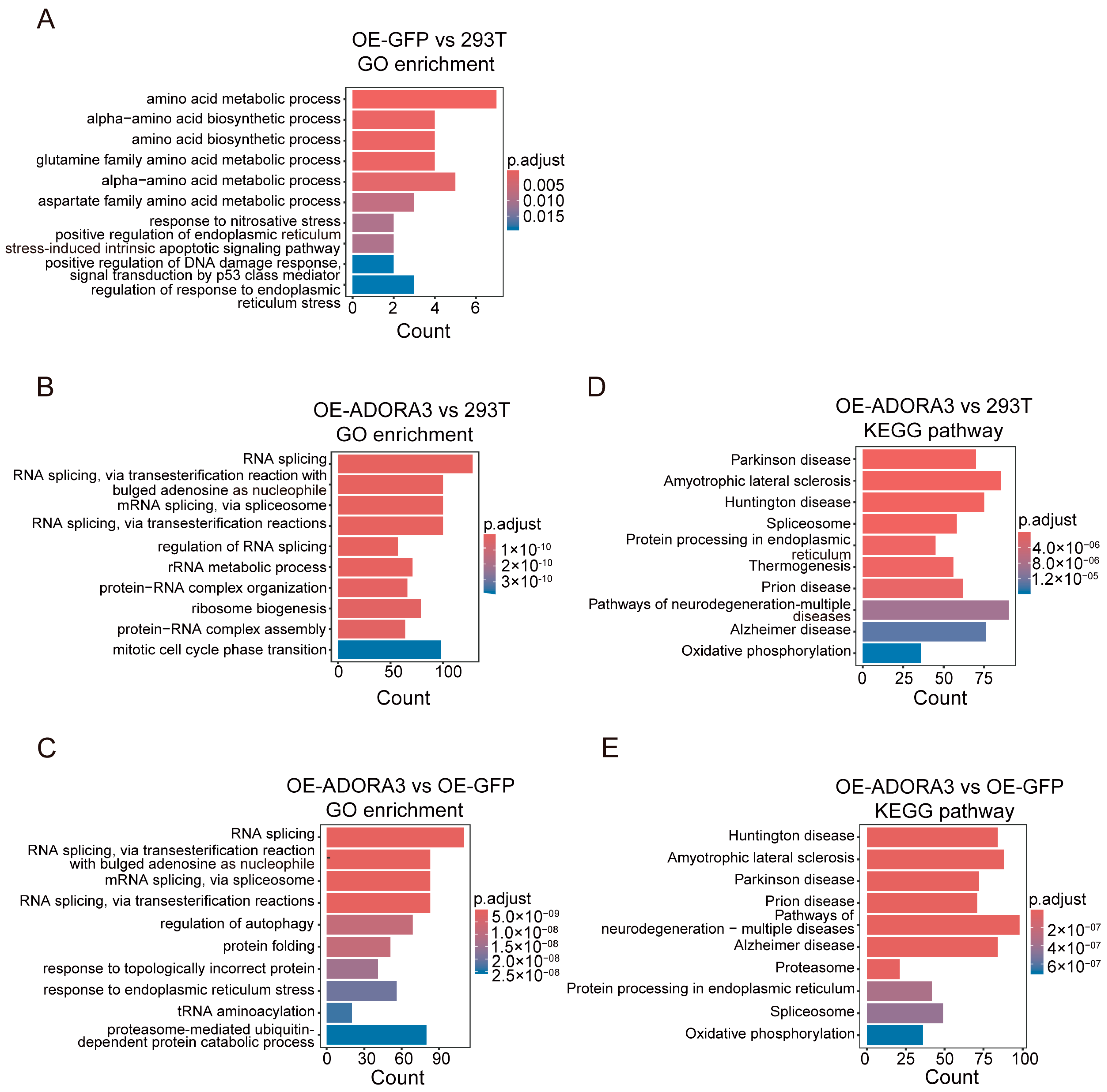
2.2. More than 2000 Genes Were Differentially Expressed in OE-ADORA3, While Only 31 Genes Were Differentially Expressed in OE-GFP
2.3. Detection of Differential Transcript Usage Among Cell Lines
2.4. The SNHG8 Gene Represents an Example of Both DEGs and gDTUs
3. Discussion
4. Materials and Methods
4.1. Generation of Genetically Modified HKE293T Cells
4.2. RNA Extraction and RT-qPCR
4.3. RNA Library Construction and Sequencing
4.4. RNA-Seq Data Analysis
4.5. Alternative Splicing Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HEK293T | Human Embryonic Kidney 293T |
| gDTUs | Genes with differential transcript usage |
| GFP | Green fluorescent protein |
| ADORA3 | Adenosine A3 receptor |
| HIV | Human immunodeficiency virus |
| DMEM | Dulbecco’s modified eagle medium |
| RT-qPCR | Quantitative reverse transcription polymerase chain reaction |
| DEGs | Differentially expressed genes |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| PCR | Polymerase chain reaction |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PCA | Principal component analysis |
| TPM | Transcripts per million |
| PSI | Percent spliced in |
| ONT | Oxford Nanopore technology |
| IGV | Integrative Genomics Viewer |
References
- Huang, Y.; Hou, R.; Lam, F.S.; Jia, Y.; Zhou, Y.; He, X.; Li, G.; Xiong, F.; Cao, Y.; Wang, D.; et al. Agonist Discovery for Membrane Proteins on Live Cells by Using DNA-encoded Libraries. J. Am. Chem. Soc. 2024, 146, 24638–24653. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, Y.; Liu, W.; He, T.; Liao, J.; Qian, Z.; Zhao, J.; Cong, Z.; Sun, D.; Liu, Z.; et al. Genome-wide pan-GPCR cell libraries accelerate drug discovery. Acta Pharm. Sin. B 2024, 14, 4296–4311. [Google Scholar] [CrossRef]
- Bi, Z.; Li, H.; Liang, Y.; Sun, D.; Liu, S.; Chen, W.; Leng, L.; Song, C.; Zhang, S.; Cong, Z.; et al. Emerging paradigms for target discovery of traditional medicines: A genome-wide pan-GPCR perspective. Innovation 2025, 6, 100774. [Google Scholar] [CrossRef]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- Tang, A.D.; Soulette, C.M.; van Baren, M.J.; Hart, K.; Hrabeta-Robinson, E.; Wu, C.J.; Brooks, A.N. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat. Commun. 2020, 11, 1438. [Google Scholar] [CrossRef] [PubMed]
- Sahlin, K.; Medvedev, P. Error correction enables use of Oxford Nanopore technology for reference-free transcriptome analysis. Nat. Commun. 2021, 12, 2. [Google Scholar] [CrossRef]
- Zufferey, R.; Nagy, D.; Mandel, R.J.; Naldini, L.; Trono, D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997, 15, 871–875. [Google Scholar] [CrossRef] [PubMed]
- Xinyue, Z.; Li, S.; Yujie, W.; Yingcai, D.; Changhao, B.; Xueli, Z. Engineering of HEK293T Cell Factory for Lentiviral Production by High-Throughput Selected Genes. Cris. J. 2024, 7, 272–282. [Google Scholar] [CrossRef]
- Uchida, N.; Washington, K.N.; Hayakawa, J.; Hsieh, M.M.; Bonifacino, A.C.; Krouse, A.E.; Metzger, M.E.; Donahue, R.E.; Tisdale, J.F. Development of a human immunodeficiency virus type 1-based lentiviral vector that allows efficient transduction of both human and rhesus blood cells. J. Virol. 2009, 83, 9854–9862. [Google Scholar] [CrossRef]
- Wolff, J.H.; Mikkelsen, J.G. Delivering genes with human immunodeficiency virus-derived vehicles: Still state-of-the-art after 25 years. J. Biomed. Sci. 2022, 29, 79. [Google Scholar] [CrossRef]
- Kutner, R.H.; Zhang, X.Y.; Reiser, J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat. Protoc. 2009, 4, 495–505. [Google Scholar] [CrossRef]
- Suleman, S.; Fawaz, S.; Roberts, T.; Ellison, S.; Bigger, B.; Themis, M. Optimised protocols to generate high titre lentiviral vectors using a novel transfection agent enabling extended HEK293T culture following transient transfection and suspension culture. J. Virol. Methods 2024, 325, 114884. [Google Scholar] [CrossRef] [PubMed]
- Meyerhof, W.; Müller-Brechlin, R.; Richter, D. Molecular cloning of a novel putative G-protein coupled receptor expressed during rat spermiogenesis. FEBS Lett. 1991, 284, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Salvatore, C.A.; Jacobson, M.A.; Taylor, H.E.; Linden, J.; Johnson, R.G. Molecular cloning and characterization of the human A3 adenosine receptor. Proc. Natl. Acad. Sci. USA 1993, 90, 10365–10369. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.G.; Auchampach, J.A.; Jacobson, K.A. Species dependence of A(3) adenosine receptor pharmacology and function. Purinergic Signal. 2023, 19, 523–550. [Google Scholar] [CrossRef]
- Borea, P.A.; Gessi, S.; Merighi, S.; Vincenzi, F.; Varani, K. Pharmacology of Adenosine Receptors: The State of the Art. Physiol. Rev. 2018, 98, 1591–1625. [Google Scholar] [CrossRef]
- Vallon, V.; Mühlbauer, B.; Osswald, H. Adenosine and kidney function. Physiol. Rev. 2006, 86, 901–940. [Google Scholar] [CrossRef]
- Cohen, S.; Fishman, P. Targeting the A(3) adenosine receptor to treat cytokine release syndrome in cancer immunotherapy. Drug Des. Dev. Ther. 2019, 13, 491–497. [Google Scholar] [CrossRef]
- Ferreira, C.B.; Sumner, R.P.; Rodriguez-Plata, M.T.; Rasaiyaah, J.; Milne, R.S.; Thrasher, A.J.; Qasim, W.; Towers, G.J. Lentiviral Vector Production Titer Is Not Limited in HEK293T by Induced Intracellular Innate Immunity. Mol. Ther. Methods Clin. Dev. 2020, 17, 209–219. [Google Scholar] [CrossRef]
- Willis, R.A.; Ramachandiran, V.; Shires, J.C.; Bai, G.; Jeter, K.; Bell, D.L.; Han, L.; Kazarian, T.; Ugwu, K.C.; Laur, O.; et al. Production of Class II MHC Proteins in Lentiviral Vector-Transduced HEK-293T Cells for Tetramer Staining Reagents. Curr. Protoc. 2021, 1, e36. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Boone, M.; Meuris, L.; Lemmens, I.; Van Roy, N.; Soete, A.; Reumers, J.; Moisse, M.; Plaisance, S.; Drmanac, R.; et al. Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat. Commun. 2014, 5, 4767. [Google Scholar] [CrossRef] [PubMed]
- Graham, F.L.; Smiley, J.; Russell, W.C.; Nairn, R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977, 36, 59–74. [Google Scholar] [CrossRef]
- Yuan, X.; Yan, Y.; Xue, M. Small nucleolar RNA host gene 8: A rising star in the targets for cancer therapy. Biomed Pharmacother. 2021, 139, 111622. [Google Scholar] [CrossRef]
- Dropulić, B. Lentiviral vectors: Their molecular design, safety, and use in laboratory and preclinical research. Hum. Gene Ther. 2011, 22, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Paugh, B.S.; Baranyi, L.; Roy, A.; He, H.-J.; Harris, L.; Cole, K.D.; Artlip, M.; Raimund, C.; Langan, P.S.; Jana, S.; et al. Reference standards for accurate validation and optimization of assays that determine integrated lentiviral vector copy number in transduced cells. Sci. Rep. 2021, 11, 389. [Google Scholar] [CrossRef] [PubMed]
- Luostarinen, A.; Kailaanmäki, A.; Turkki, V.; Köylijärvi, M.; Käyhty, P.; Leinonen, H.; Albers-Skirdenko, V.; Lipponen, E.; Ylä-Herttuala, S.; Kaartinen, T.; et al. Optimizing lentiviral vector formulation conditions for efficient ex vivo transduction of primary human T cells in chimeric antigen receptor T-cell manufacturing. Cytotherapy 2024, 26, 1084–1094. [Google Scholar] [CrossRef]
- Zufferey, R.; Dull, T.; Mandel, R.J.; Bukovsky, A.; Quiroz, D.; Naldini, L.; Trono, D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 1998, 72, 9873–9880. [Google Scholar] [CrossRef] [PubMed]
- Milone, M.C.; O’Doherty, U. Clinical use of lentiviral vectors. Leukemia 2018, 32, 1529–1541. [Google Scholar] [CrossRef]
- Moiani, A.; Paleari, Y.; Sartori, D.; Mezzadra, R.; Miccio, A.; Cattoglio, C.; Cocchiarella, F.; Lidonnici, M.R.; Ferrari, G.; Mavilio, F. Lentiviral vector integration in the human genome induces alternative splicing and generates aberrant transcripts. J. Clin. Investig. 2012, 122, 1653–1666. [Google Scholar] [CrossRef]
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef]
- Chen, C.Y.; Shyu, A.B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995, 20, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Hershey, J.W.; Sonenberg, N.; Mathews, M.B. Principles of translational control: An overview. Cold Spring Harb. Perspect. Biol. 2012, 4, a011528. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef]
- De Coster, W.; Van Broeckhoven, C. Newest Methods for Detecting Structural Variations. Trends Biotechnol. 2019, 37, 973–982. [Google Scholar] [CrossRef]
- Logsdon, G.A.; Vollger, M.R.; Eichler, E.E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 2020, 21, 597–614. [Google Scholar] [CrossRef]
- Workman, R.E.; Tang, A.D.; Tang, P.S.; Jain, M.; Tyson, J.R.; Razaghi, R.; Zuzarte, P.C.; Gilpatrick, T.; Payne, A.; Quick, J.; et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods 2019, 16, 1297–1305. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Aarthy, M.; George, A.; Ayyadurai, N. Beyond protein tagging: Rewiring the genetic code of fluorescent proteins—A review. Int. J. Biol. Macromol. 2021, 191, 840–851. [Google Scholar] [CrossRef]
- Abaandou, L.; Quan, D.; Shiloach, J. Affecting HEK293 Cell Growth and Production Performance by Modifying the Expression of Specific Genes. Cells 2021, 10, 1667. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Harla, I.; Pawluś, W.; Zarębski, M.; Dobrucki, J.W. Induction of DNA single- and double-strand breaks by excited intra- or extracellular green fluorescent protein. J. Photochem. Photobiol. B Biol. 2024, 259, 113001. [Google Scholar] [CrossRef]
- Gao, J.; Wang, L.; Liu, J.; Xie, F.; Su, B.; Wang, X. Abnormalities of Mitochondrial Dynamics in Neurodegenerative Diseases. Antioxidants 2017, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Li, H.; Liao, P.; Chen, L.; Pan, Y.; Zheng, Y.; Zhang, C.; Liu, D.; Zheng, M.; Gao, J. Mitochondrial dysfunction: Mechanisms and advances in therapy. Signal Transduct. Target. Ther. 2024, 9, 124. [Google Scholar] [CrossRef]
- Carmo, C.; Naia, L.; Lopes, C.; Rego, A.C. Mitochondrial Dysfunction in Huntington’s Disease. Adv. Exp. Med. Biol. 2018, 1049, 59–83. [Google Scholar]
- Dorn, G.W., 2nd. Reversing Dysdynamism to Interrupt Mitochondrial Degeneration in Amyotrophic Lateral Sclerosis. Cells 2023, 12, 1188. [Google Scholar] [CrossRef]
- Nikom, D.; Zheng, S. Alternative splicing in neurodegenerative disease and the promise of RNA therapies. Nat. Rev. Neurosci. 2023, 24, 457–473. [Google Scholar] [CrossRef]
- Texier, B.; Prime, M.; Atamena, D.; Belenguer, P.; Szelechowski, M. Mortalin/Hspa9 involvement and therapeutic perspective in Parkinson’s disease. Neural Regen. Res. 2023, 18, 293–298. [Google Scholar] [PubMed]
- Miyamoto, Y.; Eguchi, T.; Kawahara, K.; Hasegawa, N.; Nakamura, K.; Funakoshi-Tago, M.; Tanoue, A.; Tamura, H.; Yamauchi, J. Hypomyelinating leukodystrophy-associated missense mutation in HSPD1 blunts mitochondrial dynamics. Biochem. Biophys. Res. Commun. 2015, 462, 275–281. [Google Scholar] [CrossRef]
- Akhtar, A.; Sah, S.P. Insulin signaling pathway and related molecules: Role in neurodegeneration and Alzheimer’s disease. Neurochem. Int. 2020, 135, 104707. [Google Scholar] [CrossRef]
- Mazziotta, C.; Rotondo, J.C.; Lanzillotti, C.; Campione, G.; Martini, F.; Tognon, M. Cancer biology and molecular genetics of A(3) adenosine receptor. Oncogene 2022, 41, 301–308. [Google Scholar] [CrossRef]
- Borea, P.A.; Varani, K.; Vincenzi, F.; Baraldi, P.G.; Tabrizi, M.A.; Merighi, S.; Gessi, S. The A3 adenosine receptor: History and perspectives. Pharmacol. Rev. 2015, 67, 74–102. [Google Scholar] [CrossRef] [PubMed]
- Mlejnek, P.; Dolezel, P.; Frydrych, I. Effects of synthetic A3 adenosine receptor agonists on cell proliferation and viability are receptor independent at micromolar concentrations. J. Physiol. Biochem. 2013, 69, 405–417. [Google Scholar] [CrossRef]
- Jajoo, S.; Mukherjea, D.; Watabe, K.; Ramkumar, V. Adenosine A(3) receptor suppresses prostate cancer metastasis by inhibiting NADPH oxidase activity. Neoplasia 2009, 11, 1132–1145. [Google Scholar] [CrossRef]
- Calderwood, S.K.; Khaleque, M.A.; Sawyer, D.B.; Ciocca, D.R. Heat shock proteins in cancer: Chaperones of tumorigenesis. Trends Biochem. Sci. 2006, 31, 164–172. [Google Scholar] [CrossRef]
- Zhang, N.; Lo, C.-W.; Utsunomiya, T.; Maruyama, M.; Huang, E.; Rhee, C.; Gao, Q.; Yao, Z.; Goodman, S.B. PDGF-BB and IL-4 co-overexpression is a potential strategy to enhance mesenchymal stem cell-based bone regeneration. Stem Cell Res. Ther. 2021, 12, 40. [Google Scholar] [CrossRef]
- Elegheert, J.; Behiels, E.; Bishop, B.; Scott, S.; Woolley, R.E.; Griffiths, S.C.; Byrne, E.F.X.; Chang, V.T.; Stuart, D.I.; Jones, E.Y.; et al. Lentiviral transduction of mammalian cells for fast, scalable and high-level production of soluble and membrane proteins. Nat. Protoc. 2018, 13, 2991–3017. [Google Scholar] [CrossRef]
- Chirgwin, J.M.; Przybyla, A.E.; MacDonald, R.J.; Rutter, W.J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 1979, 18, 5294–5299. [Google Scholar] [CrossRef] [PubMed]
- Sanada, T.; Takaesu, G.; Mashima, R.; Yoshida, R.; Kobayashi, T.; Yoshimura, A. FLN29 deficiency reveals its negative regulatory role in the Toll-like receptor (TLR) and retinoic acid-inducible gene I (RIG-I)-like helicase signaling pathway. J. Biol. Chem. 2008, 283, 33858–33864. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef]
- Yu, G. Thirteen years of clusterProfiler. Innovation 2024, 5, 100722. [Google Scholar] [CrossRef] [PubMed]
- Trincado, J.L.; Entizne, J.C.; Hysenaj, G.; Singh, B.; Skalic, M.; Elliott, D.J.; Eyras, E. SUPPA2: Fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018, 19, 40. [Google Scholar] [CrossRef] [PubMed]
- Mak, Q.X.C.; Wick, R.R.; Holt, J.M.; Wang, J.R. Polishing De Novo Nanopore Assemblies of Bacteria and Eukaryotes With FMLRC. Mol. Biol. Evol. 2023, 40, msad048. [Google Scholar] [CrossRef]
- Kalikar, S.; Jain, C.; Vasimuddin, M.; Misra, S. Accelerating minimap2 for long-read sequencing applications on modern CPUs. Nat. Comput. Sci. 2022, 2, 78–83. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Hansen, A.S.; Zechner, C. Promoters adopt distinct dynamic manifestations depending on transcription factor context. Mol. Syst. Biol. 2021, 17, e9821. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Song, Y.; Fang, S.; Yan, M.; Yang, Z.; Shao, Y.; Zhao, K.; Liu, M.; Wang, Z.; Guo, Z.; et al. hnRNPU-mediated pathogenic alternative splicing drives gastric cancer progression. J. Exp. Clin. Cancer Res. 2025, 44, 8. [Google Scholar] [CrossRef]
- Sciarrillo, R.; Wojtuszkiewicz, A.; Assaraf, Y.G.; Jansen, G.; Kaspers, G.J.; Giovannetti, E.; Cloos, J. The role of alternative splicing in cancer: From oncogenesis to drug resistance. Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer Chemother. 2020, 53, 100728. [Google Scholar] [CrossRef] [PubMed]
- Bonnal, S.C.; López-Oreja, I.; Valcárcel, J. Roles and mechanisms of alternative splicing in cancer—Implications for care. Nat. Rev. Clin. Oncol. 2020, 17, 457–474. [Google Scholar] [CrossRef] [PubMed]
- Halu, A.; Chelvanambi, S.; Decano, J.L.; Matamalas, J.T.; Whelan, M.; Asano, T.; Kalicharran, N.; Singh, S.A.; Loscalzo, J.; Aikawa, M. Integrating pharmacogenomics and cheminformatics with diverse disease phenotypes for cell type-guided drug discovery. Genome Med. 2025, 17, 7. [Google Scholar] [CrossRef]
- Zhao, C.; Zhao, J.; Zhang, Y.; Zhu, Y.; Yang, Z.; Liu, S.; Tang, Q.; Yang, Y.; Wang, H.; Shu, Y.; et al. PTBP3 Mediates IL-18 Exon Skipping to Promote Immune Escape in Gallbladder Cancer. Adv. Sci. 2024, 11, e2406633. [Google Scholar] [CrossRef]





| Primer | Sequence (5′-3′) |
|---|---|
| GAPDH-F | GGAGCGAGATCCCTCCAAAAT |
| GAPDH-R | GGCTGTTGTCATACTTCTCATGG |
| ADORA3-F | GCTGGTCATCTGCGTGGTCAAG |
| ADORA3-R | GGATTGTGATGCCCAGGCTGAC |
| PMAIP1-F | ACTCACCGTGTGTAGTTGGC |
| PMAIP1-R | CACTCGACTTCCAGCTCTGCT |
| ASNS-F | GGAAGACAGCCCCGATTTACT |
| ASNS-R | AGCACGAACTGTTGTAATGTCA |
| DDIT4-F | TGAGGATGAACACTTGTGTGC |
| DDIT4-R | CCAACTGGCTAGGCATCAGC |
| DDIT3-F | GGAACCTGAGGAGAGAGTGTTC |
| DDIT3-R | CTGCCATCTCTGCAGTTGGA |
| HSPA9-F | ACCTGCTGATGAGTGCAACA |
| HSPA9-R | AGTGCCAGAACTTCCAGAGC |
| HSPD1-F | ACGACCTGTCTCGCCG |
| HSPD1-R | AATCGTAGCAACCTGTGCAA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, Y.; Liu, Z.; Liu, Q.; Tian, X.; Mo, J.; Leng, L.; Wang, C.; Xu, G.; Zhang, S.; Xie, J. Transduction of Lentiviral Vectors and ADORA3 in HEK293T Cells Modulated in Gene Expression and Alternative Splicing. Int. J. Mol. Sci. 2025, 26, 4431. https://doi.org/10.3390/ijms26094431
Qian Y, Liu Z, Liu Q, Tian X, Mo J, Leng L, Wang C, Xu G, Zhang S, Xie J. Transduction of Lentiviral Vectors and ADORA3 in HEK293T Cells Modulated in Gene Expression and Alternative Splicing. International Journal of Molecular Sciences. 2025; 26(9):4431. https://doi.org/10.3390/ijms26094431
Chicago/Turabian StyleQian, Yongqi, Zhaoyu Liu, Qingqing Liu, Xiaojuan Tian, Jing Mo, Liang Leng, Can Wang, Guoqing Xu, Sanyin Zhang, and Jiang Xie. 2025. "Transduction of Lentiviral Vectors and ADORA3 in HEK293T Cells Modulated in Gene Expression and Alternative Splicing" International Journal of Molecular Sciences 26, no. 9: 4431. https://doi.org/10.3390/ijms26094431
APA StyleQian, Y., Liu, Z., Liu, Q., Tian, X., Mo, J., Leng, L., Wang, C., Xu, G., Zhang, S., & Xie, J. (2025). Transduction of Lentiviral Vectors and ADORA3 in HEK293T Cells Modulated in Gene Expression and Alternative Splicing. International Journal of Molecular Sciences, 26(9), 4431. https://doi.org/10.3390/ijms26094431






