Association Between Vaginal Microbiota and Cervical Dysplasia Due to Persistent Human Papillomavirus Infection: A Systematic Review of Evidence from Shotgun Metagenomic Sequencing Studies
Abstract
1. Introduction
2. Methods
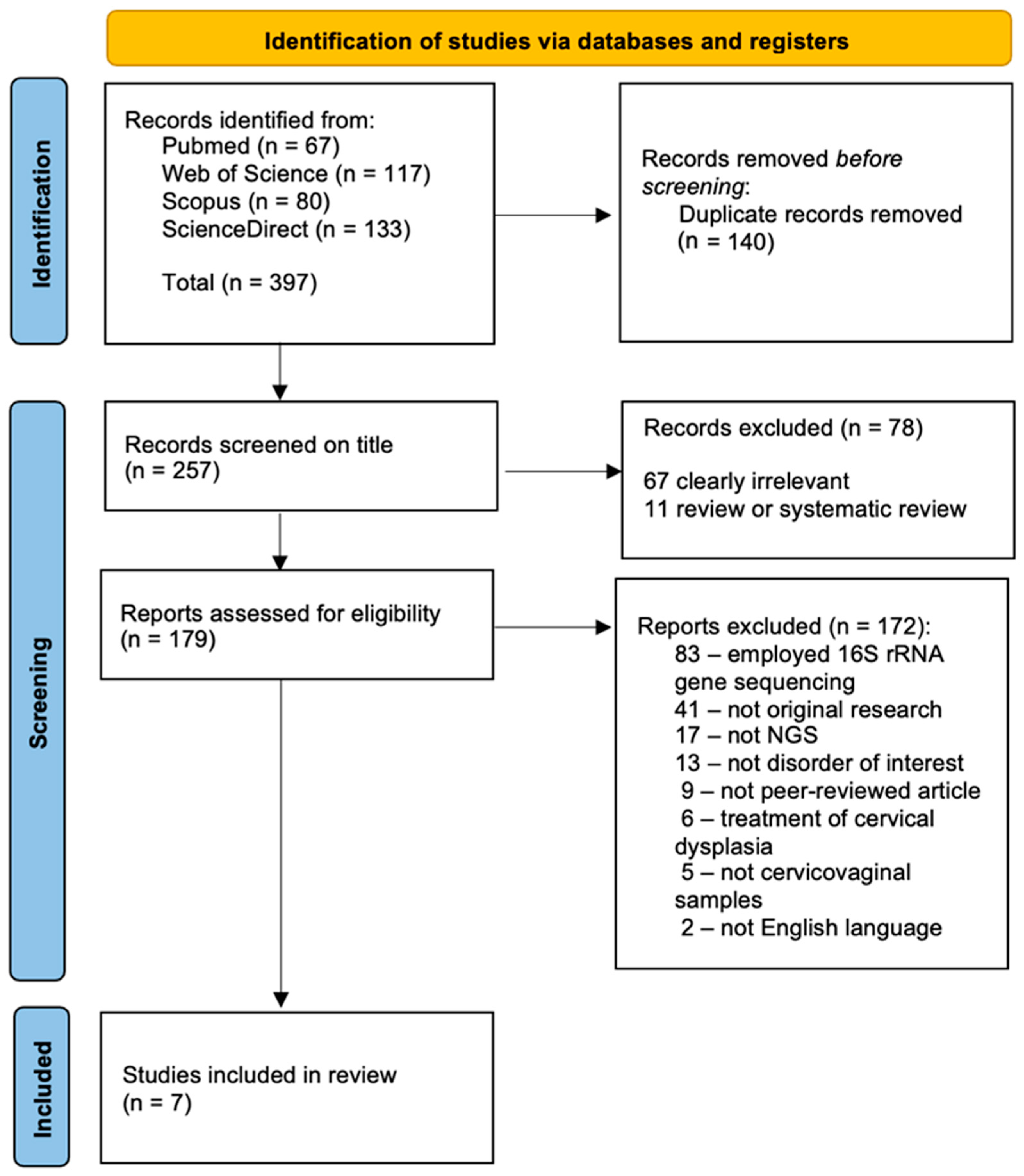
2.1. Search Strategy and Study Selection
2.2. Data Extraction
2.3. Quality Assessment
3. Results and Discussion
3.1. Study Selection and Characteristics
3.2. Differences in Microbiota Diversity
3.3. Differences at the Phylum Level
3.4. Differences at the Genus Level
3.5. Differences at Species Level
3.6. Functional Differences of Cervicovaginal Microbiome
3.7. Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| HPV | Human papilloma virus |
| CIN | Cervical intraepithelial neoplasia |
| CC | Cervical cancer |
References
- Mancilla, V.; Jimenez, N.R.; Bishop, N.S.; Flores, M.; Herbst-Kralovetz, M.M. The Vaginal Microbiota, Human Papillomavirus Infection, and Cervical Carcinogenesis: A Systematic Review in the Latina Population. J. Epidemiol. Glob. Health 2024, 14, 480–497. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Feng, Y.; Xie, L.; Ma, S.; Cai, Z.; Ma, Y. Alterations in Gut and Genital Microbiota Associated with Gynecological Diseases: A Systematic Review and Meta-Analysis. Reprod. Biol. Endocrinol. 2024, 22, 13. [Google Scholar] [CrossRef] [PubMed]
- Norenhag, J.; Du, J.; Olovsson, M.; Verstraelen, H.; Engstrand, L.; Brusselaers, N. The Vaginal Microbiota, Human Papillomavirus and Cervical Dysplasia: A Systematic Review and Network Meta-Analysis. BJOG Int. J. Obstet. Gynaecol. 2020, 127, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Brusselaers, N.; Shrestha, S.; van de Wijgert, J.; Verstraelen, H. Vaginal Dysbiosis and the Risk of Human Papillomavirus and Cervical Cancer: Systematic Review and Meta-Analysis. Am. J. Obstet. Gynecol. 2019, 221, 9–18.e8. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiang, F.; Liu, T.; Chen, Z.; Zhang, M.; Li, J.; Kang, X.; Wu, R. Leveraging Existing 16S rRNA Gene Surveys to Decipher Microbial Signatures and Dysbiosis in Cervical Carcinogenesis. Sci. Rep. 2024, 14, 11532. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, Y.; Wei, X.; Zhu, J.; Wang, X.; Xie, X.; Lu, W. The Alterations of Vaginal Microbiome in HPV16 Infection as Identified by Shotgun Metagenomic Sequencing. Front. Cell. Infect. Microbiol. 2020, 10, 286. [Google Scholar] [CrossRef]
- Hu, M.; Yang, W.; Yan, R.; Chi, J.; Xia, Q.; Yang, Y.; Wang, Y.; Sun, L.; Li, P. Co-Evolution of Vaginal Microbiome and Cervical Cancer. J. Transl. Med. 2024, 22, 559. [Google Scholar] [CrossRef]
- Liu, H.; Liang, H.; Li, D.; Wang, M.; Li, Y. Association of Cervical Dysbacteriosis, HPV Oncogene Expression, and Cervical Lesion Progression. Microbiol. Spectr. 2022, 10, e00151-22. [Google Scholar] [CrossRef]
- Onywera, H.; Williamson, A.-L.; Mbulawa, Z.Z.A.; Coetzee, D.; Meiring, T.L. Factors Associated with the Composition and Diversity of the Cervical Microbiota of Reproductive-Age Black South African Women: A Retrospective Cross-Sectional Study. PeerJ 2019, 7, e7488. [Google Scholar] [CrossRef]
- Lebeer, S.; Ahannach, S.; Gehrmann, T.; Wittouck, S.; Eilers, T.; Oerlemans, E.; Condori, S.; Dillen, J.; Spacova, I.; Vander Donck, L.; et al. A Citizen-Science-Enabled Catalogue of the Vaginal Microbiome and Associated Factors. Nat. Microbiol. 2023, 8, 2183–2195. [Google Scholar] [CrossRef]
- McKee, K.; Bassis, C.M.; Golob, J.; Palazzolo, B.; Sen, A.; Comstock, S.S.; Rosas-Salazar, C.; Stanford, J.B.; O’Connor, T.; Gern, J.E.; et al. Host Factors Are Associated with Vaginal Microbiome Structure in Pregnancy in the ECHO Cohort Consortium. Sci. Rep. 2024, 14, 11798. [Google Scholar] [CrossRef] [PubMed]
- Scillato, M.; Spitale, A.; Mongelli, G.; Privitera, G.F.; Mangano, K.; Cianci, A.; Stefani, S.; Santagati, M. Antimicrobial Properties of Lactobacillus Cell-free Supernatants against Multidrug-resistant Urogenital Pathogens. Microbiologyopen 2021, 10, e1173. [Google Scholar] [CrossRef] [PubMed]
- Glick, V.J.; Webber, C.A.; Simmons, L.E.; Martin, M.C.; Ahmad, M.; Kim, C.H.; Adams, A.N.D.; Bang, S.; Chao, M.C.; Howard, N.C.; et al. Vaginal Lactobacilli Produce Anti-Inflammatory β-Carboline Compounds. Cell Host Microbe 2024, 32, 1897–1909.e7. [Google Scholar] [CrossRef] [PubMed]
- Decout, A.; Krasias, I.; Roberts, L.; Gimeno Molina, B.; Charenton, C.; Brown Romero, D.; Tee, Q.Y.; Marchesi, J.R.; Ng, S.; Sykes, L.; et al. Lactobacillus Crispatus S-Layer Proteins Modulate Innate Immune Response and Inflammation in the Lower Female Reproductive Tract. Nat. Commun. 2024, 15, 10879. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ma, Y.; Li, R.; Chen, X.; Wan, L.; Zhao, W. Associations of Cervicovaginal Lactobacilli With High-Risk Human Papillomavirus Infection, Cervical Intraepithelial Neoplasia, and Cancer: A Systematic Review and Meta-Analysis. J. Infect. Dis. 2019, 220, 1243–1254. [Google Scholar] [CrossRef]
- Fettweis, J.M.; Serrano, M.G.; Brooks, J.P.; Edwards, D.J.; Girerd, P.H.; Parikh, H.I.; Huang, B.; Arodz, T.J.; Edupuganti, L.; Glascock, A.L.; et al. The Vaginal Microbiome and Preterm Birth. Nat. Med. 2019, 25, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Edwards, V.L.; Smith, S.B.; McComb, E.J.; Tamarelle, J.; Ma, B.; Humphrys, M.S.; Gajer, P.; Gwilliam, K.; Schaefer, A.M.; Lai, S.K.; et al. The Cervicovaginal Microbiota-Host Interaction Modulates Chlamydia Trachomatis Infection. mBio 2019, 10. [Google Scholar] [CrossRef]
- Wu, M.; Li, H.; Yu, H.; Yan, Y.; Wang, C.; Teng, F.; Fan, A.; Xue, F. Disturbances of Vaginal Microbiome Composition in Human Papillomavirus Infection and Cervical Carcinogenesis: A Qualitative Systematic Review. Front. Oncol. 2022, 12, 941741. [Google Scholar] [CrossRef]
- Fang, B.; Li, Q.; Wan, Z.; OuYang, Z.; Zhang, Q. Exploring the Association Between Cervical Microbiota and HR-HPV Infection Based on 16S rRNA Gene and Metagenomic Sequencing. Front. Cell. Infect. Microbiol. 2022, 12, 922554. [Google Scholar] [CrossRef]
- Kwon, M.; Seo, S.-S.; Kim, M.K.; Lee, D.O.; Lim, M.C. Compositional and Functional Differences between Microbiota and Cervical Carcinogenesis as Identified by Shotgun Metagenomic Sequencing. Cancers 2019, 11, 309. [Google Scholar] [CrossRef]
- Brown, D. A Review of the PubMed PICO Tool: Using Evidence-Based Practice in Health Education. Health Promot. Pract. 2020, 21, 496–498. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Norenhag, J.; Edfeldt, G.; Stålberg, K.; Garcia, F.; Hugerth, L.W.; Engstrand, L.; Fransson, E.; Du, J.; Schuppe-Koistinen, I.; Olovsson, M. Compositional and Functional Differences of the Vaginal Microbiota of Women with and without Cervical Dysplasia. Sci. Rep. 2024, 14, 11183. [Google Scholar] [CrossRef]
- Zheng, X.; Hu, N.; Liu, J.; Zhao, K.; Li, H.; Wang, J.; Zhang, M.; Zhang, L.; Song, L.; Lyu, Y.; et al. Cervicovaginal Microbiota Disorder Combined with the Change of Cytosine Phosphate Guanine Motif- Toll like Receptor 9 Axis Was Associated with Cervical Cancerization. J. Cancer Res. Clin. Oncol. 2023, 149, 17371–17381. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhao, X.; Wu, F.; Chen, J.; Luo, J.; Wu, C.; Chen, T. Effectiveness of Vaginal Probiotics Lactobacillus Crispatus Chen-01 in Women with High-Risk HPV Infection: A Prospective Controlled Pilot Study. Aging (Albany NY) 2024, 16, 11446–11459. [Google Scholar] [CrossRef]
- Genital Tract Microbiota Composition Profiles and Use of Prebiotics and Probiotics in Gynaecological Cancer Prevention: Review of the Current Evidence, the European Society of Gynaecological Oncology Prevention Committee Statement—The Lancet Microbe. Available online: https://www.thelancet.com/journals/lanmic/article/PIIS2666-5247(23)00257-4/fulltext (accessed on 2 March 2025).
- Usyk, M.; Zolnik, C.P.; Castle, P.E.; Porras, C.; Herrero, R.; Gradissimo, A.; Gonzalez, P.; Safaeian, M.; Schiffman, M.; Burk, R.D. Cervicovaginal Microbiome and Natural History of HPV in a Longitudinal Study. PLoS Pathog. 2020, 16, e1008376. [Google Scholar] [CrossRef]
- Banister, C.E.; Messersmith, A.R.; Cai, B.; Spiryda, L.B.; Glover, S.H.; Pirisi, L.; Creek, K.E. Disparity in the Persistence of High-Risk Human Papillomavirus Genotypes Between African American and European American Women of College Age. J. Infect. Dis. 2015, 211, 100–108. [Google Scholar] [CrossRef]
| 1st Author, Year | Country | Study Design | Population | HPV Genotypes | N | Groups | Primary Comparator | Type of Sequencing |
|---|---|---|---|---|---|---|---|---|
| Liu et al., 2022 [8] | China | Observational, cross-sectional | Patients diagnosed with HPV, CIN, or CC (histologically verified) for the first time. | High risk—16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 82 | 115 | HR-HPV without CIN (n = 34) CIN with HR-HPV (n = 40) CC (n = 41) | HR-HPV group | Metagenomic shotgun sequencing |
| Zheng et al., 2023 [24] | China | Observational, cross-sectional | Patients with histologically verified CIN1, CIN2/3, squamous CC, and healthy controls. | 16 | 341 | CIN1 (n = 90) CIN2/3 (n = 78) CC (n = 49) NC (n = 124) Subdivision to HPV16-positive (n = 128), other HPV-positive (n = 34) and HPV-negative (n = 179) | NC group | Metagenomic shotgun sequencing |
| Hu et al., 2024 [7] | China | Retrospective observational cohort | HPV-positive patients and patients with histologically verified CIN, CC, and healthy controls. | Not specified | 151 | CC (n = 42) CIN (n = 43) HPV+ (n = 34) NC (n = 32) | NC group | Metagenomic shotgun sequencing |
| Kwon et al., 2019 [20] | South Korea | Observational, cross-sectional | Patients with histologically verified CIN or CC and healthy controls. | - | 47 | CIN 2/3 (n = 17) CC (n = 12) NC (n = 18) | NC group | Metagenomic shotgun sequencing |
| Norenhag et al., 2024 [23] | Sweden | Observational, cross-sectional | Patients with histologically verified dysplasia and cancer (LSIL, HSIL, CC) and healthy controls. | Low risk—6, 11, 32, 34, 37, 40, 42, 43, 44, 54, 61, 62, 70, 71, 72, 74, 80, 81, 83, 84, 87, 89, 90, 91, 98, 101, 103, 106, 107, 108, 114, 115, 118, 119, 121, 124, 129, 149, 155, 163, 168 High risk—16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, 82 | 354 | Dysplasia and cancer (n = 177; (LSIL n = 81, HSIL n = 94, cancer n = 2)) NC (n = 177) Subdivision to HPV-negative, (n = 35), LR-HPV (n = 26), HR-HPV (n = 126) | NC group | Metagenomic shotgun sequencing |
| Fang et al., 2022 [19] | China | Observational, cross-sectional | Reproductive-age women with HR-HPV and healthy controls. | Not specified | 40 | HR-HPV (n = 20) NC (n = 20) | NC group | 16S rRNA gene and shotgun metagenomic sequencing |
| Yang et al., 2020 [6] | China | Observational, cross-sectional | Reproductive-age women with HPV16 and healthy controls. | 16 | 57 | HPV16 positive (n = 27) NC (n = 25) | NC group | Metagenomic shotgun sequencing |
| Species | HPV | Dysplasia | CC | Aerobic/Anaerobic Status | BV-Related Organisms | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author, Year | Author, Year | Author, Year | ||||||||||||
| Zheng et al., 2023 [24] | Hu et al., 2024 [7] | Norenhag et al., 2024 [23] | Fang et al., 2022 [19] | Yang et al., 2020 [6] | Liu et al., 2022 [8] ** | Zheng et al., 2023 [24] | Hu et al., 2024 [7] | Norenhag et al., 2024 [23] | Liu et al., 2022 ** [8] | Zheng et al., 2023 [24] | Hu et al., 2024 [7] | |||
| Lactobacillus crispatus | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | Fan | ||||
| Lactobacillus iners | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | Fan | |||||
| Lactobacillus gasseri | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | Fan | |||||||
| Lactobacillus jensenii | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | Fan | ||||||
| Lactobacillus helveticus | ↓ | Fan | ||||||||||||
| Lactobacillus acidophilus | ↓ | ↓ | Fan | |||||||||||
| Lactobacillus johnsonii | ↑ | Fan | ||||||||||||
| Gardnerella vaginalis | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↓ | ↓ | An | BV |
| Gardnerellasp_304 | ↑ | An | BV | |||||||||||
| Gardnerella sp_2612 | ↑ | An | BV | |||||||||||
| Bifidobacterium breve | ↓ | ↑ | ↑ | ↑ | ↑ | An | ||||||||
| Bifidobacterium longum | ↑ | ↑ | An | |||||||||||
| Bifidobacterium bifidum | ↑ | An | ||||||||||||
| Prevotella bivia | ↓ | ↑ | ↑ | ↑ | ↑ | ↑ | An | BV | ||||||
| Prevotella amnii | ↓ | ↑ | ↑ | ↓ | ↑ | ↓ | An | BV | ||||||
| Prevotella corporis | ↑ | An | ||||||||||||
| Prevotella disiens | ↑ | An | BV | |||||||||||
| Prevotella timonensis | ↓ | ↓ | ↓ | An | ||||||||||
| Peptoniphilus lacrimalis | ↑ | An | ||||||||||||
| Peptoniphilus harei | ↑ | An | BV | |||||||||||
| Mageeibacillus indolicus | ↑ | An | ||||||||||||
| Atopobium vaginae/Fannyhessea vaginae | ↓ | ↑ | ↓ | ↑ | ↓ | An | BV | |||||||
| Mobiluncus curtisii | ↑ | An | BV | |||||||||||
| Coriobacteriales bacterium DNF00809 | ↑ | An | ||||||||||||
| Peptostreptococcus anaerobius | ↑ | An | BV | |||||||||||
| Veillonella montpellierensis | ↑ | ↑ | ↑ | An | ||||||||||
| Megasphaera sp. UPII_135E | ↑ | An | BV | |||||||||||
| Fusobacterium nucleatum | ↑ | An | BV | |||||||||||
| Methanobrevibacter oralis | ↑ | An | ||||||||||||
| Finegoldia magna | ↑ | An | ||||||||||||
| Porphyromonas uneanis | ↓ | ↑ | ↑ | An | ||||||||||
| Porphyromonas asaccharolytica | ↑ | ↑ | An | |||||||||||
| Snethia amnii | ↑ | ↑ | An | BV | ||||||||||
| Phocaeicola vulgatus | ↑ | An | ||||||||||||
| Bacteroides fragilis | ↑ | ↑ | ↑ | ↑ | An | |||||||||
| Bacteroides thetaiotaomicron | ↑ | An | ||||||||||||
| Clostridium botulinum | ↑ | An | ||||||||||||
| Anaerococcus lactolyticus | ↑ | ↑ | An | |||||||||||
| Anaerococcus tetradius | ↑ | ↑ | An | |||||||||||
| Peptoniphilus lacrimalis | ↑ | An | ||||||||||||
| Burkholderia pseudomallei | ↑ | A | ||||||||||||
| Ureaplasma parvum | ↑ | A | ||||||||||||
| Neisseria gonorrhoeae | ↑ | A | ||||||||||||
| Bacillus velezensis | ↑ | ↑ | A | |||||||||||
| Aerococcus christensenii | ↑ | A | ||||||||||||
| Klebsiella pneumonia | ↑ | Fan | ||||||||||||
| Escherichia coli | ↓ | ↓ | ↓ | ↓ | ↑ | ↓ | Fan | |||||||
| Streptococcus agalacitae | ↓ | ↑ | ↓ | Fan | ||||||||||
| Streptococcus mitis oralis pneumoniae | ↑ | ↑ | ↑ | Fan | ||||||||||
| Staphylococcus aureus | ↑ | ↑ | ↑ | Fan | ||||||||||
| Salmonela enterica | ↑ | ↑ | Fan | |||||||||||
| Streptococcus mitis | ↓ | ↓ | Fan | |||||||||||
| Candida albicans | ↑ | A, fungus | ||||||||||||
| Alpha papillomavirus 9 | ↑ | Virus | ||||||||||||
| Group | Enriched Pathway | Contributing Species |
|---|---|---|
| Healthy | L. crispatus (mainly), L. jensenii, L. iners, L. rhamnosus, G. vaginalis, F. vaginae [23] | |
| HPV | ||
| CIN | G. vaginalis, B. longum, F. vaginae, P. bivia, P. timonensis, S. agalactiae, S. anginosus [23] | |
| Cervical cancer |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Žukienė, G.; Narutytė, R.; Rudaitis, V. Association Between Vaginal Microbiota and Cervical Dysplasia Due to Persistent Human Papillomavirus Infection: A Systematic Review of Evidence from Shotgun Metagenomic Sequencing Studies. Int. J. Mol. Sci. 2025, 26, 4258. https://doi.org/10.3390/ijms26094258
Žukienė G, Narutytė R, Rudaitis V. Association Between Vaginal Microbiota and Cervical Dysplasia Due to Persistent Human Papillomavirus Infection: A Systematic Review of Evidence from Shotgun Metagenomic Sequencing Studies. International Journal of Molecular Sciences. 2025; 26(9):4258. https://doi.org/10.3390/ijms26094258
Chicago/Turabian StyleŽukienė, Guoda, Ramunė Narutytė, and Vilius Rudaitis. 2025. "Association Between Vaginal Microbiota and Cervical Dysplasia Due to Persistent Human Papillomavirus Infection: A Systematic Review of Evidence from Shotgun Metagenomic Sequencing Studies" International Journal of Molecular Sciences 26, no. 9: 4258. https://doi.org/10.3390/ijms26094258
APA StyleŽukienė, G., Narutytė, R., & Rudaitis, V. (2025). Association Between Vaginal Microbiota and Cervical Dysplasia Due to Persistent Human Papillomavirus Infection: A Systematic Review of Evidence from Shotgun Metagenomic Sequencing Studies. International Journal of Molecular Sciences, 26(9), 4258. https://doi.org/10.3390/ijms26094258







