Placental Protein Citrullination Signatures Are Modified in Early- and Late-Onset Fetal Growth Restriction
Abstract
1. Introduction
2. Results
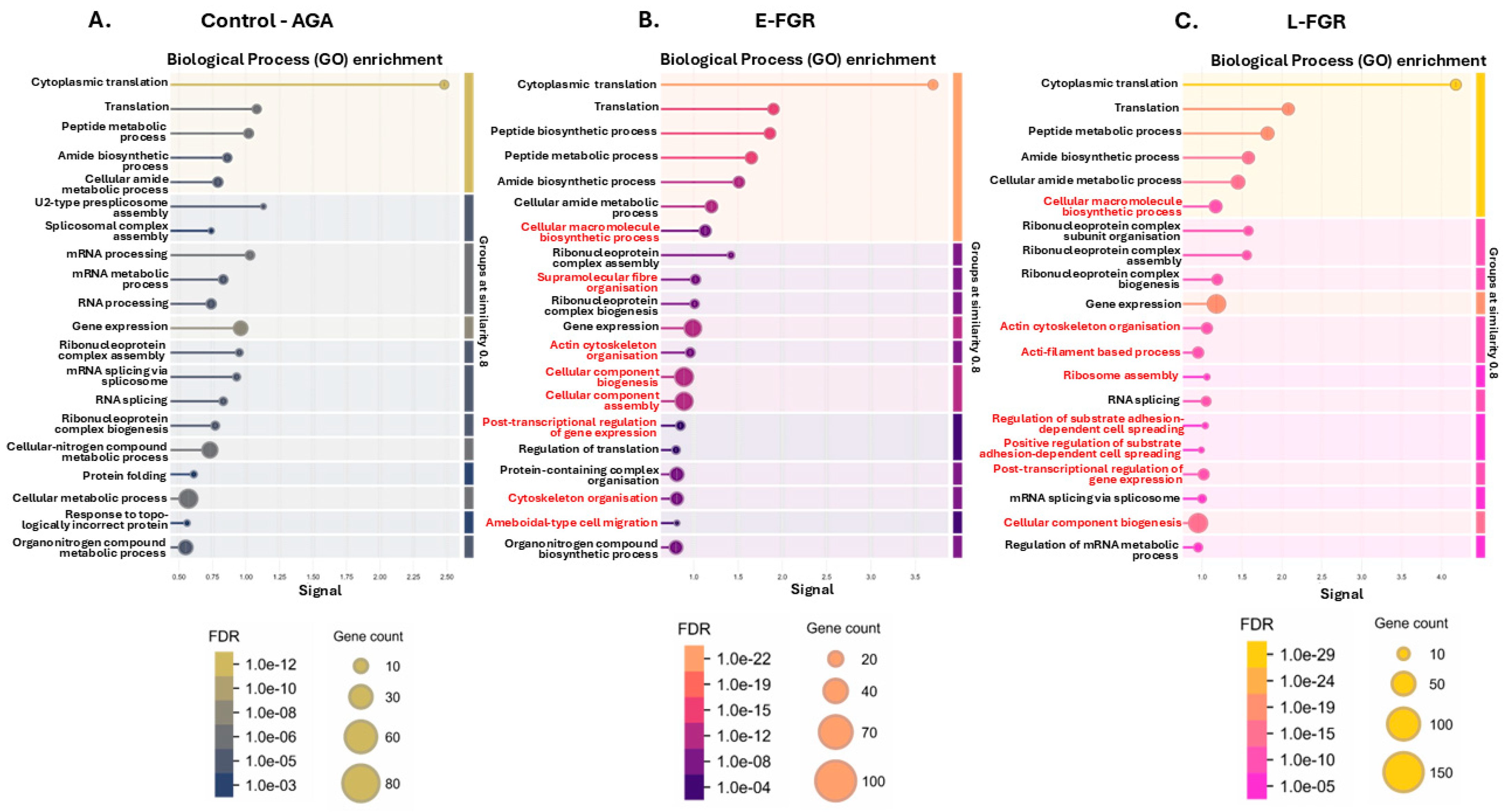
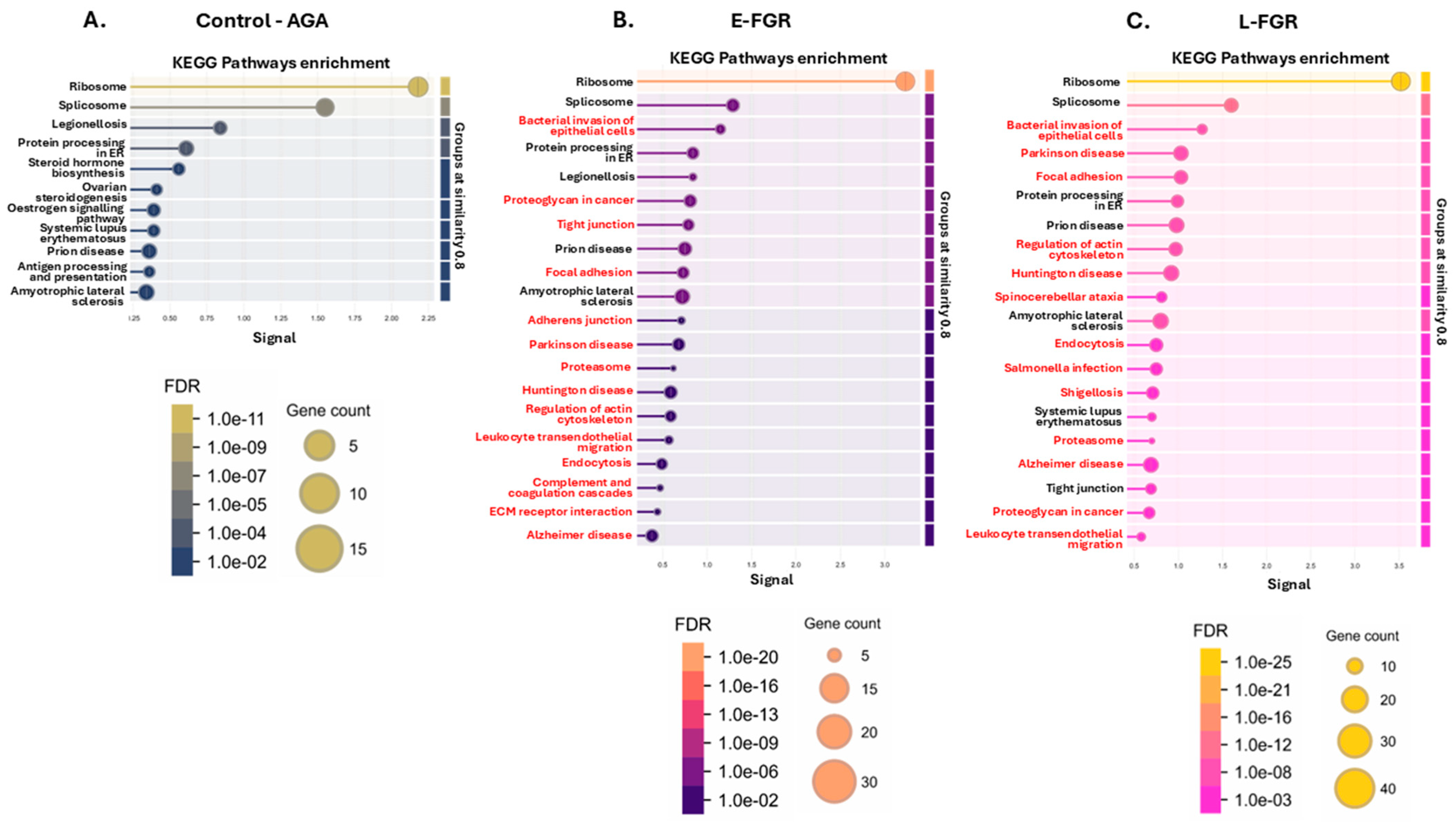
2.1. The Placental Protein Citrullinome Is Modified in FGR Compared with Control AGA Placentas
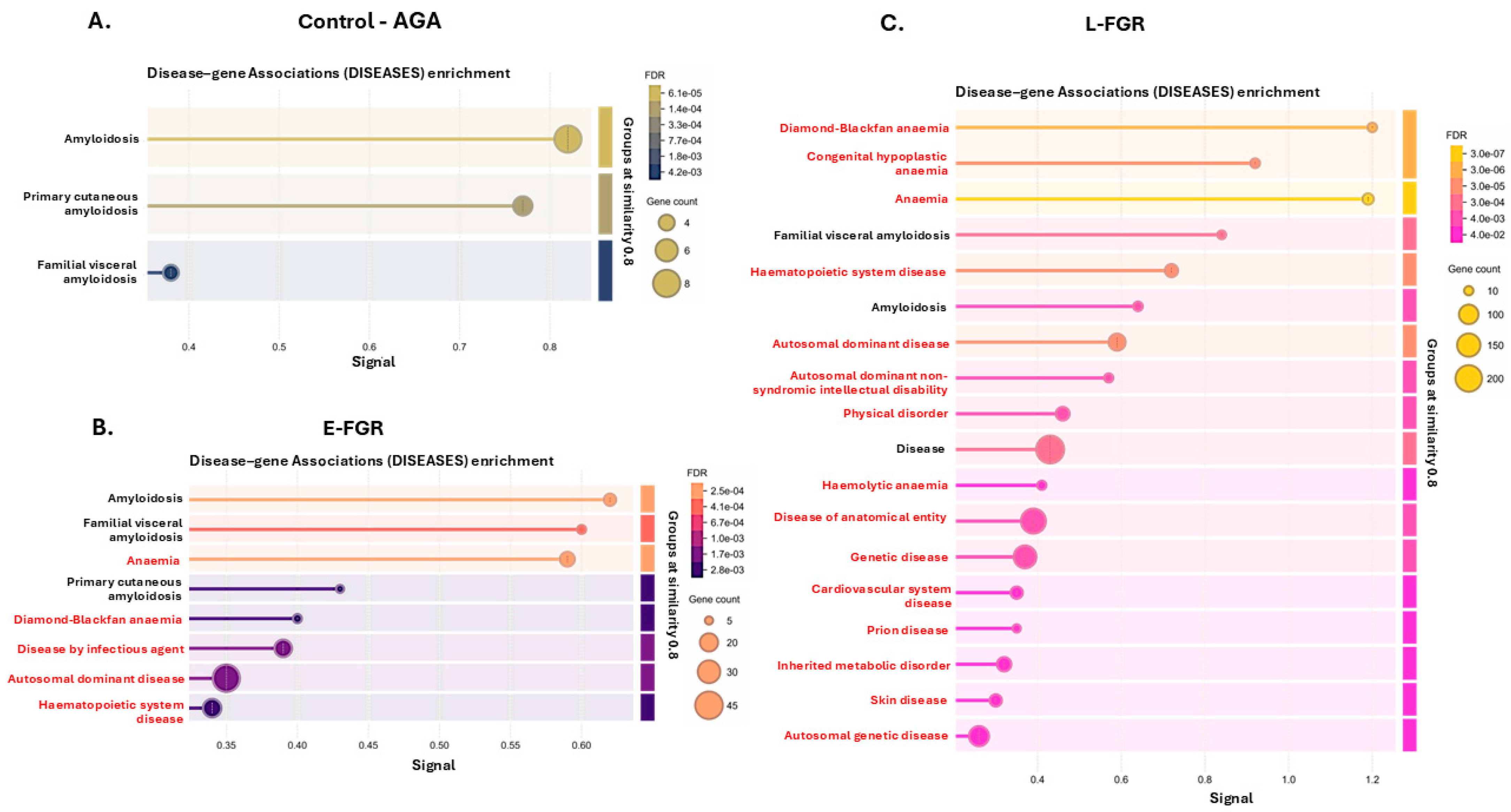
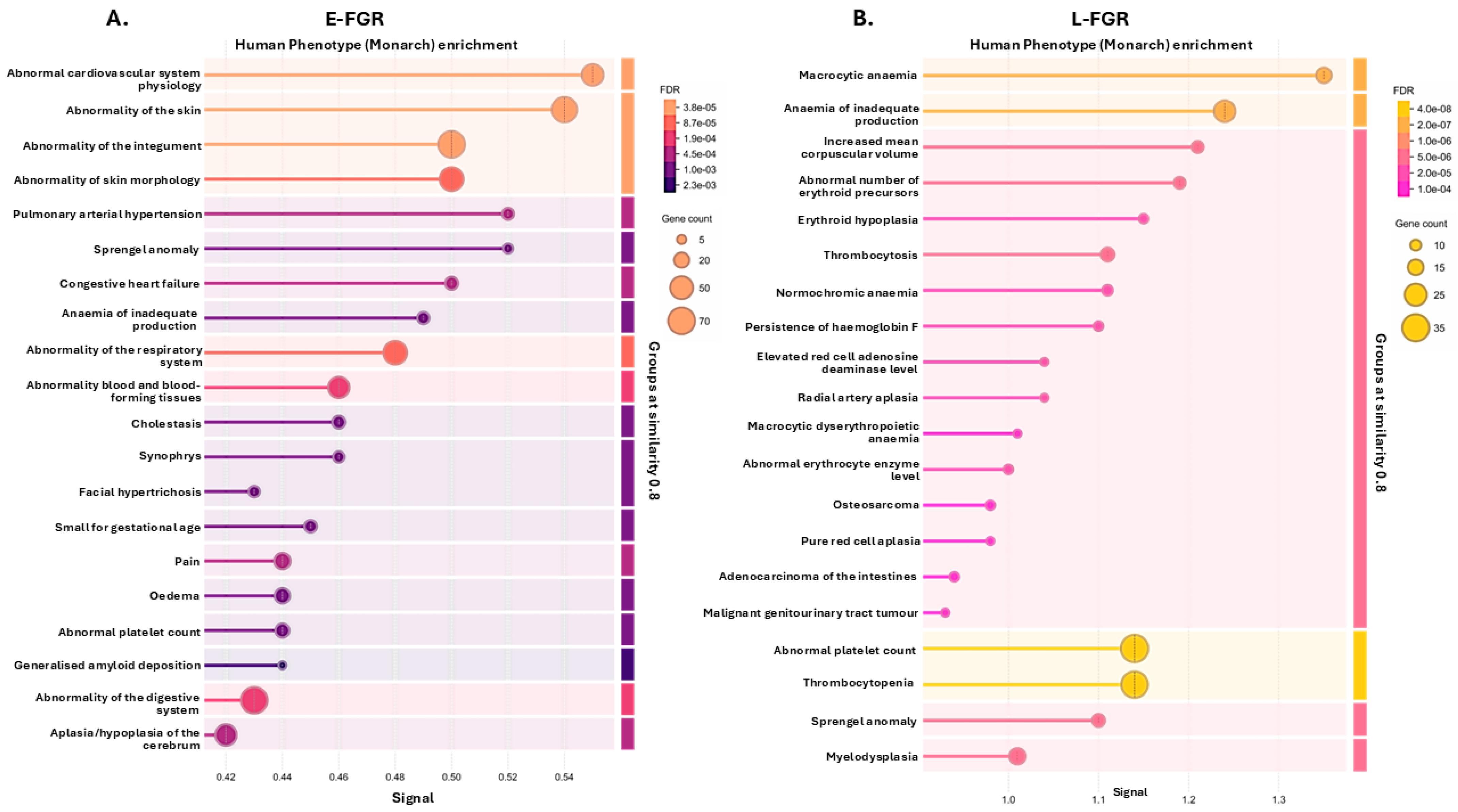
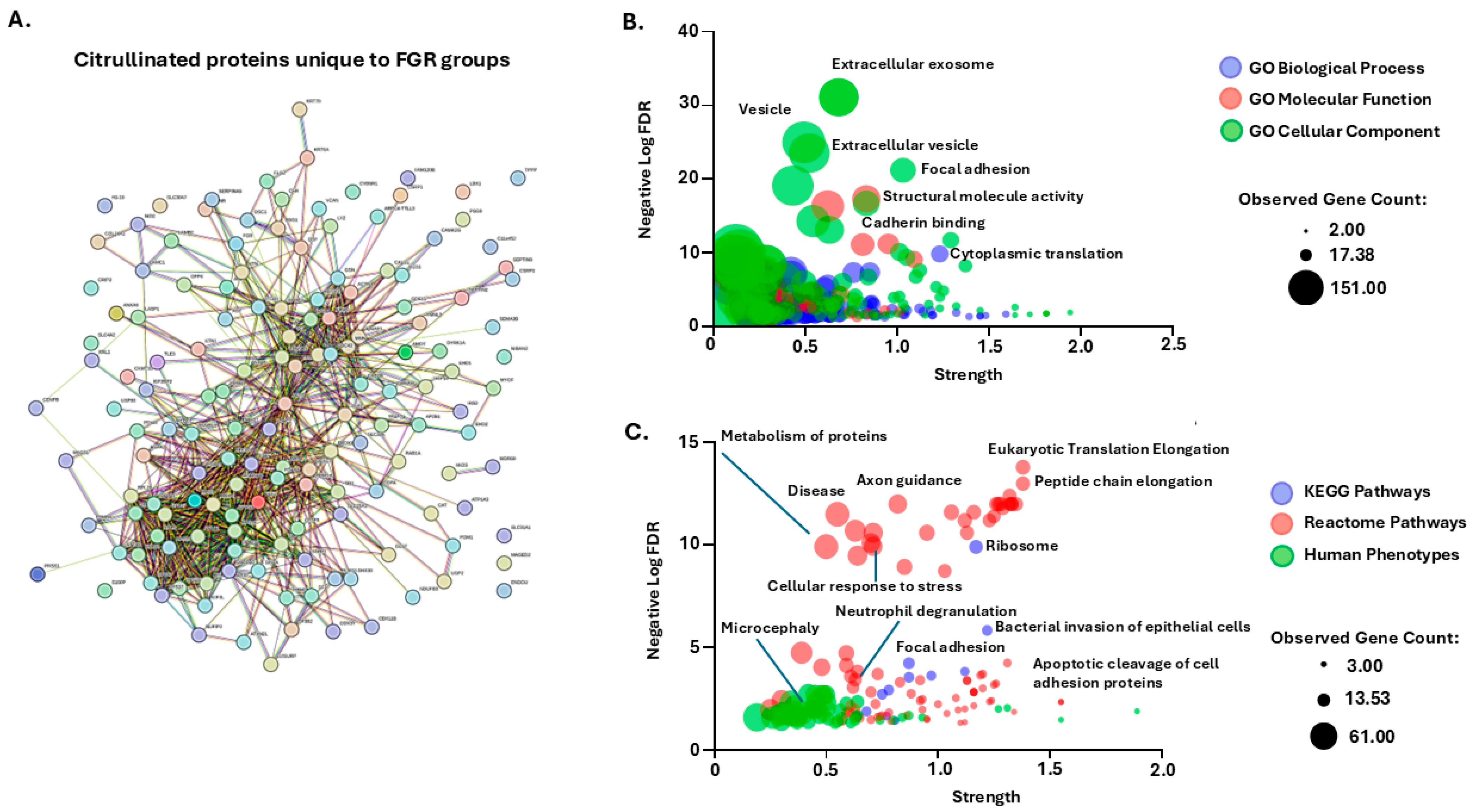
2.2. Citrullinated Proteins and Functional Enrichment Associations Identified in FGR Placentas Only
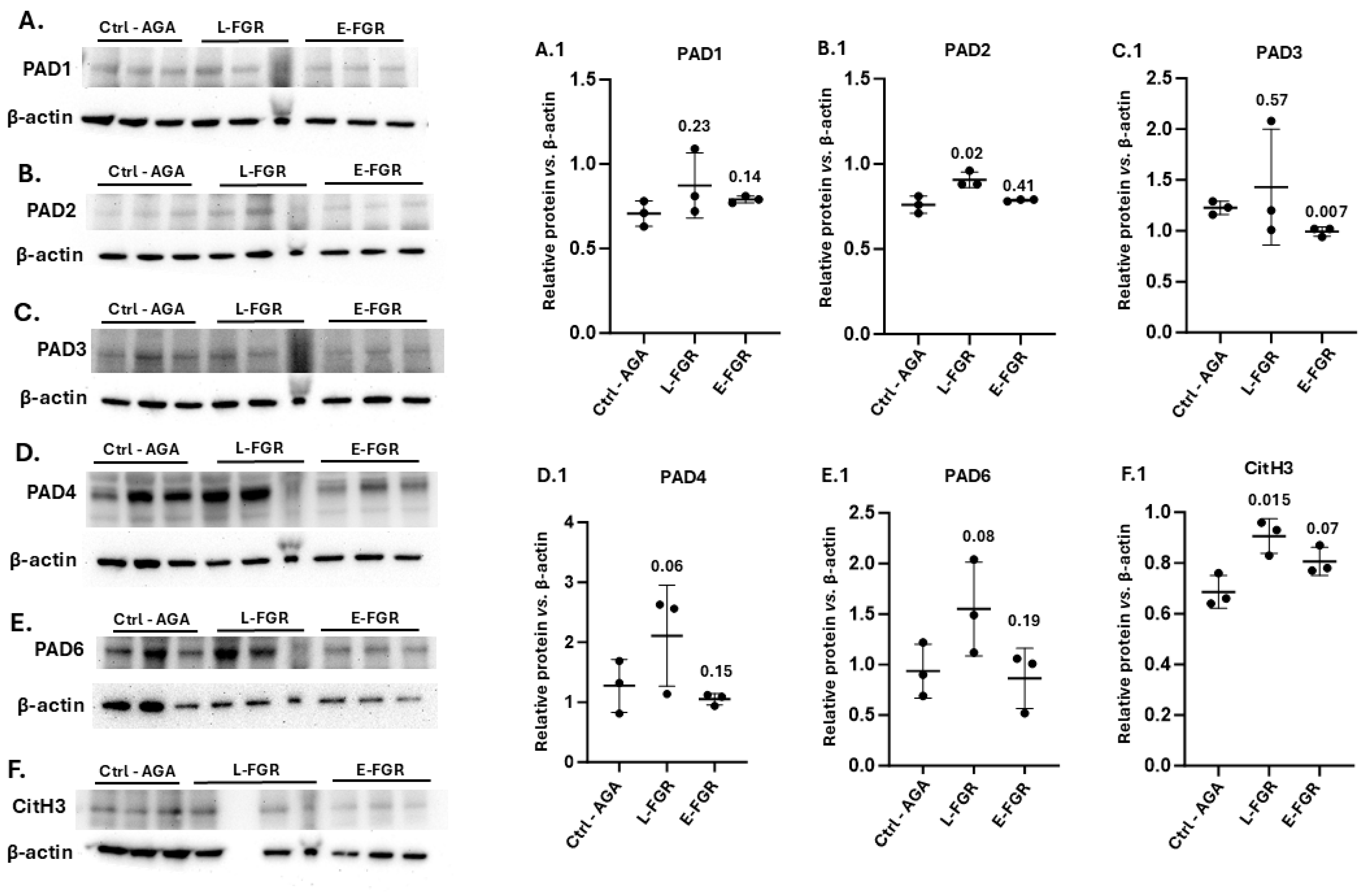
2.3. Placental PAD Isozyme Abundance and Histone H3 Citrullination Are Modified in FGR
3. Discussion
3.1. Possible Roles for Citrullination in Placental Function and Implications in FGR
3.2. Placental Citrullinome Associations to Disease and Phenotype Pathways in FGR
3.3. Histone H3 Citrullination in FGR
4. Materials and Methods
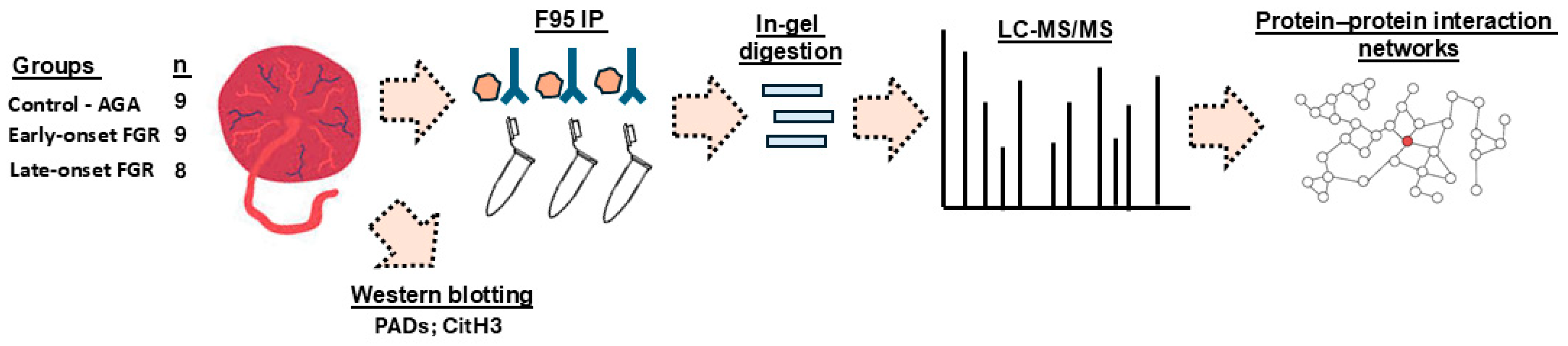
4.1. Samples and Sample Preparation
4.2. Isolation of Citrullinated Proteins from Placental Samples
4.3. Liquid Chromatography with Tandem Mass Spectrometry (LC-MS/MS)
4.4. Functional Enrichment and Protein–Protein Interaction (PPI) Network Analysis
4.5. Western Blotting
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Romo, A.; Carceller, R.; Tobajas, J. Intrauterine growth retardation (IUGR): Epidemiology and etiology. Pediatr. Endocrinol. Rev. 2009, 6 (Suppl. S3), 332–336. [Google Scholar] [PubMed]
- Sharma, D.; Shastri, S.; Sharma, P. Intrauterine Growth Restriction: Antenatal and Postnatal Aspects. Clin. Med. Insights Pediatr. 2016, 10, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Chew, L.C.; Obiyo, O.; Reed, D.J.; Verma, R.P. Fetal Growth Restriction. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2024; [Updated 8 August 2023]. Available online: https://www.ncbi.nlm.nih.gov/books/NBK562268/ (accessed on 22 March 2025).
- Groom, K.M.; David, A.L. The role of aspirin, heparin, and other interventions in the prevention and treatment of fetal growth restriction. Am. J. Obstet. Gynecol. 2018, 218, S829–S840. [Google Scholar] [CrossRef] [PubMed]
- Gordijn, S.J.; Beune, I.M.; Thilaganathan, B.; Papageorghiou, A.; Baschat, A.A.; Baker, P.N.; Silver, R.M.; Wynia, K.; Ganzevoort, W. Consensus definition of fetal growth restriction: A Delphi procedure. Ultrasound Obstet. Gynecol. 2016, 48, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Figueras, F.; Gratacós, E. Update on the diagnosis and classification of fetal growth restriction and proposal of a stage-based management protocol. Fetal Diagn. Ther. 2014, 36, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Aviram, A.; Sherman, C.; Kingdom, J.; Zaltz, A.; Barrett, J.; Melamed, N. Defining early vs late fetal growth restriction by placental pathology. Acta Obstet. Gynecol. Scand. 2019, 98, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Lingam, I.; Okell, J.; Maksym, K.; Spencer, R.; Peebles, D.; Buquis, G.; Ambler, G.; Morsing, E.; Ley, D.; Singer, D.; et al. Neonatal outcomes following early fetal growth restriction: A subgroup analysis of the EVERREST study. Arch. Dis. Child. Fetal Neonatal Ed. 2023, 108, 599–606. [Google Scholar] [CrossRef]
- Vaughan, O.R.; Maksym, K.; Silva, E.; Barentsen, K.; Anthony, R.V.; Brown, T.L.; Hillman, S.L.; Spencer, R.; David, A.L.; Rosario, F.J.; et al. Placenta-specific Slc38a2/SNAT2 knockdown causes fetal growth restriction in mice. Clin. Sci. 2021, 135, 2049–2066. [Google Scholar] [CrossRef]
- Mondal, S.; Thompson, P.R. Protein Arginine Deiminases (PADs): Biochemistry and Chemical Biology of Protein Citrullination. Acc. Chem. Res. 2019, 52, 818–832. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y. Peptidylarginine deiminases in citrullination, gene regulation, health and pathogenesis. Biochim. Biophys. Acta 2013, 1829, 1126–1135. [Google Scholar] [CrossRef]
- Witalison, E.E.; Thompson, P.R.; Hofseth, L.J. Protein Arginine Deiminases and Associated Citrullination: Physiological Functions and Diseases Associated with Dysregulation. Curr. Drug Targets 2015, 16, 700–710. [Google Scholar] [CrossRef] [PubMed]
- Alghamdi, M.; Alasmari, D.; Assiri, A.; Mattar, E.; Aljaddawi, A.A.; Alattas, S.G.; Redwan, E.M. An Overview of the Intrinsic Role of Citrullination in Autoimmune Disorders. J. Immunol. Res. 2019, 2019, 7592851. [Google Scholar] [CrossRef] [PubMed]
- Magnadóttir, B.; Hayes, P.; Hristova, M.; Bragason, B.T.; Nicholas, A.P.; Dodds, A.W.; Guðmundsdóttir, S.; Lange, S. Post-translational protein deimination in cod (Gadus morhua L.) ontogeny novel roles in tissue remodelling and mucosal immune defences? Dev. Comp. Immunol. 2018, 87, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Lange, S. Peptidylarginine deiminases and extracellular vesicles: Prospective drug targets and biomarkers in central nervous system diseases and repair. Neural Regen. Res. 2021, 16, 934–938. [Google Scholar] [CrossRef]
- Ciesielski, O.; Biesiekierska, M.; Panthu, B.; Soszyński, M.; Pirola, L.; Balcerczyk, A. Citrullination in the pathology of inflammatory and autoimmune disorders: Recent advances and future perspectives. Cell Mol. Life Sci. 2022, 79, 94. [Google Scholar] [CrossRef] [PubMed]
- Yuzhalin, A.E. Citrullination in Cancer. Cancer Res. 2019, 79, 1274–1284. [Google Scholar] [CrossRef]
- Barasa, L.; Thompson, P.R. Protein citrullination: Inhibition, identification and insertion. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2023, 378, 20220240. [Google Scholar] [CrossRef]
- Harada, K.; Carr, S.M.; Shrestha, A.; La Thangue, N.B. Citrullination and the protein code: Crosstalk between post-translational modifications in cancer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2023, 378, 20220243. [Google Scholar] [CrossRef] [PubMed]
- Yurttas, P.; Vitale, A.M.; Fitzhenry, R.J.; Cohen-Gould, L.; Wu, W.; Gossen, J.A.; Coonrod, S.A. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development 2008, 135, 2627–2636. [Google Scholar] [CrossRef]
- Xu, Y.; Shi, Y.; Fu, J.; Yu, M.; Feng, R.; Sang, Q.; Liang, B.; Chen, B.; Qu, R.; Li, B.; et al. Mutations in PADI6 Cause Female Infertility Characterized by Early Embryonic Arrest. Am. J. Hum. Genet. 2016, 99, 744–752. [Google Scholar] [CrossRef]
- Qian, J.; Nguyen, N.M.P.; Rezaei, M.; Huang, B.; Tao, Y.; Zhang, X.; Cheng, Q.; Yang, H.; Asangla, A.; Majewski, J.; et al. Biallelic PADI6 variants linking infertility, miscarriages, and hydatidiform moles. Eur. J. Hum. Genet. 2018, 26, 1007–1013. [Google Scholar] [CrossRef]
- Guerrin, M.; Ishigami, A.; Méchin, M.C.; Nachat, R.; Valmary, S.; Sebbag, M.; Simon, M.; Senshu, T.; Serre, G. cDNA cloning, gene organization and expression analysis of human peptidylarginine deiminase type I. Biochem. J. 2003, 370 Pt 1, 167–174. [Google Scholar] [CrossRef]
- Suryawanshi, H.; Morozov, P.; Straus, A.; Sahasrabudhe, N.; Max, K.E.A.; Garzia, A.; Kustagi, M.; Tuschl, T.; Williams, Z. A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 2018, 4, eaau4788. [Google Scholar] [CrossRef]
- Pique-Regi, R.; Romero, R.; Tarca, A.L.; Sendler, E.D.; Xu, Y.; Garcia-Flores, V.; Leng, Y.; Luca, F.; Hassan, S.S.; Gomez-Lopez, N. Single cell transcriptional signatures of the human placenta in term and preterm parturition. Elife 2019, 8, e52004. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.O.; Li, G.; Young, C.H.; Snow, B.; Khan, S.A.; DeVore, S.B.; Edwards, S.; Bouma, G.J.; Navratil, A.M.; Cherrington, B.D.; et al. Peptidylarginine deiminase enzymes and citrullinated proteins in female reproductive physiology and associated diseases. Biol. Reprod. 2022, 107, 1395–1410. [Google Scholar] [CrossRef] [PubMed]
- Erpenbeck, L.; Chowdhury, C.S.; Zsengellér, Z.K.; Gallant, M.; Burke, S.D.; Cifuni, S.; Hahn, S.; Wagner, D.D.; Karumanchi, S.A. PAD4 Deficiency Decreases Inflammation and Susceptibility to Pregnancy Loss in a Mouse Model. Biol. Reprod. 2016, 95, 132. [Google Scholar] [CrossRef]
- Lin, X.; Zhang, Y.; He, X.; Chen, Y.; Chen, N.; Liu, J.; Wang, M.; Li, Y.; Yang, H.; Fan, L.; et al. The Choline Metabolite TMAO Inhibits NETosis and Promotes Placental Development in GDM of Humans and Mice. Diabetes 2021, 70, 2250–2263. [Google Scholar] [CrossRef]
- Maronek, M.; Gardlik, R. The Citrullination-Neutrophil Extracellular Trap Axis in Chronic Diseases. J. Innate Immun. 2022, 14, 393–417. [Google Scholar] [CrossRef] [PubMed]
- Le Plénier, S.; Goron, A.; Sotiropoulos, A.; Archambault, E.; Guihenneuc, C.; Walrand, S.; Salles, J.; Jourdan, M.; Neveux, N.; Cynober, L.; et al. Citrulline directly modulates muscle protein synthesis via the PI3K/MAPK/4E-BP1 pathway in a malnourished state: Evidence from in vivo, ex vivo, and in vitro studies. Am. J. Physiol. Endocrinol. Metab. 2017, 312, E27–E36. [Google Scholar] [CrossRef]
- Vlaardingerbroek, H.; Van den Akker, C.H.P.; Schierbeek, H.; Vermes, A.; Duvekot, J.J.; Steegers, E.A.P.; Van Goudoever, J.B. Protein Synthesis Rates of Human Placentas Studied In Vivo at Different Gestational Ages Using Stable Isotopes. Arch. Dis. Child. 2008, 93, p113. [Google Scholar]
- Yung, H.W.; Calabrese, S.; Hynx, D.; Hemmings, B.A.; Cetin, I.; Charnock-Jones, D.S.; Burton, G.J. Evidence of placental translation inhibition and endoplasmic reticulum stress in the etiology of human intrauterine growth restriction. Am. J. Pathol. 2008, 173, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.F.; Fik, E.; Zamudio, S.; Illsley, N.P. Global protein synthesis in human trophoblast is resistant to inhibition by hypoxia. Placenta 2012, 33, 31–38. [Google Scholar] [CrossRef]
- Yung, H.W.; Cox, M.; Tissot van Patot, M.; Burton, G.J. Evidence of endoplasmic reticulum stress and protein synthesis inhibition in the placenta of non-native women at high altitude. FASEB J. 2012, 26, 1970–1981. [Google Scholar] [CrossRef] [PubMed]
- Quigley, E.B.; DeVore, S.B.; Khan, S.A.; Geisterfer, Z.M.; Rothfuss, H.M.; Sequoia, A.O.; Thompson, P.R.; Gatlin, J.C.; Cherrington, B.D.; Navratil, A.M. GnRH Induces Citrullination of the Cytoskeleton in Murine Gonadotrope Cells. Int. J. Mol. Sci. 2024, 25, 3181. [Google Scholar] [CrossRef]
- Jansson, T.; Castillo-Castrejon, M.; Gupta, M.B.; Powell, T.L.; Rosario, F.J. Down-regulation of placental Cdc42 and Rac1 links mTORC2 inhibition to decreased trophoblast amino acid transport in human intrauterine growth restriction. Clin. Sci. 2020, 134, 53–70. [Google Scholar] [CrossRef]
- Riquelme, G.; Vallejos, C.; de Gregorio, N.; Morales, B.; Godoy, V.; Berrios, M.; Bastías, N.; Rodríguez, C. Lipid rafts and cytoskeletal proteins in placental microvilli membranes from preeclamptic and IUGR pregnancies. J. Membr. Biol. 2011, 241, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.C.; Jansson, T.; Bale, T.L.; Powell, T.L. Maternal-fetal cross-talk via the placenta: Influence on offspring development and metabolism. Development 2023, 150, dev202088. [Google Scholar] [CrossRef]
- Hess, P.E.; Winn, V.D. Extracellular Vesicles in Preeclampsia: Can Small Packages Carry a Big Punch? Anesth. Analg. 2022, 134, 710–712. [Google Scholar] [CrossRef]
- Paul, N.; Sultana, Z.; Fisher, J.J.; Maiti, K.; Smith, R. Extracellular vesicles- crucial players in human pregnancy. Placenta 2023, 140, 30–38. [Google Scholar] [CrossRef]
- Liu, X.; Fei, H.; Yang, C.; Wang, J.; Zhu, X.; Yang, A.; Shi, Z.; Jin, X.; Yang, F.; Wu, D.; et al. Trophoblast-Derived Extracellular Vesicles Promote Preeclampsia by Regulating Macrophage Polarization. Hypertension 2022, 79, 2274–2287. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, C.S. Placenta Extracellular Vesicles: Messengers Connecting Maternal and Fetal Systems. Biomolecules 2024, 14, 995. [Google Scholar] [CrossRef]
- Miranda, J.; Paules, C.; Nair, S.; Lai, A.; Palma, C.; Scholz-Romero, K.; Rice, G.E.; Gratacos, E.; Crispi, F.; Salomon, C. Placental exosomes profile in maternal and fetal circulation in intrauterine growth restriction—Liquid biopsies to monitoring fetal growth. Placenta 2018, 64, 34–43. [Google Scholar] [CrossRef]
- Ariyakumar, G.; Morris, J.M.; McKelvey, K.J.; Ashton, A.W.; McCracken, S.A. NF-κB regulation in maternal immunity during normal and IUGR pregnancies. Sci. Rep. 2021, 11, 20971. [Google Scholar] [CrossRef] [PubMed]
- Villalobos-Labra, R.; Liu, R.; Spaans, F.; Sáez, T.; Semeria Maitret, T.; Quon, A.; Sawamura, T.; Cooke, C.M.; Davidge, S.T. Placenta-Derived Extracellular Vesicles From Preeclamptic Pregnancies Impair Vascular Endothelial Function via Lectin-Like Oxidized LDL Receptor-1. Hypertension 2023, 80, 2226–2238. [Google Scholar] [CrossRef] [PubMed]
- Villalobos-Labra, R.; Liu, R.; Spaans, F.; Sáez, T.; Semeria Maitret, T.; Quon, A.; Sawamura, T.; Cooke, C.M.; Davidge, S.T. Placenta-derived extracellular vesicles from preeclamptic and healthy pregnancies impair ex vivo vascular endothelial function. Biosci. Rep. 2022, 42, BSR20222185. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhang, Y.; Cai, X.; Zhang, Y.; Yan, S.; Wang, J.; Zhang, S.; Yin, T.; Yang, C.; Yang, J. Extracellular vesicles derived from M1 macrophages deliver miR-146a-5p and miR-146b-5p to suppress trophoblast migration and invasion by targeting TRAF6 in recurrent spontaneous abortion. Theranostics 2021, 11, 5813–5830. [Google Scholar] [CrossRef]
- Chaemsaithong, P.; Luewan, S.; Taweevisit, M.; Chiangjong, W.; Pongchaikul, P.; Thorner, P.S.; Tongsong, T.; Chutipongtanate, S. Placenta-Derived Extracellular Vesicles in Pregnancy Complications and Prospects on a Liquid Biopsy for Hemoglobin Bart’s Disease. Int. J. Mol. Sci. 2023, 24, 5658. [Google Scholar] [CrossRef] [PubMed]
- Farrelly, R.; Kennedy, M.G.; Spencer, R.; Forbes, K. Extracellular vesicles as markers and mediators of pregnancy complications: Gestational diabetes, pre-eclampsia, preterm birth and fetal growth restriction. J. Physiol. 2023, 601, 4973–4988. [Google Scholar] [CrossRef]
- Paul, N.; Maiti, K.; Sultana, Z.; Fisher, J.J.; Zhang, H.; Cole, N.; Morgan, T.; Smith, R. Human placenta releases extracellular vesicles carrying corticotrophin releasing hormone mRNA into the maternal blood. Placenta 2024, 146, 71–78. [Google Scholar] [CrossRef]
- Kholia, S.; Jorfi, S.; Thompson, P.R.; Causey, C.P.; Nicholas, A.P.; Inal, J.M.; Lange, S. A novel role for peptidylarginine deiminases in microvesicle release reveals therapeutic potential of PAD inhibition in sensitizing prostate cancer cells to chemotherapy. J. Extracell. Vesicles 2015, 4, 26192. [Google Scholar] [CrossRef]
- Kosgodage, U.S.; Trindade, R.P.; Thompson, P.R.; Inal, J.M.; Lange, S. Chloramidine/Bisindolylmaleimide-I-Mediated Inhibition of Exosome and Microvesicle Release and Enhanced Efficacy of Cancer Chemotherapy. Int. J. Mol. Sci. 2017, 18, 1007. [Google Scholar] [CrossRef] [PubMed]
- Kosgodage, U.S.; Uysal-Onganer, P.; MacLatchy, A.; Kraev, I.; Chatterton, N.P.; Nicholas, A.P.; Inal, J.M.; Lange, S. Peptidylarginine Deiminases Post-Translationally Deiminate Prohibitin and Modulate Extracellular Vesicle Release and MicroRNAs in Glioblastoma Multiforme. Int. J. Mol. Sci. 2018, 20, 103. [Google Scholar] [CrossRef] [PubMed]
- Lange, S.; Gallagher, M.; Kholia, S.; Kosgodage, U.S.; Hristova, M.; Hardy, J.; Inal, J.M. Peptidylarginine Deiminases-Roles in Cancer and Neurodegeneration and Possible Avenues for Therapeutic Intervention via Modulation of Exosome and Microvesicle (EMV) Release? Int. J. Mol. Sci. 2017, 18, 1196. [Google Scholar] [CrossRef] [PubMed]
- Sancandi, M.; Uysal-Onganer, P.; Kraev, I.; Mercer, A.; Lange, S. Protein Deimination Signatures in Plasma and Plasma-EVs and Protein Deimination in the Brain Vasculature in a Rat Model of Pre-Motor Parkinson’s Disease. Int. J. Mol. Sci. 2020, 21, 2743. [Google Scholar] [CrossRef] [PubMed]
- Uysal-Onganer, P.; MacLatchy, A.; Mahmoud, R.; Kraev, I.; Thompson, P.R.; Inal, J.M.; Lange, S. Peptidylarginine Deiminase Isozyme-Specific PAD2, PAD3 and PAD4 Inhibitors Differentially Modulate Extracellular Vesicle Signatures and Cell Invasion in Two Glioblastoma Multiforme Cell Lines. Int. J. Mol. Sci. 2020, 21, 1495. [Google Scholar] [CrossRef]
- Uysal-Onganer, P.; D’Alessio, S.; Mortoglou, M.; Kraev, I.; Lange, S. Peptidylarginine Deiminase Inhibitor Application, Using Cl-Amidine, PAD2, PAD3 and PAD4 Isozyme-Specific Inhibitors in Pancreatic Cancer Cells, Reveals Roles for PAD2 and PAD3 in Cancer Invasion and Modulation of Extracellular Vesicle Signatures. Int. J. Mol. Sci. 2021, 22, 1396. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, B.; Haider, S.; Meinhardt, G.; Pollheimer, J.; Knöfler, M. WNT and NOTCH signaling in human trophoblast development and differentiation. Cell Mol. Life Sci. 2022, 79, 292. [Google Scholar] [CrossRef]
- Huang, Z.; Huang, S.; Song, T.; Yin, Y.; Tan, C. Placental Angiogenesis in Mammals: A Review of the Regulatory Effects of Signaling Pathways and Functional Nutrients. Adv. Nutr. 2021, 12, 2415–2434. [Google Scholar] [CrossRef]
- Sipilä, K.; Haag, S.; Denessiouk, K.; Käpylä, J.; Peters, E.C.; Denesyuk, A.; Hansen, U.; Konttinen, Y.; Johnson, M.S.; Holmdahl, R.; et al. Citrullination of collagen II affects integrin-mediated cell adhesion in a receptor-specific manner. FASEB J. 2014, 28, 3758–3768. [Google Scholar] [CrossRef] [PubMed]
- Stefanelli, V.L.; Choudhury, S.; Hu, P.; Liu, Y.; Schwenzer, A.; Yeh, C.R.; Chambers, D.M.; von Beck, K.; Li, W.; Segura, T.; et al. Citrullination of fibronectin alters integrin clustering and focal adhesion stability promoting stromal cell invasion. Matrix Biol. 2019, 82, 86–104. [Google Scholar] [CrossRef] [PubMed]
- Adu-Gyamfi, E.A.; Czika, A.; Gorleku, P.N.; Ullah, A.; Panhwar, Z.; Ruan, L.L.; Ding, Y.B.; Wang, Y.X. The Involvement of Cell Adhesion Molecules, Tight Junctions, and Gap Junctions in Human Placentation. Reprod. Sci. 2021, 28, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Firan, M.; Bawdon, R.; Radu, C.; Ober, R.J.; Eaken, D.; Antohe, F.; Ghetie, V.; Ward, E.S. The MHC class I-related receptor, FcRn, plays an essential role in the maternofetal transfer of gamma-globulin in humans. Int. Immunol. 2001, 13, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, R.; Zampieri, M.; Mechelli, R.; Annibali, V.; Guastafierro, T.; Ciccarone, F.; Coarelli, G.; Umeton, R.; Salvetti, M.; Caiafa, P. Methylation-dependent PAD2 upregulation in multiple sclerosis peripheral blood. Mult. Scler. 2012, 18, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.S.; Rong, C.S.; Xu, H.M.; Sun, T.; Hou, J.; Xu, Y. Peptidyl Arginine Deiminase, Type II (PADI2) Is Involved in Urothelial Bladder Cancer. Pathol. Oncol. Res. 2020, 26, 1279–1285. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, I.O.; Parsi, S.; Ostrow, L.W.; Brown, R.H.; Thompson, P.R.; Xu, Z. PAD2 dysregulation and aberrant protein citrullination feature prominently in reactive astrogliosis and myelin protein aggregation in sporadic ALS. Neurobiol. Dis. 2024, 192, 106414. [Google Scholar] [CrossRef]
- Ballasy, N.N.; Bering, E.A.; Kokorudz, C.; Radford, B.N.; Zhao, X.; Dean, W.; Hemberger, M. Padi2/3 Deficiency Alters the Epigenomic Landscape and Causes Premature Differentiation of Mouse Trophoblast Stem Cells. Cells 2022, 11, 2466. [Google Scholar] [CrossRef]
- Hasmasanu, M.G.; Bolboaca, S.D.; Baizat, M.I.; Drugan, T.C.; Zaharie, G.C. Neonatal short-term outcomes in infants with intrauterine growth restriction. Saudi Med. J. 2015, 36, 947–953. [Google Scholar] [CrossRef]
- Lange, S.; Rocha-Ferreira, E.; Thei, L.; Mawjee, P.; Bennett, K.; Thompson, P.R.; Subramanian, V.; Nicholas, A.P.; Peebles, D.; Hristova, M.; et al. Peptidylarginine deiminases: Novel drug targets for prevention of neuronal damage following hypoxic ischemic insult (HI) in neonates. J. Neurochem. 2014, 130, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Lange, S. Peptidylarginine Deiminases as Drug Targets in Neonatal Hypoxic-Ischemic Encephalopathy. Front. Neurol. 2016, 7, 22. [Google Scholar] [CrossRef]
- Mitra, S.; Harvey-Jones, K.; Kraev, I.; Verma, V.; Meehan, C.; Mintoft, A.; Norris, G.; Campbell, E.; Tucker, K.; Robertson, N.J.; et al. The Extracellular Vesicle Citrullinome and Signature in a Piglet Model of Neonatal Seizures. Int. J. Mol. Sci. 2023, 24, 11529. [Google Scholar] [CrossRef] [PubMed]
- Aughwane, R.; Mufti, N.; Flouri, D.; Maksym, K.; Spencer, R.; Sokolska, M.; Kendall, G.; Atkinson, D.; Bainbridge, A.; Deprest, J.; et al. Magnetic resonance imaging measurement of placental perfusion and oxygen saturation in early-onset fetal growth restriction. BJOG 2021, 128, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Witters, G.; Van Robays, J.; Willekes, C.; Coumans, A.; Peeter, s.H.; Gyselaers, W.; Fryns, J.P. Trisomy 13, 18, 21, Triploidy and Turner syndrome: The 5T’s. Look at the hands. Facts Views Vis. Obgyn. 2011, 3, 15–21. [Google Scholar] [PubMed]
- Valsamakis, G.; Kanaka-Gantenbein, C.; Malamitsi-Puchner, A.; Mastorakos, G. Causes of intrauterine growth restriction and the postnatal development of the metabolic syndrome. Ann. N. Y. Acad. Sci. 2006, 1092, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Crispi, F.; Miranda, J.; Gratacós, E. Long-term cardiovascular consequences of fetal growth restriction: Biology, clinical implications, and opportunities for prevention of adult disease. Am. J. Obstet. Gynecol. 2018, 218, S869–S879. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.K.; Brown, L.D.; Rozance, P.J.; Serkova, N.J.; Hay, W.W.; Friedman, J.E.; Wesolowski, S.R. Differential effects of intrauterine growth restriction and a hypersinsulinemic-isoglycemic clamp on metabolic pathways and insulin action in the fetal liver. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2019, 316, R427–R440. [Google Scholar] [CrossRef] [PubMed]
- Buhimschi, I.A.; Nayeri, U.A.; Zhao, G.; Shook, L.L.; Pensalfini, A.; Funai, E.F.; Bernstein, I.M.; Glabe, C.G.; Buhimschi, C.S. Protein misfolding, congophilia, oligomerization, and defective amyloid processing in preeclampsia. Sci. Transl. Med. 2014, 6, 245ra92. [Google Scholar] [CrossRef]
- Venturi, G.; Montanaro, L. How Altered Ribosome Production Can Cause or Contribute to Human Disease: The Spectrum of Ribosomopathies. Cells 2020, 9, 2300. [Google Scholar] [CrossRef]
- Melamed, N.; Baschat, A.; Yinon, Y.; Athanasiadis, A.; Mecacci, F.; Figueras, F.; Berghella, V.; Nazareth, A.; Tahlak, M.; McIntyre, H.D.; et al. FIGO (international Federation of Gynecology and obstetrics) initiative on fetal growth: Best practice advice for screening, diagnosis, and management of fetal growth restriction. Int. J. Gynaecol. Obstet. 2021, 152 (Suppl. S1), 3–57. [Google Scholar] [CrossRef]
- Hokken-Koelega, A.C.S.; van der Steen, M.; Boguszewski, M.C.S.; Cianfarani, S.; Dahlgren, J.; Horikawa, R.; Mericq, V.; Rapaport, R.; Alherbish, A.; Braslavsky, D.; et al. International Consensus Guideline on Small for Gestational Age: Etiology and Management From Infancy to Early Adulthood. Endocr. Rev. 2023, 44, 539–565. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, X.; Yu, F.L.; Kong, X.M.; Lin, N.; Liu, C.S.; Liu, T.T.; Guan, J.C. Teratogenicity of Staphylococcus aureus L-forms using a mouse whole-embryo culture model. J. Med. Microbiol. 2013, 62 Pt 5, 677–682. [Google Scholar] [CrossRef]
- Flannery, D.D.; Edwards, E.M.; Coggins, S.A.; Horbar, J.D.; Puopolo, K.M. Late-Onset Sepsis Among Very Preterm Infants. Pediatrics 2022, 150, e2022058813. [Google Scholar] [CrossRef]
- Huncikova, Z.; Vatne, A.; Stensvold, H.J.; Lang, A.M.; Støen, R.; Brigtsen, A.K.; Salvesen, B.; Øymar, K.A.A.; Rønnestad, A.; Klingenberg, C.; et al. Late-onset sepsis in very preterm infants in Norway in 2009–2018: A population-based study. Arch. Dis. Child. Fetal Neonatal Ed. 2023, 108, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Madhi, S.A.; Briner, C.; Maswime, S.; Mose, S.; Mlandu, P.; Chawana, R.; Wadula, J.; Adam, Y.; Izu, A.; Cutland, C.L. Causes of stillbirths among women from South Africa: A prospective, observational study. Lancet Glob. Health 2019, 7, e503–e512. [Google Scholar] [CrossRef] [PubMed]
- Betancourt, D.M.; Llana, M.N.; Sarnacki, S.H.; Cerquetti, M.C.; Monzalve, L.S.; Pustovrh, M.C.; Giacomodonato, M.N. Salmonella enteritidis foodborne infection induces altered placental morphometrics in the murine model. Placenta 2021, 109, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Noto Llana, M.; Sarnacki, S.H.; Aya Castañeda, M.delR.; Pustovrh, M.C.; Gartner, A.S.; Buzzola, F.R.; Cerquetti, M.C.; Giacomodonato, M.N. Salmonella enterica serovar Enteritidis enterocolitis during late stages of gestation induces an adverse pregnancy outcome in the murine model. PLoS ONE 2014, 9, e111282. [Google Scholar] [CrossRef]
- Rebarber, A.; Star Hampton, B.; Lewis, V.; Bender, S. Shigellosis complicating preterm premature rupture of membranes resulting in congenital infection and preterm delivery. Obstet. Gynecol. 2002, 100 Pt 2, 1063–1065. [Google Scholar] [CrossRef]
- Parisot, M.; Jolivet, A.; Boukhari, R.; Carles, G. Shigellosis and Pregnancy in French Guiana: Obstetric and Neonatal Complications. Am. J. Trop. Med. Hyg. 2016, 95, 26–30. [Google Scholar] [CrossRef]
- Platt-Samoraj, A.; Szweda, W.; Ugorski, M. Isolation of Yersinia enterocolitica from aborted fetuses and sows in pig farms with reproductive disturbances. Pol. J. Vet. Sci. 2009, 12, 189–193. [Google Scholar]
- Asarkua, H.; Fukami, T.; Inagaki, T.; Tateyama, N. Serious Influence of Yersinia Enterocolitis on Pregnancy in a Woman Complicated With Chronic Hypertension and Gestational Diabetes Mellitus: A Case Report. J. Preg Child. Health 2015, 2, 135. [Google Scholar] [CrossRef]
- Zhao, T.; Pan, B.; Alam, H.B.; Liu, B.; Bronson, R.T.; Deng, Q.; Wu, E.; Li, Y. Protective effect of Cl-amidine against CLP-induced lethal septic shock in mice. Sci. Rep. 2016, 6, 36696. [Google Scholar] [CrossRef]
- Biron, B.M.; Chung, C.S.; O’Brien, X.M.; Chen, Y.; Reichner, J.S.; Ayala, A. Cl-Amidine Prevents Histone 3 Citrullination and Neutrophil Extracellular Trap Formation, and Improves Survival in a Murine Sepsis Model. J. Innate Immun. 2017, 9, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Pan, B.; Alam, H.B.; Deng, Q.; Wang, Y.; Chen, E.; Liu, B.; Tian, Y.; Williams, A.M.; Duan, X.; et al. Inhibition of peptidylarginine deiminase alleviates LPS-induced pulmonary dysfunction and improves survival in a mouse model of lethal endotoxemia. Eur. J. Pharmacol. 2018, 833, 432–440. [Google Scholar] [CrossRef]
- Kosgodage, U.S.; Matewele, P.; Mastroianni, G.; Kraev, I.; Brotherton, D.; Awamaria, B.; Nicholas, A.P.; Lange, S.; Inal, J.M. Peptidylarginine Deiminase Inhibitors Reduce Bacterial Membrane Vesicle Release and Sensitize Bacteria to Antibiotic Treatment. Front. Cell Infect. Microbiol. 2019, 9, 227. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.J.; Cooper, C. Early environmental factors and rheumatoid arthritis. Clin. Exp. Immunol. 2006, 143, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.J.; Cooper, C. Early environmental exposure and the development of lupus. Lupus 2006, 15, 814–819. [Google Scholar] [CrossRef]
- Mosing, M.A.; Lundholm, C.; Cnattingius, S.; Gatz, M.; Pedersen, N.L. Associations between birth characteristics and age-related cognitive impairment and dementia: A registry-based cohort study. PLoS Med. 2018, 15, e1002609. [Google Scholar] [CrossRef]
- Shah, D.K.; Pereira, S.; Lodygensky, G.A. Long-Term Neurologic Consequences following Fetal Growth Restriction: The Impact on Brain Reserve. Dev. Neurosci. 2024, 47, 139–146. [Google Scholar] [CrossRef]
- Mercer, A.; Sancandi, M.; Maclatchy, A.; Lange, S. Brain-Region-Specific Differences in Protein Citrullination/Deimination in a Pre-Motor Parkinson’s Disease Rat Model. Int. J. Mol. Sci. 2024, 25, 11168. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Wambua, S.; Lee, S.I.; Okoth, K.; Wang, Z.; Fazla, F.; Fayaz, A.; Eastwood, K.A.; Nelson-Piercy, C.; Nirantharakumar, K.; et al. Autoimmune diseases and adverse pregnancy outcomes: An umbrella review. Lancet 2023, 402 (Suppl. S1), S84. [Google Scholar] [CrossRef] [PubMed]
- Bruggeman, Y.; Sodré, F.M.C.; Buitinga, M.; Mathieu, C.; Overbergh, L.; Kracht, M.J.L. Targeting citrullination in autoimmunity: Insights learned from preclinical mouse models. Expert Opin. Ther. Targets 2021, 25, 269–281. [Google Scholar] [CrossRef]
- Ghozzi, M.; Mankai, A.; Melayah, S.; Mechi, F.; Chedly, Z.; Trabelsi, A.; Ghedira, I. Frequency of serological markers of rheumatoid arthritis in patients with Hashimoto’s thyroiditis. Lab. Med. 2024, 55, 433–438. [Google Scholar] [CrossRef]
- Prasad, D.; Prasad, R.V.; Choudhary, M.K.; Kumari, K. Cardiovascular disease in pregnancy and its outcome: A prospective study. J. Fam. Med. Prim. Care 2023, 12, 2714–2720. [Google Scholar] [CrossRef]
- Poon, L.C.; Shennan, A.; Hyett, J.A.; Kapur, A.; Hadar, E.; Divakar, H.; McAuliffe, F.; da Silva Costa, F.; von Dadelszen, P.; McIntyre, H.D.; et al. The International Federation of Gynecology and Obstetrics (FIGO) initiative on pre-eclampsia: A pragmatic guide for first-trimester screening and prevention. Int. J. Gynaecol. Obstet. 2019, 145 (Suppl. S1), 1–33. [Google Scholar] [CrossRef]
- Wu, Z.; Deng, Q.; Pan, B.; Alam, H.B.; Tian, Y.; Bhatti, U.F.; Liu, B.; Mondal, S.; Thompson, P.R.; Li, Y. Inhibition of PAD2 Improves Survival in a Mouse Model of Lethal LPS-Induced Endotoxic Shock. Inflammation 2020, 43, 1436–1445. [Google Scholar] [CrossRef] [PubMed]
- Carnevale, R.; Leopizzi, M.; Dominici, M.; d’Amati, G.; Bartimoccia, S.; Nocella, C.; Cammisotto, V.; D’Amico, A.; Castellani, V.; Baratta, F.; et al. PAD4-Induced NETosis Via Cathepsin G-Mediated Platelet-Neutrophil Interaction in ChAdOx1 Vaccine-Induced Thrombosis-Brief Report. Arterioscler. Thromb. Vasc. Biol. 2023, 43, e396–e403. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, U.M.; Hall, D.L.; Rawls, A.Z.; Alexander, B.T. Epigenetic processes during preeclampsia and effects on fetal development and chronic health. Clin. Sci. 2021, 135, 2307–2327. [Google Scholar] [CrossRef] [PubMed]
- Salmeri, N.; Carbone, I.F.; Cavoretto, P.I.; Farina, A.; Morano, D. Epigenetics Beyond Fetal Growth Restriction: A Comprehensive Overview. Mol. Diagn. Ther. 2022, 26, 607–626. [Google Scholar] [CrossRef]
- Wang, J.; Wu, Z.; Li, D.; Li, N.; Dindot, S.V.; Satterfield, M.C.; Bazer, F.W.; Wu, G. Nutrition, epigenetics, and metabolic syndrome. Antioxid. Redox Signal 2012, 17, 282–301. [Google Scholar] [CrossRef]
- Nelissen, E.C.; van Montfoort, A.P.; Dumoulin, J.C.; Evers, J.L. Epigenetics and the placenta. Hum. Reprod. Update 2011, 17, 397–417. [Google Scholar] [CrossRef]
- Ji, Y.; Wu, Z.; Dai, Z.; Wang, X.; Li, J.; Wang, B.; Wu, G. Fetal and neonatal programming of postnatal growth and feed efficiency in swine. J. Anim. Sci. Biotechnol. 2017, 8, 42. [Google Scholar] [CrossRef]
- Goyal, D.; Limesand, S.W.; Goyal, R. Epigenetic responses and the developmental origins of health and disease. J. Endocrinol. 2019, 242, T105–T119. [Google Scholar] [CrossRef]
- Meister, S.; Kolben, T.; Beyer, S.; Hutter, S.; Hofmann, S.; Kuhn, C.; Messner, J.; Mayr, D.; Solano, M.E.; Jegen, M.; et al. Sex-specific epigenetic gene activation profiles are differentially modulated in human placentas affected by intrauterine growth restriction. J. Reprod. Immunol. 2020, 139, 103124. [Google Scholar] [CrossRef]
- Stoikou, M.; Grimolizzi, F.; Giaglis, S.; Schäfer, G.; van Breda, S.V.; Hoesli, I.M.; Lapaire, O.; Huhn, E.A.; Hasler, P.; Rossi, S.W.; et al. Gestational Diabetes Mellitus Is Associated with Altered Neutrophil Activity. Front. Immunol. 2017, 8, 702. [Google Scholar] [CrossRef] [PubMed]
- Hahn, S.; Giaglis, S.; Hoesli, I.; Hasler, P. Neutrophil NETs in reproduction: From infertility to preeclampsia and the possibility of fetal loss. Front. Immunol. 2012, 3, 362. [Google Scholar] [CrossRef] [PubMed]
- Moodley, M.; Moodley, J.; Naicker, T. Placental neutrophil reverse trans-migration and maternal serum neutrophil extracellular trap expression in HIV infection co-morbid pre-eclampsia in women of African ancestry. Histochem. Cell Biol. 2024, 162, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Arisan, E.D.; Uysal-Onganer, P.; Lange, S. Putative Roles for Peptidylarginine Deiminases in COVID-19. Int. J. Mol. Sci. 2020, 21, 4662. [Google Scholar] [CrossRef] [PubMed]
- Veras, F.P.; Pontelli, M.C.; Silva, C.M.; Toller-Kawahisa, J.E.; de Lima, M.; Nascimento, D.C.; Schneider, A.H.; Caetité, D.; Tavares, L.A.; Paiva, I.M.; et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 2020, 217, e20201129. [Google Scholar] [CrossRef] [PubMed]
- Auriti, C.; De Rose, D.U.; Santisi, A.; Martini, L.; Piersigilli, F.; Bersani, I.; Ronchetti, M.P.; Caforio, L. Pregnancy and viral infections: Mechanisms of fetal damage, diagnosis and prevention of neonatal adverse outcomes from cytomegalovirus to SARS-CoV-2 and Zika virus. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166198. [Google Scholar] [CrossRef]
- Moltner, S.; de Vrijer, B.; Banner, H. Placental infarction and intrauterine growth restriction following SARS-CoV-2 infection. Arch. Gynecol. Obstet. 2021, 304, 1621–1622. [Google Scholar] [CrossRef]
- Smith, E.R.; Oakley, E.; Grandner, G.W.; Ferguson, K.; Farooq, F.; Afshar, Y.; Ahlberg, M.; Ahmadzia, H.; Akelo, V.; Aldrovandi, G.; et al. Adverse maternal, fetal, and newborn outcomes among pregnant women with SARS-CoV-2 infection: An individual participant data meta-analysis. BMJ Glob. Health 2023, 8, e009495. [Google Scholar] [CrossRef]
- Mitta, M.; Holt, L.; Chandrasekaran, S.; Dude, C. The association between parental SARS-CoV-2 infection in pregnancy and fetal growth restriction. J. Perinat. Med. 2024, 52, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Hu, X.; Wang, X. Crossing epigenetic frontiers: The intersection of novel histone modifications and diseases. Signal Transduct. Target. Ther. 2024, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Spencer, R.; Ambler, G.; Brodszki, J.; Diemert, A.; Figueras, F.; Gratacós, E.; Hansson, S.R.; Hecher, K.; Huertas-Ceballos, A.; Marlow, N.; et al. EVERREST prospective study: A 6-year prospective study to define the clinical and biological characteristics of pregnancies affected by severe early onset fetal growth restriction. BMC Pregnancy Childbirth 2017, 17, 43. [Google Scholar] [CrossRef] [PubMed]
- D’Alessio, S.; Cheng, H.; Eaton, L.; Kraev, I.; Pamenter, M.E.; Lange, S. Acute Hypoxia Alters Extracellular Vesicle Signatures and the Brain Citrullinome of Naked Mole-Rats (Heterocephalus glaber). Int. J. Mol. Sci. 2022, 23, 4683. [Google Scholar] [CrossRef] [PubMed]
- Jha, P.; Rajoria, P.; Poonia, P.; Chopra, M. Identification of novel PAD2 inhibitors using pharmacophore-based virtual screening, molecular docking, and MD simulation studies. Sci. Rep. 2024, 14, 28097. [Google Scholar] [CrossRef]

| Control | n | Early-Onset FGR | n | Late-Onset FGR | n | p Value | |
|---|---|---|---|---|---|---|---|
| Ethnicity (white) | 7 (78%) | 9 | 5 (56%) | 9 | 3 (38%) | 8 | 0.291 |
| Age (year) | 35.8 ± 4.6 | 9 | 31.8 ± 4.6 | 9 | 31.0 ± 5.7 | 8 | 0.119 |
| Body mass index (kg/m2) | 25.4 ± 5.1 | 9 | 29.7 ± 12.7 | 9 | 27.4 ± 8.2 | 8 | 0.627 |
| Gestation at delivery (wk) | 39.4 ± 0.8 a | 9 | 30.8 ± 5.2 b | 9 | 37.4 ± 1.7 a | 8 | <0.001 |
| Delivery mode (c-section) | 8 (89%) | 9 | 9 (100%) | 9 | 7 (88%) | 8 | 0.751 |
| Infant sex (female) | 4 (44%) | 9 | 5 (56%) | 9 | 3 (38%) | 8 | 0.885 |
| Birth weight (g) | 3411 ± 386 a | 9 | 1040 ± 679 b | 9 | 2151 ± 467 c | 8 | <0.001 |
| Placenta weight (g) | 472 ± 44 a | 5 | 219 ± 108 b | 4 | 330 ± 84 b | 6 | 0.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vaughan, O.R.; Maksym, K.; Hillman, S.; Spencer, R.N.; Hristova, M.; David, A.L.; Lange, S. Placental Protein Citrullination Signatures Are Modified in Early- and Late-Onset Fetal Growth Restriction. Int. J. Mol. Sci. 2025, 26, 4247. https://doi.org/10.3390/ijms26094247
Vaughan OR, Maksym K, Hillman S, Spencer RN, Hristova M, David AL, Lange S. Placental Protein Citrullination Signatures Are Modified in Early- and Late-Onset Fetal Growth Restriction. International Journal of Molecular Sciences. 2025; 26(9):4247. https://doi.org/10.3390/ijms26094247
Chicago/Turabian StyleVaughan, Owen R., Kasia Maksym, Sara Hillman, Rebecca N. Spencer, Mariya Hristova, Anna L. David, and Sigrun Lange. 2025. "Placental Protein Citrullination Signatures Are Modified in Early- and Late-Onset Fetal Growth Restriction" International Journal of Molecular Sciences 26, no. 9: 4247. https://doi.org/10.3390/ijms26094247
APA StyleVaughan, O. R., Maksym, K., Hillman, S., Spencer, R. N., Hristova, M., David, A. L., & Lange, S. (2025). Placental Protein Citrullination Signatures Are Modified in Early- and Late-Onset Fetal Growth Restriction. International Journal of Molecular Sciences, 26(9), 4247. https://doi.org/10.3390/ijms26094247







