The 2SP Site Mutation in the Bovine Natural Resistance-Associated Macrophage 1 Promoter Exhibits Antituberculosis Potential
Abstract
1. Introduction
2. Results
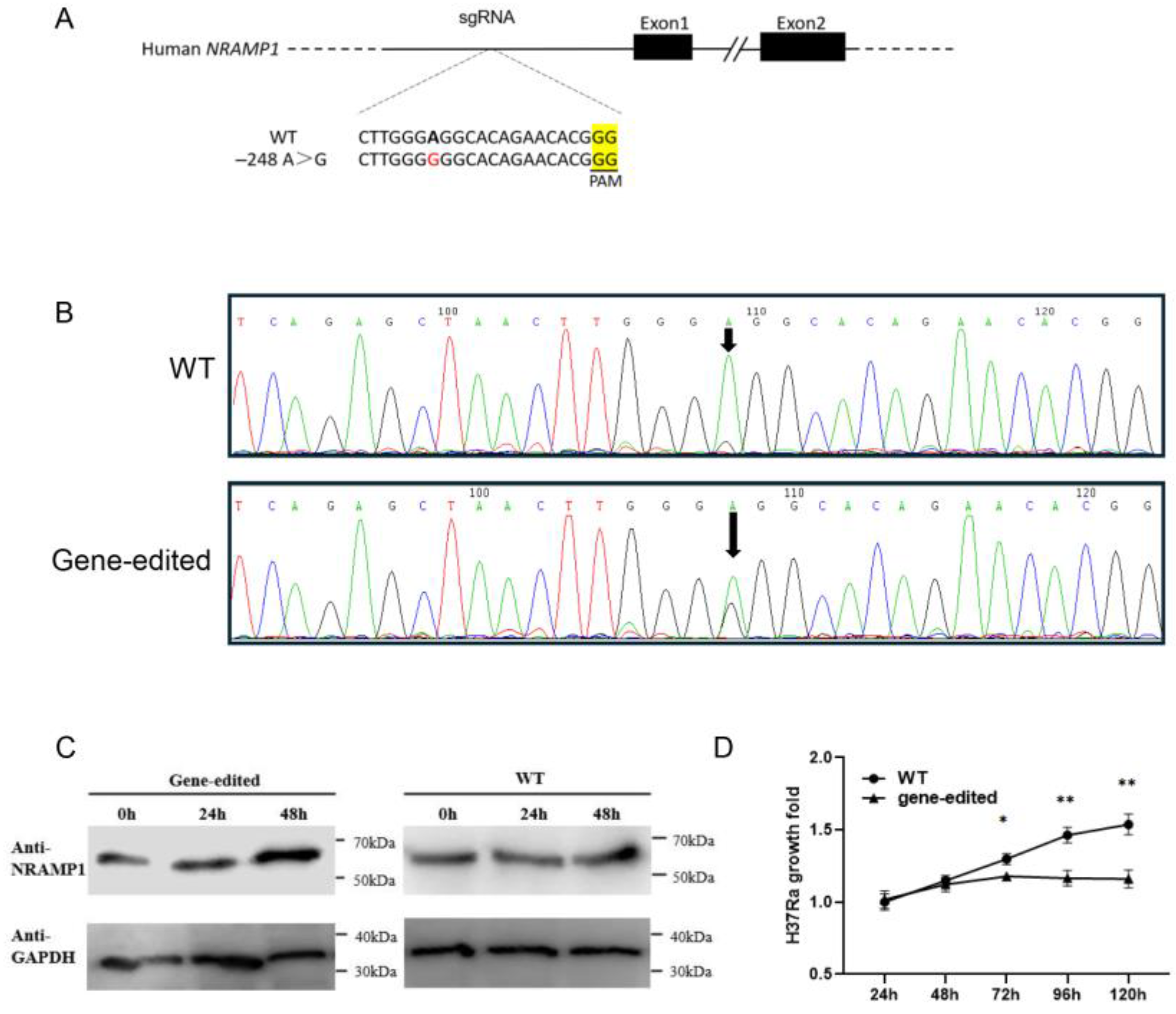
2.1. Detection of the Core Promoter of Bovine NRAMP1 and Comparative Analysis Between Mouse and Cattle
2.2. Mutation at the 2SP Site Specifically Increases NRAMP1 Promoter Activity After H37Ra Infection
2.3. Altering the Affinity of the 2SP Site for SP1/SP3 Affects NRAMP1 Promoter Activity After H37Ra Infection
2.4. SP1 and SP3 Play Different Roles in Regulating the Activity of the Nramp1 Promoter
2.5. Validating the Impact of the 2SP Site Mutations on the SP1/SP3-Mediated Regulation of NRAMP1 Expression
2.6. Base Editing of the Homologous Region of the 2SP Site in THP-1 Cells and Phenotypic Verification
3. Discussion
4. Materials and Methods
4.1. qPCR
4.2. Construction of Vectors
4.3. Cell Culture
4.4. H37Ra Infection
4.5. Luciferase Assays
4.6. Editing of NRAMP1 in THP-1 Cells
4.7. Western Blot Analyses
4.8. SP1/SP3 3D Structure Modeling and Ligand Docking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| TB | tuberculosis |
| Mtb | Mycobacterium tuberculosis |
| M. bovis | Mycobacterium bovis |
| NRAMP1 | natural resistance-associated macrophage 1 |
| MAF | minor allele frequency |
| TF | transcription factor |
| Gapdh | glyceraldehyde 3-phosphate dehydrogenase |
| MOI | multiplicity of infection |
| hpi | hours post-infection |
| HADDOCK | High Ambiguity Driven DOCKing |
| SP | specificity protein |
| BFFs | bovine fetal fibroblasts |
References
- Zumla, A.; Raviglione, M.; Hafner, R.; von Reyn, C.F. Tuberculosis. N. Engl. J. Med. 2013, 368, 745–755. [Google Scholar] [CrossRef]
- Comstock, G.W. Tuberculosis in twins: A re-analysis of the Prophit survey. Am. Rev. Respir. Dis. 1978, 117, 621–624. [Google Scholar] [CrossRef]
- Wiart, A.; Jepson, A.; Banya, W.; Bennett, S.; Whittle, H.; Martin, N.G.; Hill, A.V. Quantitative association tests of immune responses to antigens of Mycobacterium tuberculosis: A study of twins in West Africa. Twin. Res. 2004, 7, 578–588. [Google Scholar] [CrossRef] [PubMed]
- Abel, L.; El-Baghdadi, J.; Bousfiha, A.A.; Casanova, J.L.; Schurr, E. Human genetics of tuberculosis: A long and winding road. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130428. [Google Scholar] [CrossRef] [PubMed]
- Bellamy, R.; Beyers, N.; McAdam, K.P.; Ruwende, C.; Gie, R.; Samaai, P.; Bester, D.; Meyer, M.; Corrah, T.; Collin, M.; et al. Genetic susceptibility to tuberculosis in Africans: A genome-wide scan. Proc. Natl. Acad. Sci. USA 2000, 97, 8005–8009. [Google Scholar] [CrossRef] [PubMed]
- Cooke, G.S.; Campbell, S.J.; Bennett, S.; Lienhardt, C.; McAdam, K.P.; Sirugo, G.; Sow, O.; Gustafson, P.; Mwangulu, F.; van Helden, P.; et al. Mapping of a novel susceptibility locus suggests a role for MC3R and CTSZ in human tuberculosis. Am. J. Respir. Crit. Care Med. 2008, 178, 203–207. [Google Scholar] [CrossRef]
- Ghanavi, J.; Farnia, P.; Farnia, P.; Velayati, A.A. Human genetic background in susceptibility to tuberculosis. Int. J. Mycobacteriol. 2020, 9, 239–247. [Google Scholar] [CrossRef]
- Hoal, E.G.; Lewis, L.A.; Jamieson, S.E.; Tanzer, F.; Rossouw, M.; Victor, T.; Hillerman, R.; Beyers, N.; Blackwell, J.M.; Van Helden, P.D. SLC11A1 (NRAMP1) but not SLC11A2 (NRAMP2) polymorphisms are associated with susceptibility to tuberculosis in a high-incidence community in South Africa. Int. J. Tuberc. Lung Dis. 2004, 8, 1464–1471. [Google Scholar]
- Meyer, C.G.; Thye, T. Host genetic studies in adult pulmonary tuberculosis. Semin. Immunol. 2014, 26, 445–453. [Google Scholar] [CrossRef]
- Cosivi, O.; Grange, J.M.; Daborn, C.J.; Raviglione, M.C.; Fujikura, T.; Cousins, D.; Robinson, R.A.; Huchzermeyer, H.F.; de Kantor, I.; Meslin, F.X. Zoonotic tuberculosis due to Mycobacterium bovis in developing countries. Emerg. Infect. Dis. 1998, 4, 59–70. [Google Scholar] [CrossRef]
- O’Reilly, L.M.; Daborn, C.J. The epidemiology of Mycobacterium bovis infections in animals and man: A review. Tuber. Lung Dis. 1995, 76 (Suppl. 1), 1–46. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Sun, L.; Jiao, W.; Zhao, S.; Li, H.; Guan, X.; Jiao, A.; Jiang, Z.; Shen, A. SLC11A1 (Formerly NRAMP1) gene polymorphisms associated with pediatric tuberculosis in China. Clin. Infect. Dis. 2009, 48, 733–738. [Google Scholar] [CrossRef]
- Vidal, S.M.; Malo, D.; Vogan, K.; Skamene, E.; Gros, P. Natural resistance to infection with intracellular parasites: Isolation of a candidate for Bcg. Cell 1993, 73, 469–485. [Google Scholar] [CrossRef] [PubMed]
- Amer, A.O.; Swanson, M.S. A phagosome of one’s own: A microbial guide to life in the macrophage. Curr. Opin. Microbiol. 2002, 5, 56–61. [Google Scholar] [CrossRef]
- Wessling-Resnick, M. Nramp1 and Other Transporters Involved in Metal Withholding during Infection. J. Biol. Chem. 2015, 290, 18984–18990. [Google Scholar] [CrossRef]
- Forbes, J.R.; Gros, P. Iron, manganese, and cobalt transport by Nramp1 (Slc11a1) and Nramp2 (Slc11a2) expressed at the plasma membrane. Blood 2003, 102, 1884–1892. [Google Scholar] [CrossRef]
- Gruenheid, S.; Pinner, E.; Desjardins, M.; Gros, P. Natural resistance to infection with intracellular pathogens: The Nramp1 protein is recruited to the membrane of the phagosome. J. Exp. Med. 1997, 185, 717–730. [Google Scholar] [CrossRef]
- Archer, N.S.; Nassif, N.T.; O’Brien, B.A. Genetic variants of SLC11A1 are associated with both autoimmune and infectious diseases: Systematic review and meta-analysis. Genes Immun. 2015, 16, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Azad, A.K.; Sadee, W.; Schlesinger, L.S. Innate immune gene polymorphisms in tuberculosis. Infect. Immun. 2012, 80, 3343–3359. [Google Scholar] [CrossRef]
- Brites, D.; Gagneux, S. Old and new selective pressures on Mycobacterium tuberculosis. Infect. Genet. Evol. 2012, 12, 678–685. [Google Scholar] [CrossRef]
- Moller, M.; Hoal, E.G. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis 2010, 90, 71–83. [Google Scholar] [CrossRef]
- Shahzad, F.; Bashir, N.; Ali, A.; Nadeem, A.; Ammar, A.; Kashif, M.; Javaid, K.; Jahan, S.; Tahir, R.; Rizwan, M.; et al. SLC11A1 genetic variation and low expression may cause immune response impairment in TB patients. Genes Immun 2022, 23, 85–92. [Google Scholar] [CrossRef]
- Cellier, M.F. Nutritional immunity: Homology modeling of Nramp metal import. Adv. Exp. Med. Biol. 2012, 946, 335–351. [Google Scholar] [CrossRef] [PubMed]
- Cellier, M.F. Cell-Type Specific Determinants of NRAMP1 Expression in Professional Phagocytes. Biology 2013, 2, 233–283. [Google Scholar] [CrossRef] [PubMed]
- Peracino, B.; Wagner, C.; Balest, A.; Balbo, A.; Pergolizzi, B.; Noegel, A.A.; Steinert, M.; Bozzaro, S. Function and mechanism of action of Dictyostelium Nramp1 (Slc11a1) in bacterial infection. Traffic 2006, 7, 22–38. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Wang, Y.; Wang, Z.; He, X.; Wang, L.; Yuan, D.; He, Y.; Jin, T.; He, S. Association of SLC11A1 Polymorphisms with Tuberculosis Susceptibility in the Chinese Han Population. Front. Genet. 2022, 13, 899124. [Google Scholar] [CrossRef]
- Vidal, S.M.; Pinner, E.; Lepage, P.; Gauthier, S.; Gros, P. Natural resistance to intracellular infections: Nramp1 encodes a membrane phosphoglycoprotein absent in macrophages from susceptible (Nramp1 D169) mouse strains. J. Immunol. 1996, 157, 3559–3568. [Google Scholar] [CrossRef]
- Liu, K.; Zhang, B.; Teng, Z.; Wang, Y.; Dong, G.; Xu, C.; Qin, B.; Song, C.; Chai, J.; Li, Y.; et al. Association between SLC11A1 (NRAMP1) polymorphisms and susceptibility to tuberculosis in Chinese Holstein cattle. Tuberculosis 2017, 103, 10–15. [Google Scholar] [CrossRef]
- Malik, S.; Abel, L.; Tooker, H.; Poon, A.; Simkin, L.; Girard, M.; Adams, G.J.; Starke, J.R.; Smith, K.C.; Graviss, E.A.; et al. Alleles of the NRAMP1 gene are risk factors for pediatric tuberculosis disease. Proc. Natl. Acad. Sci. USA 2005, 102, 12183–12188. [Google Scholar] [CrossRef]
- Barnes, I.; Duda, A.; Pybus, O.G.; Thomas, M.G. Ancient urbanization predicts genetic resistance to tuberculosis. Evolution 2011, 65, 842–848. [Google Scholar] [CrossRef]
- Zaahl, M.G.; Robson, K.J.; Warnich, L.; Kotze, M.J. Expression of the SLC11A1 (NRAMP1) 5′-(GT)n repeat: Opposite effect in the presence of –237C→T. Blood Cells Mol. Dis. 2004, 33, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Searle, S.; Blackwell, J.M. Evidence for a functional repeat polymorphism in the promoter of the human NRAMP1 gene that correlates with autoimmune versus infectious disease susceptibility. J. Med. Genet. 1999, 36, 295–299. [Google Scholar] [CrossRef]
- Yuan, M.; Zhang, J.; Gao, Y.; Yuan, Z.; Zhu, Z.; Wei, Y.; Wu, T.; Han, J.; Zhang, Y. HMEJ-based safe-harbor genome editing enables efficient generation of cattle with increased resistance to tuberculosis. J. Biol. Chem. 2021, 296, 100497. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, H.; Wang, Y.; Liu, X.; Chen, L.; Li, Q.; Cui, C.; Liu, X.; Zhang, J.; Zhang, Y. Single Cas9 nickase induced generation of NRAMP1 knockin cattle with reduced off-target effects. Genome. Biol. 2017, 18, 13. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, J.; O’Connor, L.; Middleton, C.P.; Keane, P.; Gillemans, N.; Cazier, J.B.; Philipsen, S.; Bonifer, C. Robust hematopoietic specification requires the ubiquitous Sp1 and Sp3 transcription factors. Epigenetics Chromatin 2019, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Fang, J.; Jia, Q.; Meng, H.; Liu, F.; Mao, J. Transcription factor specificity protein (SP) family in renal physiology and diseases. PeerJ 2025, 13, e18820. [Google Scholar] [CrossRef]
- Safe, S. Specificity Proteins (Sp) and Cancer. Int. J. Mol. Sci. 2023, 24, 5164. [Google Scholar] [CrossRef]
- Li, L.; He, S.; Sun, J.M.; Davie, J.R. Gene regulation by Sp1 and Sp3. Biochem. Cell Biol. 2004, 82, 460–471. [Google Scholar] [CrossRef]
- Letovsky, J.; Dynan, W.S. Measurement of the binding of transcription factor Sp1 to a single GC box recognition sequence. Nucleic Acids Res. 1989, 17, 2639–2653. [Google Scholar] [CrossRef]
- Richer, E.; Campion, C.G.; Dabbas, B.; White, J.H.; Cellier, M.F. Transcription factors Sp1 and C/EBP regulate NRAMP1 gene expression. FEBS J. 2008, 275, 5074–5089. [Google Scholar] [CrossRef]
- Resendes, K.K.; Rosmarin, A.G. Sp1 control of gene expression in myeloid cells. Crit. Rev. Eukaryot. Gene. Expr. 2004, 14, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Hata, Y.; Duh, E.; Zhang, K.; Robinson, G.S.; Aiello, L.P. Transcription factors Sp1 and Sp3 alter vascular endothelial growth factor receptor expression through a novel recognition sequence. J. Biol. Chem. 1998, 273, 19294–19303. [Google Scholar] [CrossRef] [PubMed]
- Ward, S.V.; Samuel, C.E. The PKR kinase promoter binds both Sp1 and Sp3, but only Sp3 functions as part of the interferon-inducible complex with ISGF-3 proteins. Virology 2003, 313, 553–566. [Google Scholar] [CrossRef]
- Su, W.; Jackson, S.; Tjian, R.; Echols, H. DNA looping between sites for transcriptional activation: Self-association of DNA-bound Sp1. Genes Dev. 1991, 5, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, I.A.; Courey, A.J.; Wall, J.S.; Jackson, S.P.; Hough, P.V. DNA looping and Sp1 multimer links: A mechanism for transcriptional synergism and enhancement. Proc. Natl. Acad. Sci. USA 1991, 88, 5670–5674. [Google Scholar] [CrossRef]
- Chackerian, A.A.; Behar, S.M. Susceptibility to Mycobacterium tuberculosis: Lessons from inbred strains of mice. Tuberculosis 2003, 83, 279–285. [Google Scholar] [CrossRef]
- Shepelkova, G.; Pommerenke, C.; Alberts, R.; Geffers, R.; Evstifeev, V.; Apt, A.; Schughart, K.; Wilk, E. Analysis of the lung transcriptome in Mycobacterium tuberculosis-infected mice reveals major differences in immune response pathways between TB-susceptible and resistant hosts. Tuberculosis 2013, 93, 263–269. [Google Scholar] [CrossRef]
- LoBue, P.A.; Enarson, D.A.; Thoen, C.O. Tuberculosis in humans and animals: An overview. Int. J. Tuberc. Lung Dis. 2010, 14, 1075–1078. [Google Scholar]
- Sato, M.P.; Makino, T.; Kawata, M. Natural selection in a population of Drosophila melanogaster explained by changes in gene expression caused by sequence variation in core promoter regions. BMC Evol. Biol. 2016, 16, 35. [Google Scholar] [CrossRef]
- Li, X.; Qian, X.; Wang, B.; Xia, Y.; Zheng, Y.; Du, L.; Xu, D.; Xing, D.; DePinho, R.A.; Lu, Z. Programmable base editing of mutated TERT promoter inhibits brain tumour growth. Nat. Cell Biol. 2020, 22, 282–288. [Google Scholar] [CrossRef]
- Liao, J.; Chen, S.; Hsiao, S.; Jiang, Y.; Yang, Y.; Zhang, Y.; Wang, X.; Lai, Y.; Bauer, D.E.; Wu, Y. Therapeutic adenine base editing of human hematopoietic stem cells. Nat. Commun. 2023, 14, 207. [Google Scholar] [CrossRef]
- Ravi, N.S.; Wienert, B.; Wyman, S.K.; Bell, H.W.; George, A.; Mahalingam, G.; Vu, J.T.; Prasad, K.; Bandlamudi, B.P.; Devaraju, N.; et al. Identification of novel HPFH-like mutations by CRISPR base editing that elevate the expression of fetal hemoglobin. eLife 2022, 11, e65421. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Tong, Q.; Ge, H.; Li, W.; Liu, J.; Wang, Y.; Guo, Z.; Quan, F.; Zhang, Y. Identification of SP110 in horse (Equus caballus): Isolation of novel splice variants and evidence of activation effects on macrophages. Tuberculosis 2016, 101, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.F.; Gamage, A.M.; Chan, W.O.Y.; Hiller, M.; Teeling, E.C. Decoding bat immunity: The need for a coordinated research approach. Nat. Rev. Immunol. 2021, 21, 269–271. [Google Scholar] [CrossRef]
- Friedrichs, V.; Toussaint, C.; Schafer, A.; Rissmann, M.; Dietrich, O.; Mettenleiter, T.C.; Pei, G.; Balkema-Buschmann, A.; Saliba, A.E.; Dorhoi, A. Landscape and age dynamics of immune cells in the Egyptian rousette bat. Cell Rep. 2022, 40, 111305. [Google Scholar] [CrossRef]
- LeVan, T.D.; Bloom, J.W.; Bailey, T.J.; Karp, C.L.; Halonen, M.; Martinez, F.D.; Vercelli, D. A common single nucleotide polymorphism in the CD14 promoter decreases the affinity of Sp protein binding and enhances transcriptional activity. J. Immunol. 2001, 167, 5838–5844. [Google Scholar] [CrossRef]
- Qi, T.; Wu, F.; Xie, Y.; Gao, S.; Li, M.; Pu, J.; Li, D.; Lan, F.; Wang, Y. Base Editing Mediated Generation of Point Mutations Into Human Pluripotent Stem Cells for Modeling Disease. Front. Cell Dev. Biol. 2020, 8, 590581. [Google Scholar] [CrossRef]
- Hwang, G.H.; Park, J.; Lim, K.; Kim, S.; Yu, J.; Yu, E.; Kim, S.T.; Eils, R.; Kim, J.S.; Bae, S. Web-based design and analysis tools for CRISPR base editing. BMC Bioinform. 2018, 19, 542. [Google Scholar] [CrossRef]
- Bae, S.; Park, J.; Kim, J.S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 2014, 30, 1473–1475. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Krivak, R.; Hoksza, D. P2Rank: Machine learning based tool for rapid and accurate prediction of ligand binding sites from protein structure. J. Cheminform. 2018, 10, 39. [Google Scholar] [CrossRef] [PubMed]
- Honorato, R.V.; Trellet, M.E.; Jimenez-Garcia, B.; Schaarschmidt, J.J.; Giulini, M.; Reys, V.; Koukos, P.I.; Rodrigues, J.; Karaca, E.; van Zundert, G.C.P.; et al. The HADDOCK2.4 web server for integrative modeling of biomolecular complexes. Nat. Protoc. 2024, 19, 3219–3241. [Google Scholar] [CrossRef] [PubMed]
- Honorato, R.V.; Koukos, P.I.; Jimenez-Garcia, B.; Tsaregorodtsev, A.; Verlato, M.; Giachetti, A.; Rosato, A.; Bonvin, A. Structural Biology in the Clouds: The WeNMR-EOSC Ecosystem. Front. Mol. Biosci. 2021, 8, 729513. [Google Scholar] [CrossRef] [PubMed]
- Abdelaal, H.F.M.; Thacker, T.C.; Wadie, B.; Palmer, M.V.; Talaat, A.M. Transcriptional Profiling of Early and Late Phases of Bovine Tuberculosis. Infect. Immun. 2022, 90, e0031321. [Google Scholar] [CrossRef]
- Nalpas, N.C.; Magee, D.A.; Conlon, K.M.; Browne, J.A.; Healy, C.; McLoughlin, K.E.; Rue-Albrecht, K.; McGettigan, P.A.; Killick, K.E.; Gormley, E.; et al. RNA sequencing provides exquisite insight into the manipulation of the alveolar macrophage by tubercle bacilli. Sci. Rep. 2015, 5, 13629. [Google Scholar] [CrossRef]
- Nalpas, N.C.; Park, S.D.; Magee, D.A.; Taraktsoglou, M.; Browne, J.A.; Conlon, K.M.; Rue-Albrecht, K.; Killick, K.E.; Hokamp, K.; Lohan, A.J.; et al. Whole-transcriptome, high-throughput RNA sequence analysis of the bovine macrophage response to Mycobacterium bovis infection in vitro. BMC Genom. 2013, 14, 230. [Google Scholar] [CrossRef]
- Olson, G.S.; Murray, T.A.; Jahn, A.N.; Mai, D.; Diercks, A.H.; Gold, E.S.; Aderem, A. Type I interferon decreases macrophage energy metabolism during mycobacterial infection. Cell Rep. 2021, 35, 109195. [Google Scholar] [CrossRef]
- Rothchild, A.C.; Olson, G.S.; Nemeth, J.; Amon, L.M.; Mai, D.; Gold, E.S.; Diercks, A.H.; Aderem, A. Alveolar macrophages generate a noncanonical NRF2-driven transcriptional response to Mycobacterium tuberculosis in vivo. Sci. Immunol. 2019, 4, eaaw6693. [Google Scholar] [CrossRef]



| Binding Pocket Prediction | ||||||||
| P2Rank | Bovine SP1 | Bovine SP3 | ||||||
| Rank * | 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 |
| Score | 5.72 | 4.11 | 2.33 | 2.00 | 7.14 | 5.23 | 3.65 | 1.09 |
| Probability | 0.245 | 0.146 | 0.049 | 0.036 | 0.341 | 0.215 | 0.064 | 0.007 |
| Number of residues | 9 | 8 | 5 | 5 | 10 | 12 | 6 | 6 |
| Average conservation score | 78.637 | 84.98 | 80.862 | 88.962 | 77.02 | 80.285 | 81.635 | 65.845 |
| Energy of interaction (docking with bovine SP1) | ||||||||
| 2SP (cattle) | h2SPM (humans) | p2SPM (pigs) | m2SPM (mice) | 2SPM2 (mutation) | ||||
| HADDOCK score ** | −88.9 ± 14.3 | −72.0 ± 15.8 | −65.5 ± 10.4 | −64.8 ± 13.5 | −37.5 ± 7.9 | |||
| Van der Waals energy | −91.4 ± 6.2 | −72.5 ± 12.0 | −80.1 ± 9.1 | −74.7 ± 16.7 | −70.4 ± 3.8 | |||
| Electrostatic energy | −379.3± 35.3 | −335.1 ± 32.6 | −198.2 ± 46.8 | −215.1 ± 28.9 | −272.3 ± 4.3 | |||
| Desolvation energy | 32.9 ± 2.5 | 28.0 ± 2.3 | 17.5 ± 1.7 | 18.9 ± 1.4 | 27.5 ± 1.3 | |||
| Sum of energies | −526.7 | −451.6 | −326.3 | −335.6 | −352.7 | |||
| Buried surface area | 2487.8 ± 121.2 | 2114.5 ± 303.0 | 1881.0 ± 238.7 | 1930.4 ± 319.8 | 2019.9 ± 96.7 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Y.; Yuan, M.; Zhang, Y.; Gao, Y. The 2SP Site Mutation in the Bovine Natural Resistance-Associated Macrophage 1 Promoter Exhibits Antituberculosis Potential. Int. J. Mol. Sci. 2025, 26, 4229. https://doi.org/10.3390/ijms26094229
Wei Y, Yuan M, Zhang Y, Gao Y. The 2SP Site Mutation in the Bovine Natural Resistance-Associated Macrophage 1 Promoter Exhibits Antituberculosis Potential. International Journal of Molecular Sciences. 2025; 26(9):4229. https://doi.org/10.3390/ijms26094229
Chicago/Turabian StyleWei, Yongke, Mengke Yuan, Yong Zhang, and Yuanpeng Gao. 2025. "The 2SP Site Mutation in the Bovine Natural Resistance-Associated Macrophage 1 Promoter Exhibits Antituberculosis Potential" International Journal of Molecular Sciences 26, no. 9: 4229. https://doi.org/10.3390/ijms26094229
APA StyleWei, Y., Yuan, M., Zhang, Y., & Gao, Y. (2025). The 2SP Site Mutation in the Bovine Natural Resistance-Associated Macrophage 1 Promoter Exhibits Antituberculosis Potential. International Journal of Molecular Sciences, 26(9), 4229. https://doi.org/10.3390/ijms26094229






