Structural Insights into the Dynamics of Water in SOD1 Catalysis and Drug Interactions
Abstract
1. Introduction
2. Results
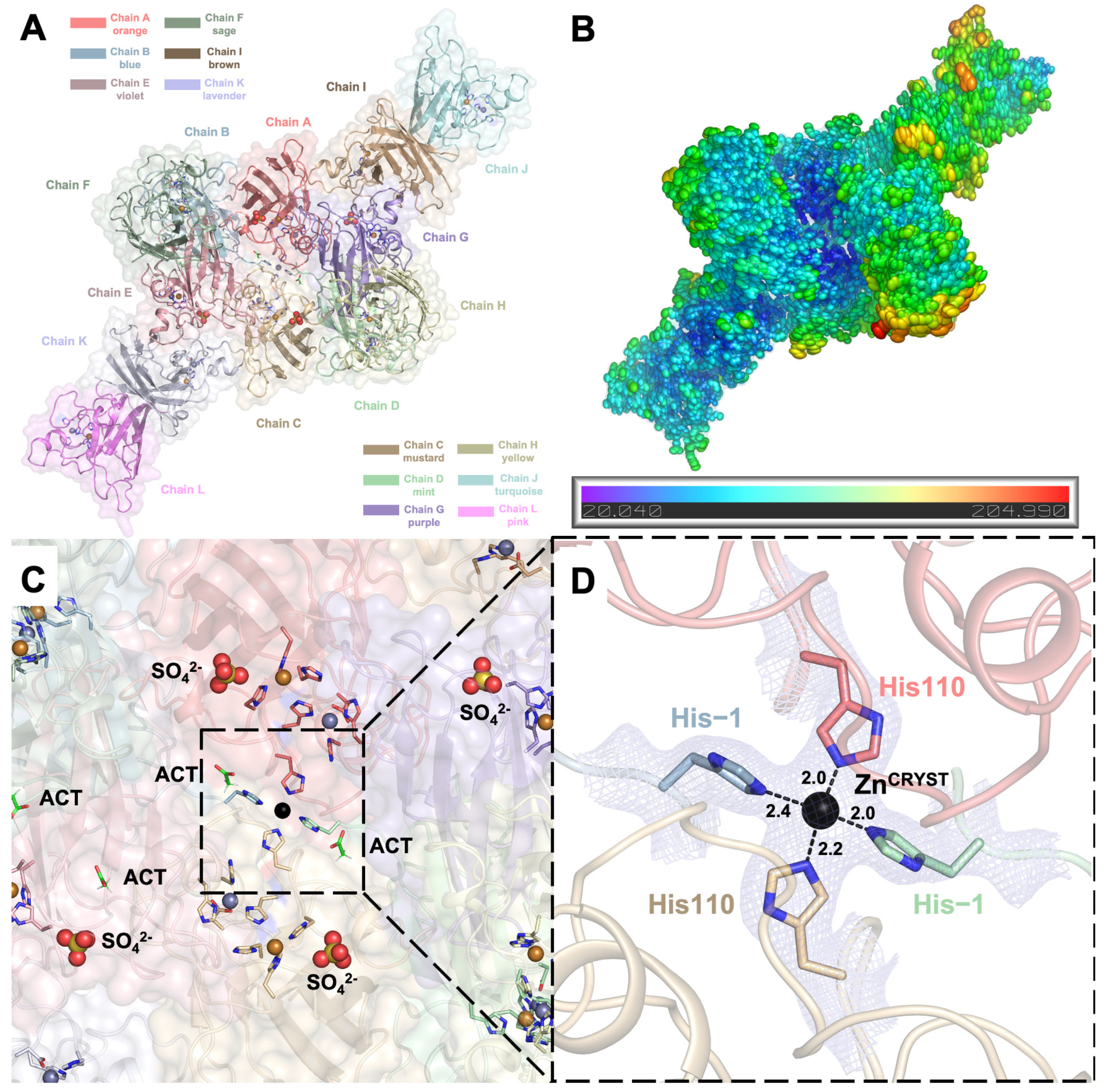
2.1. Crystal Structure of hSOD1 in a New Crystal Form
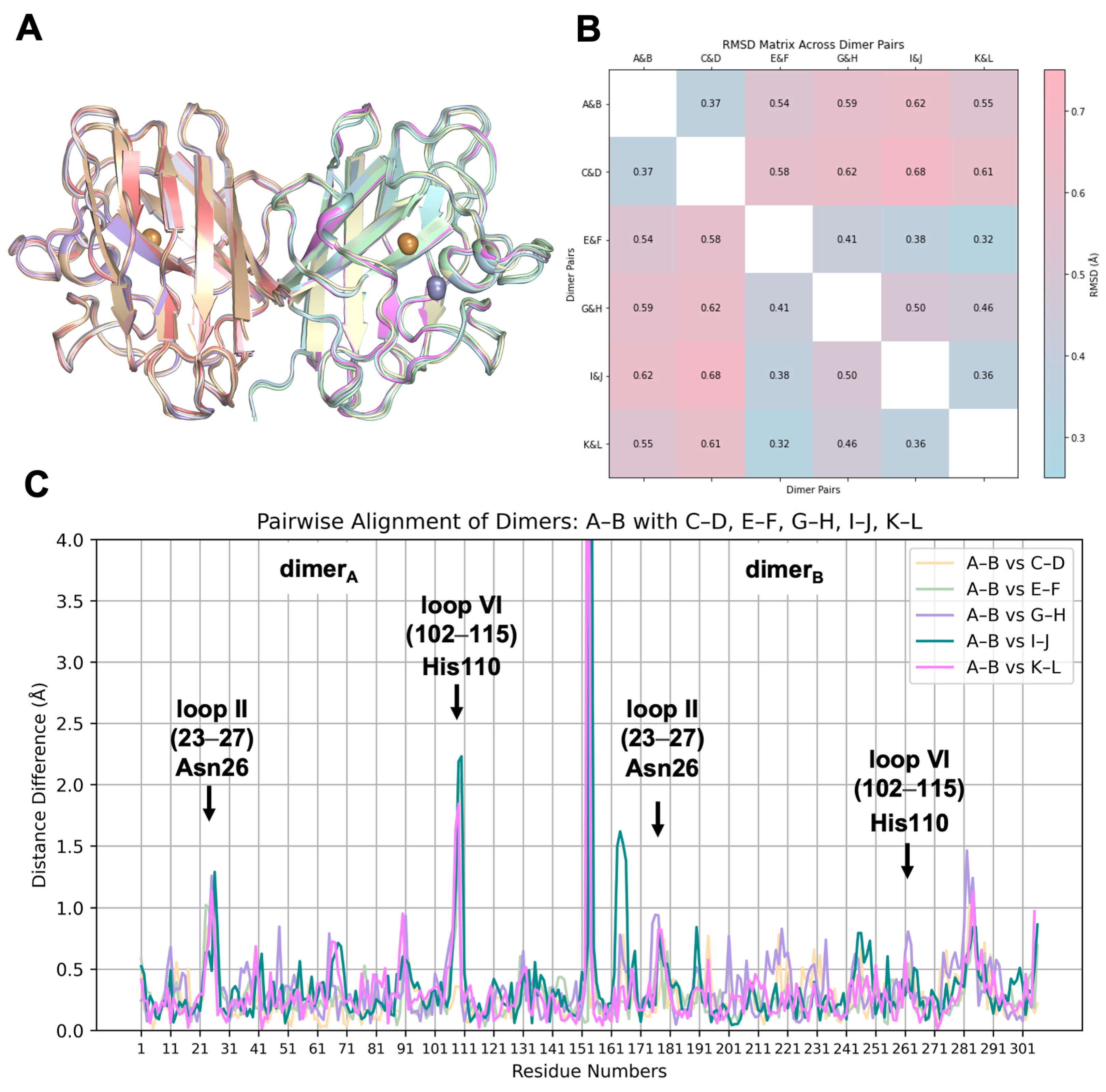
2.2. hSOD1 Monomers and Dimers Display Modest Conformational Changes
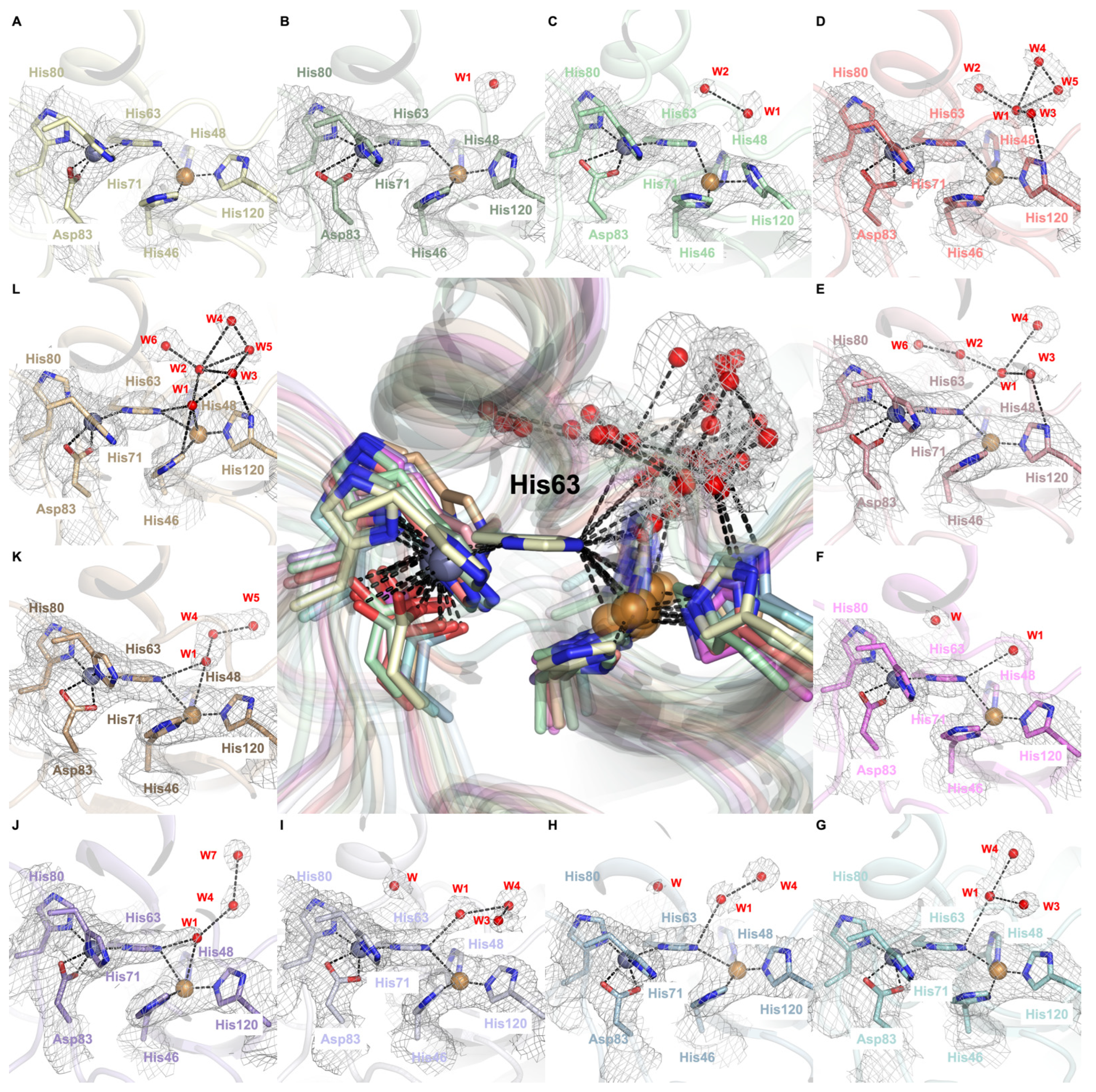
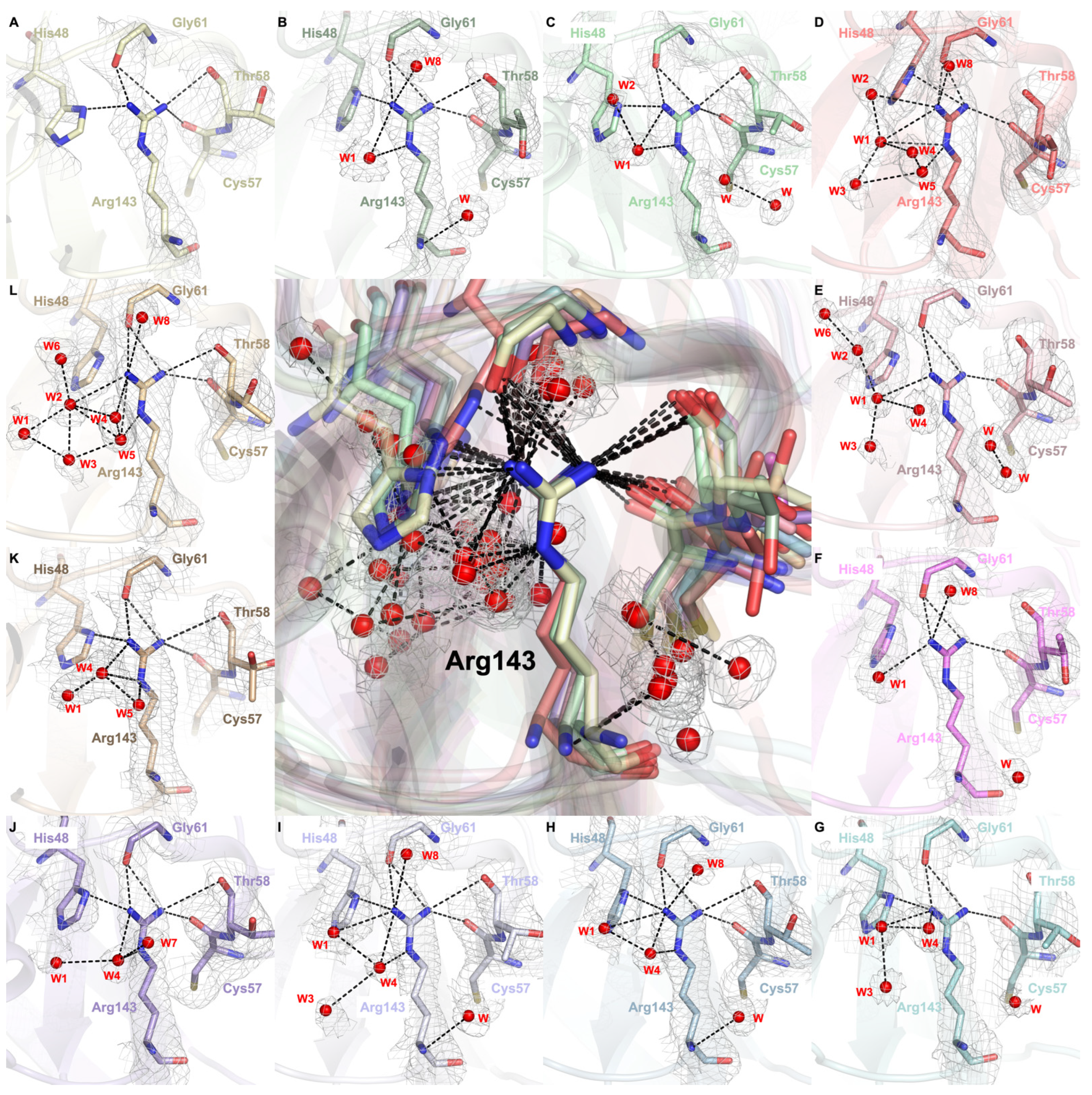
2.3. Dynamic Water Molecules Reveal New Conformations of the Active Site
2.4. Arg143 Dynamics Reveal a Key Allosteric State
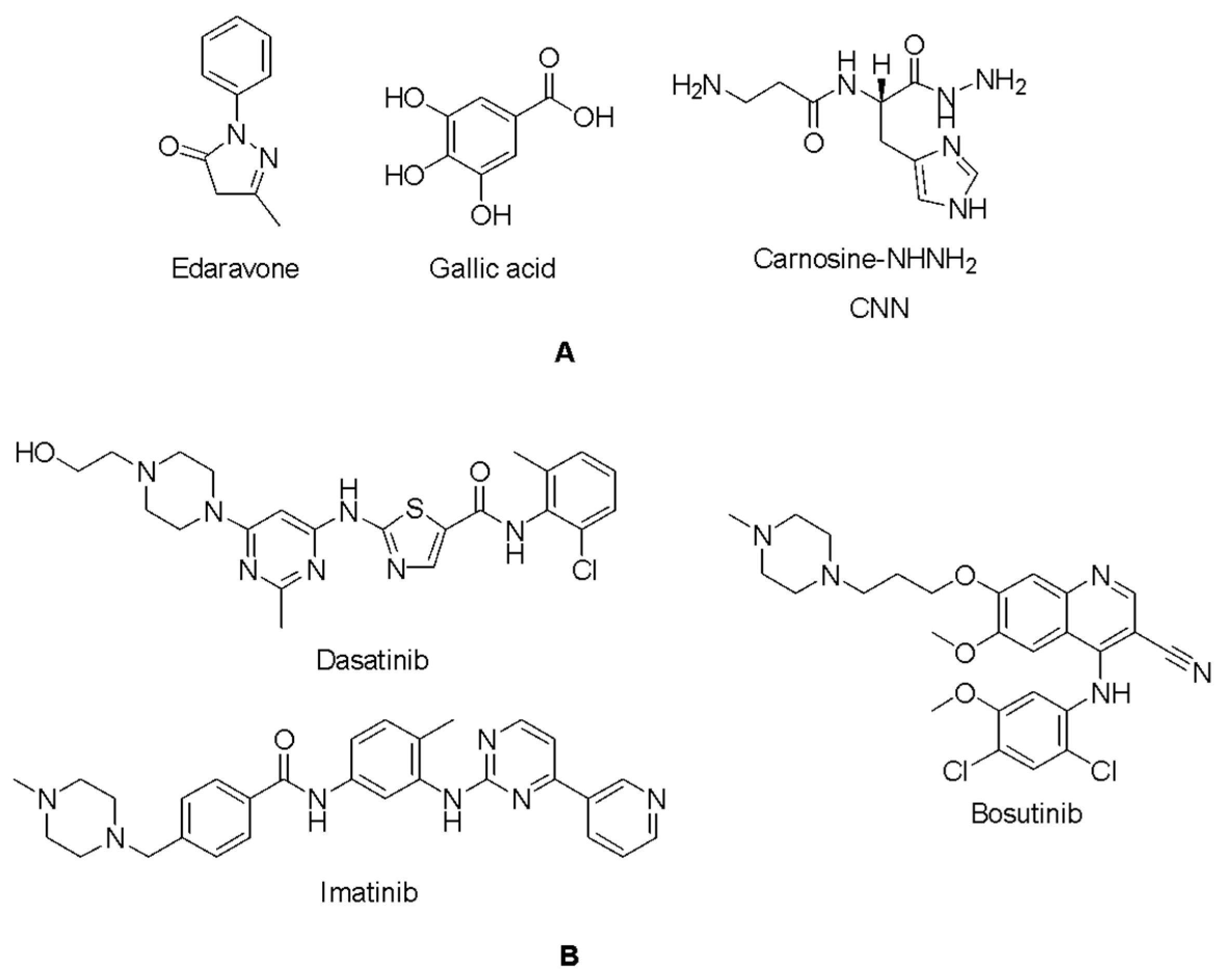
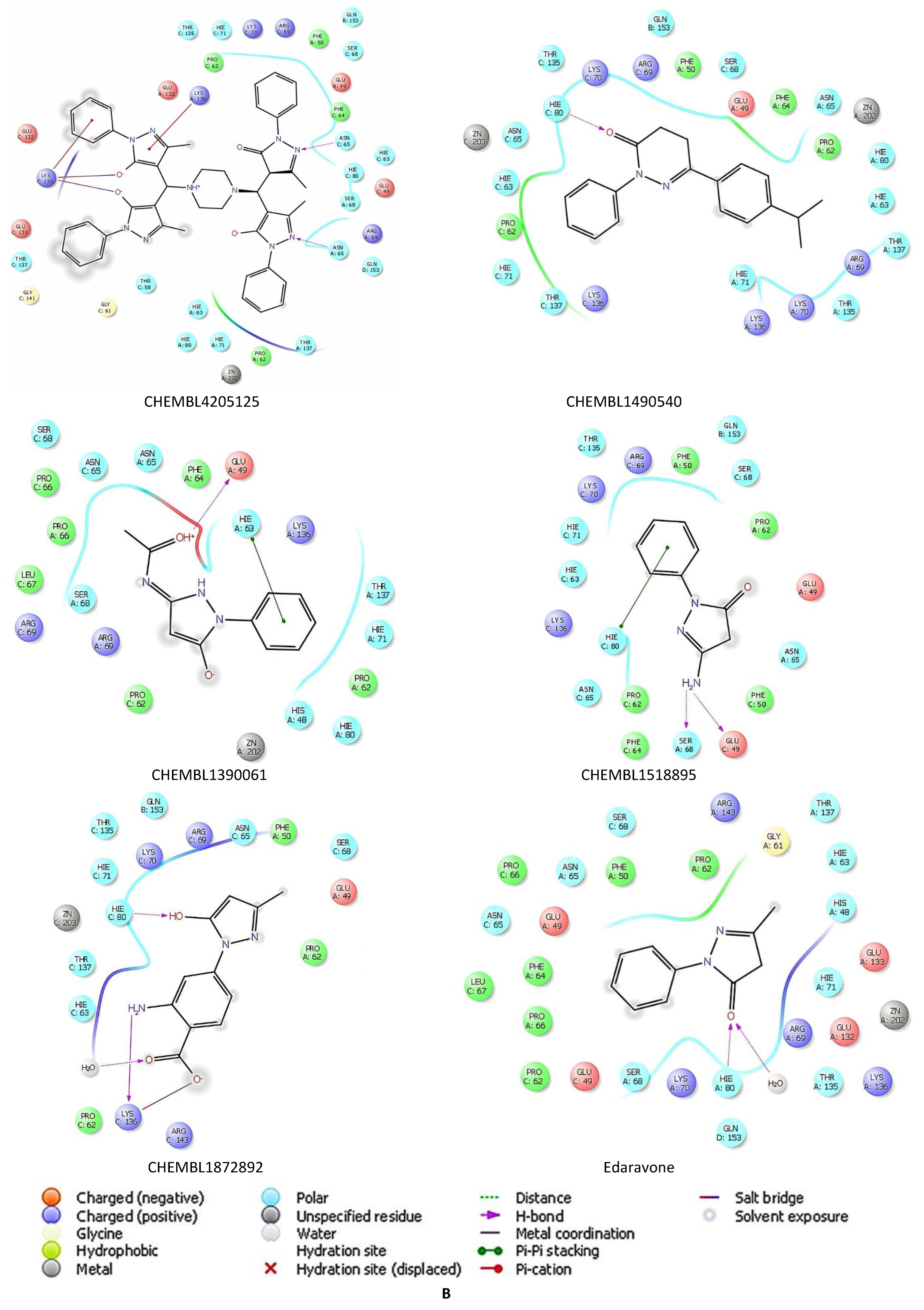
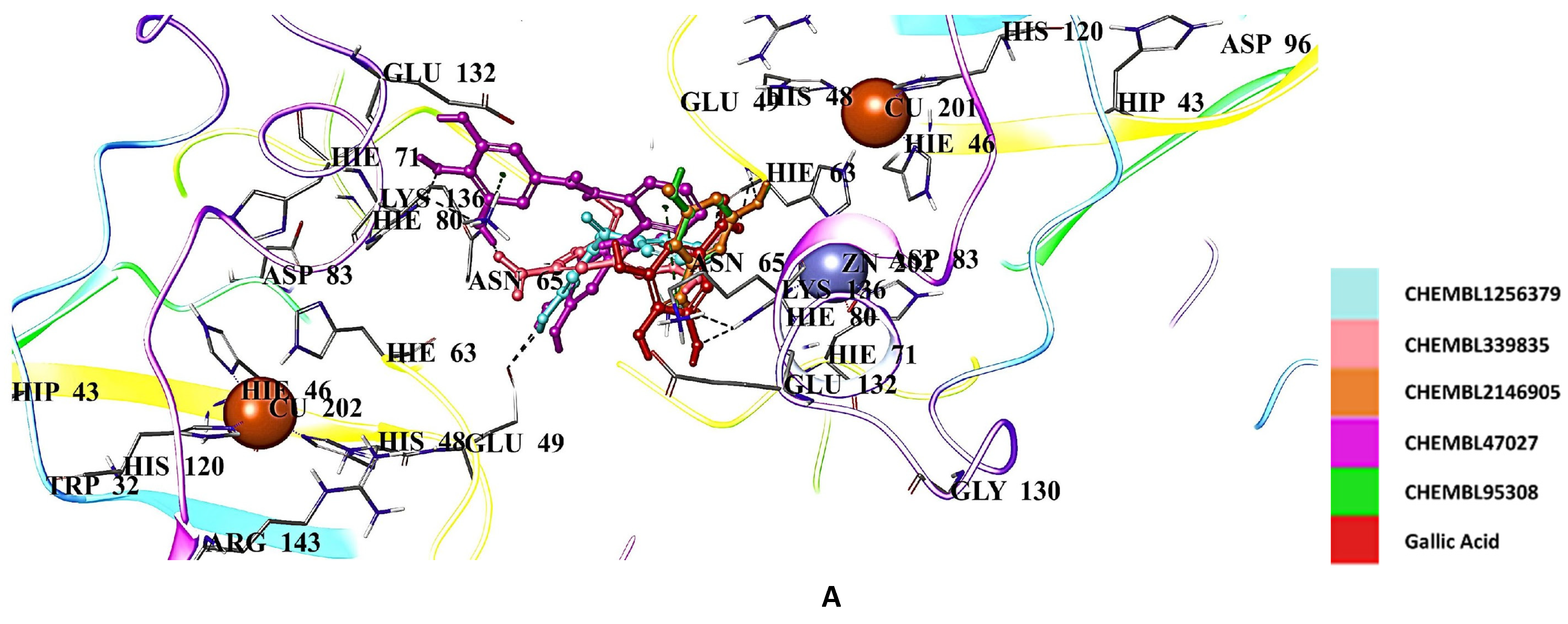
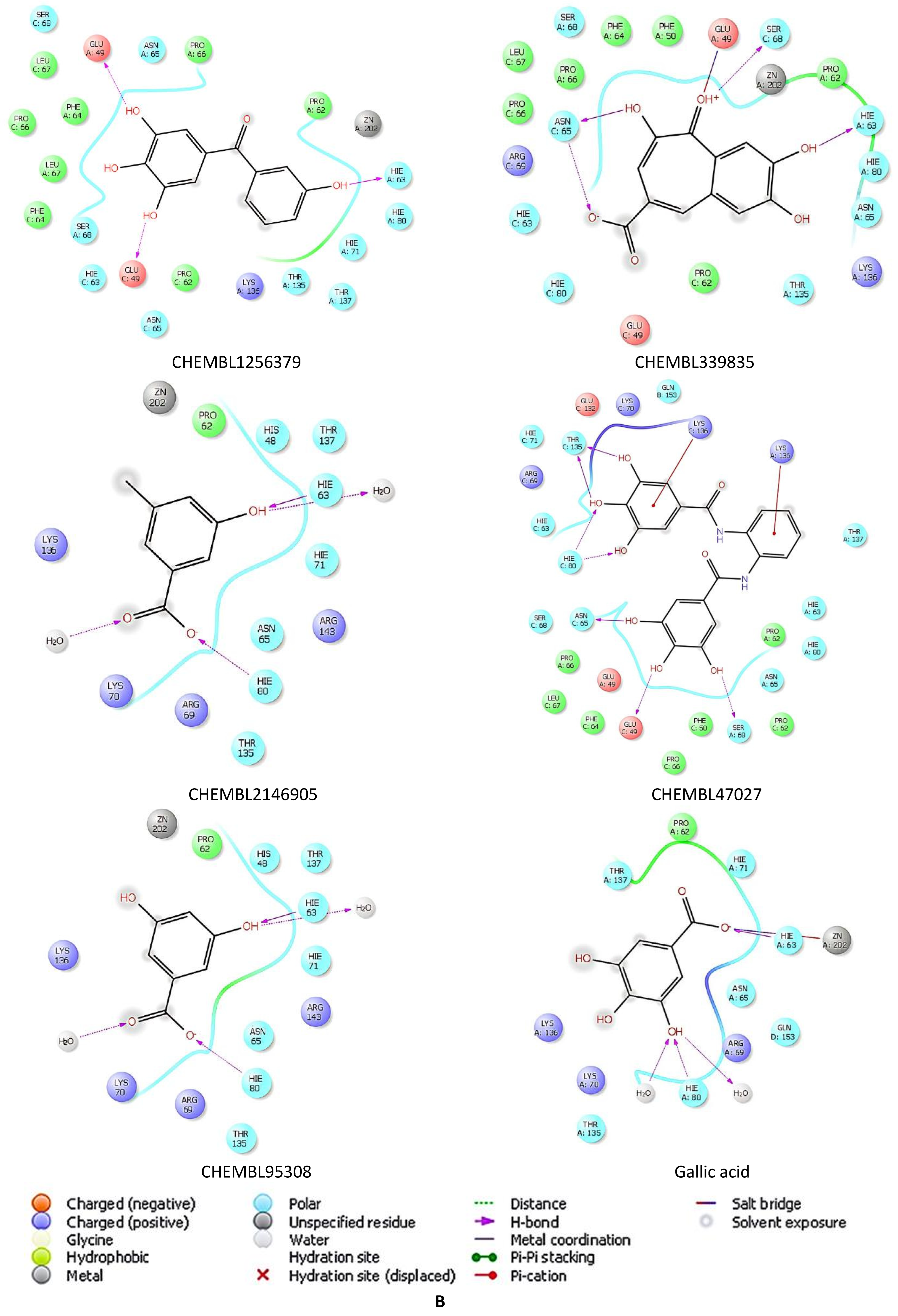
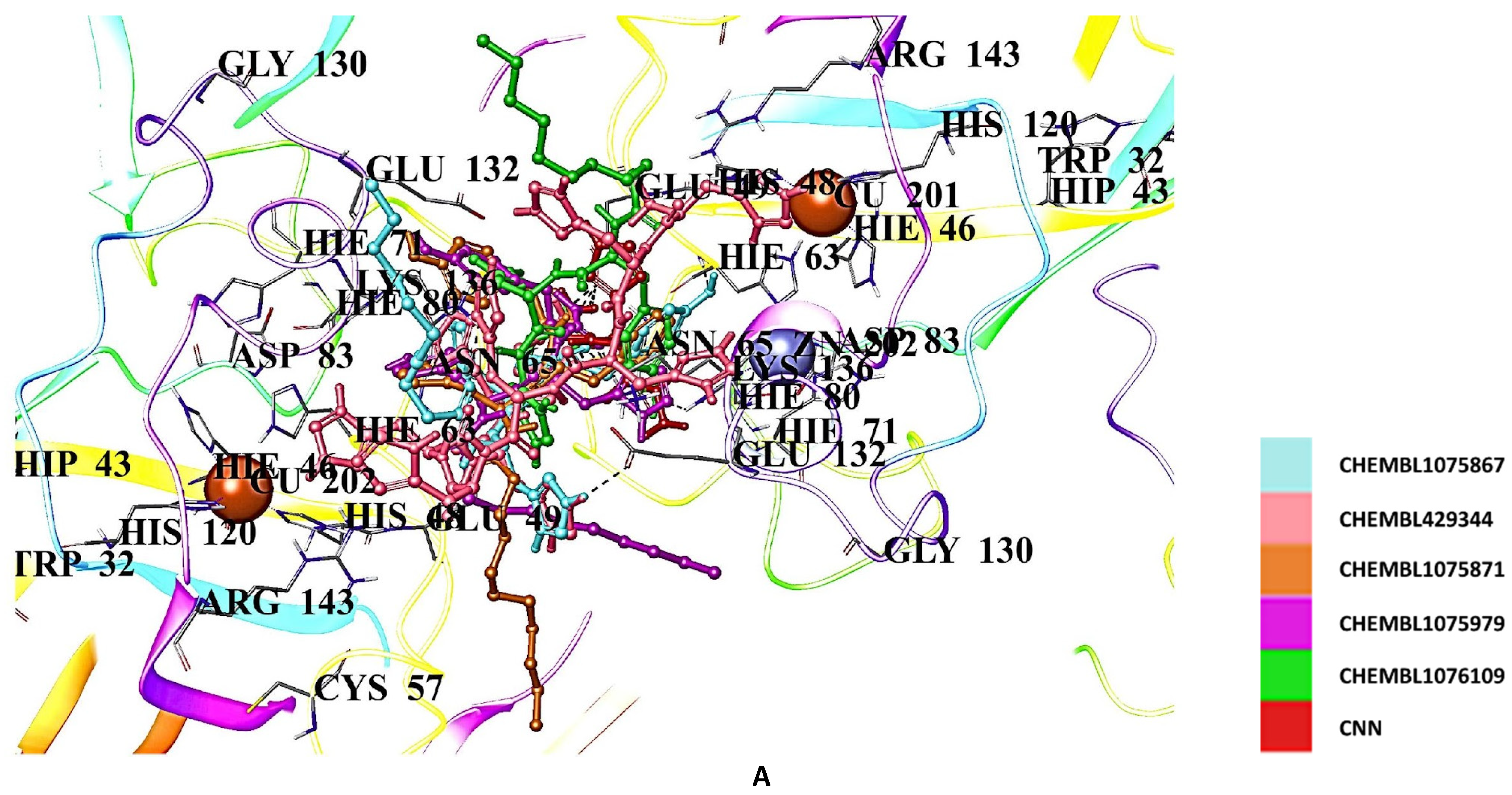
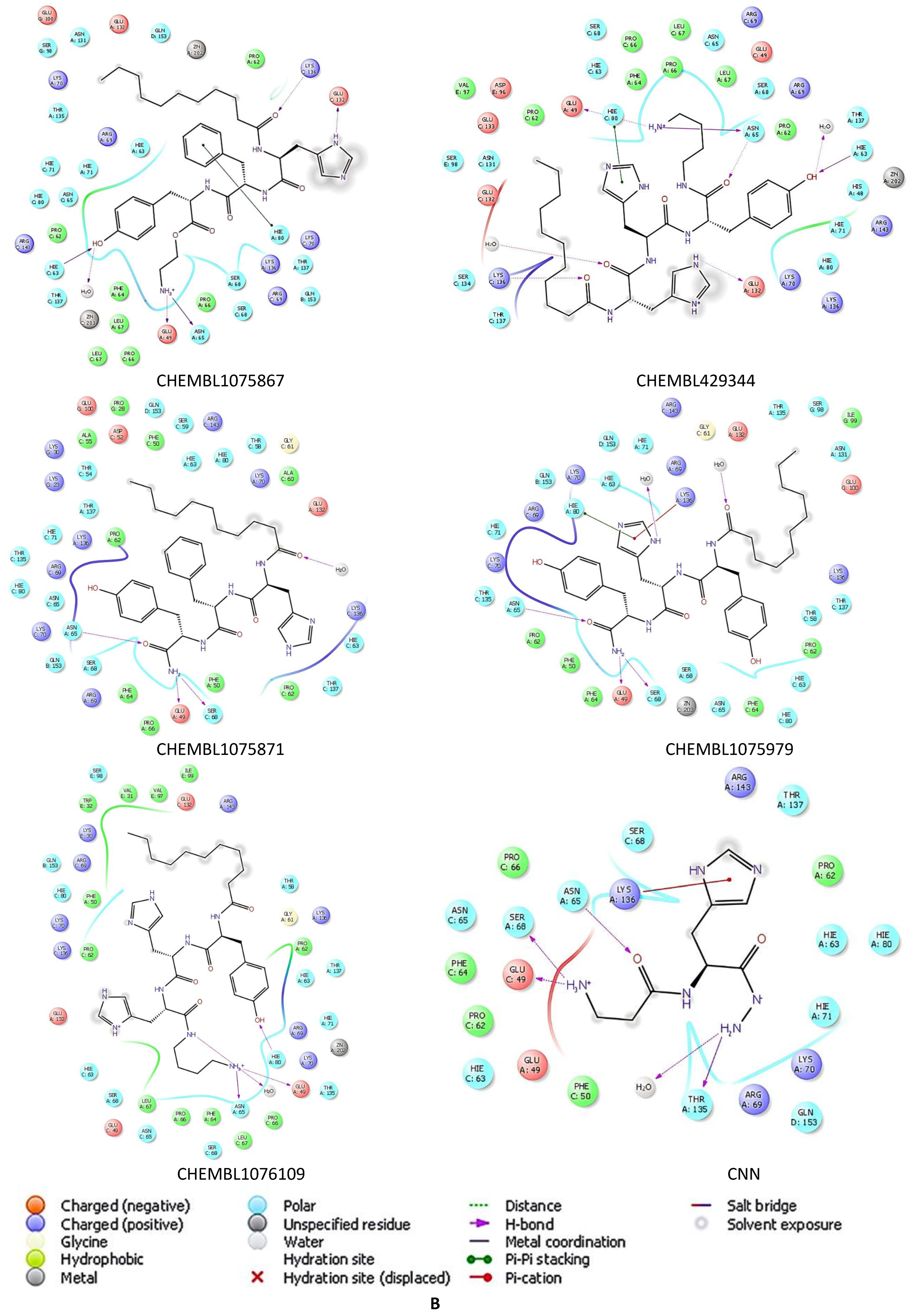
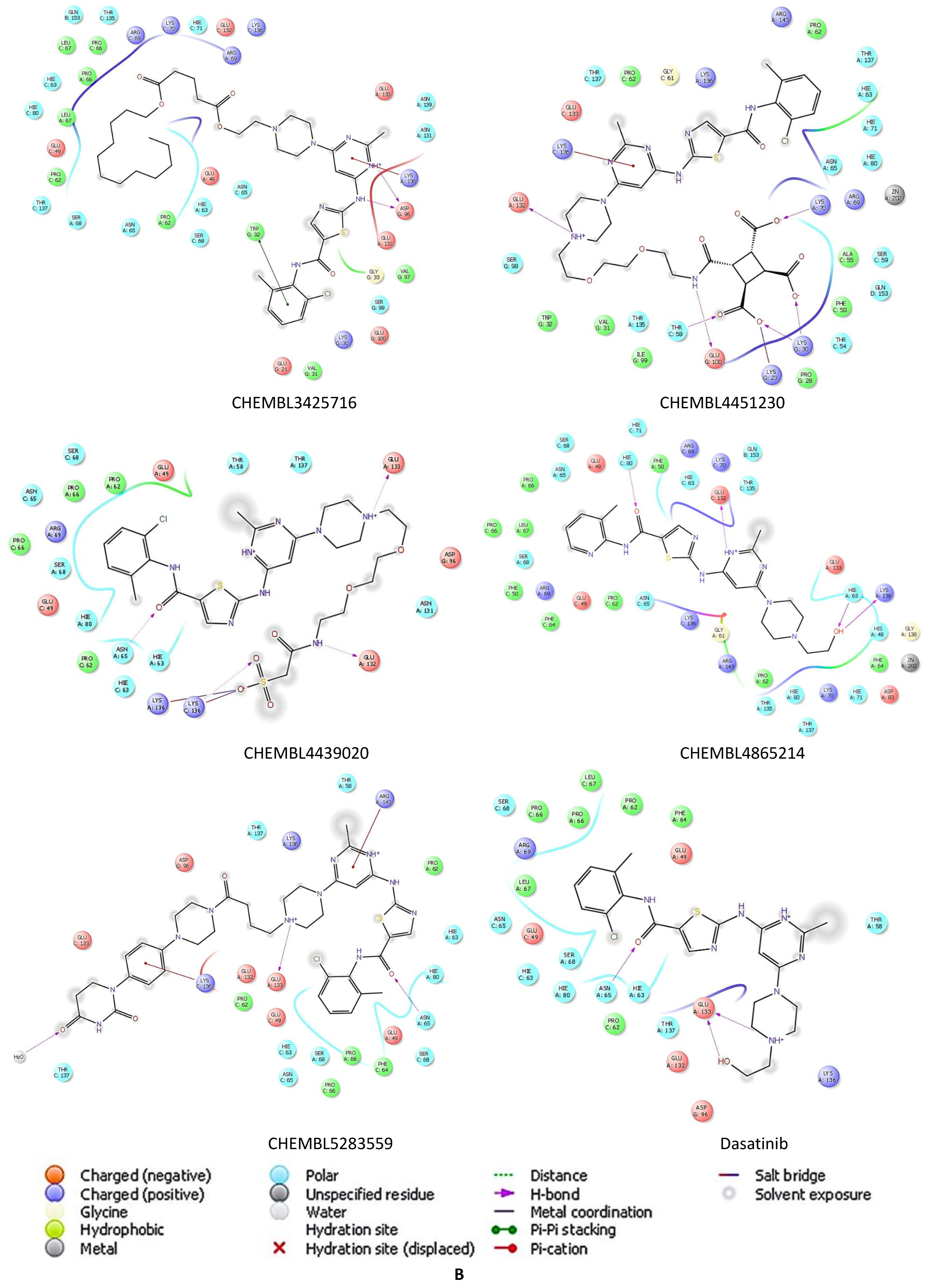
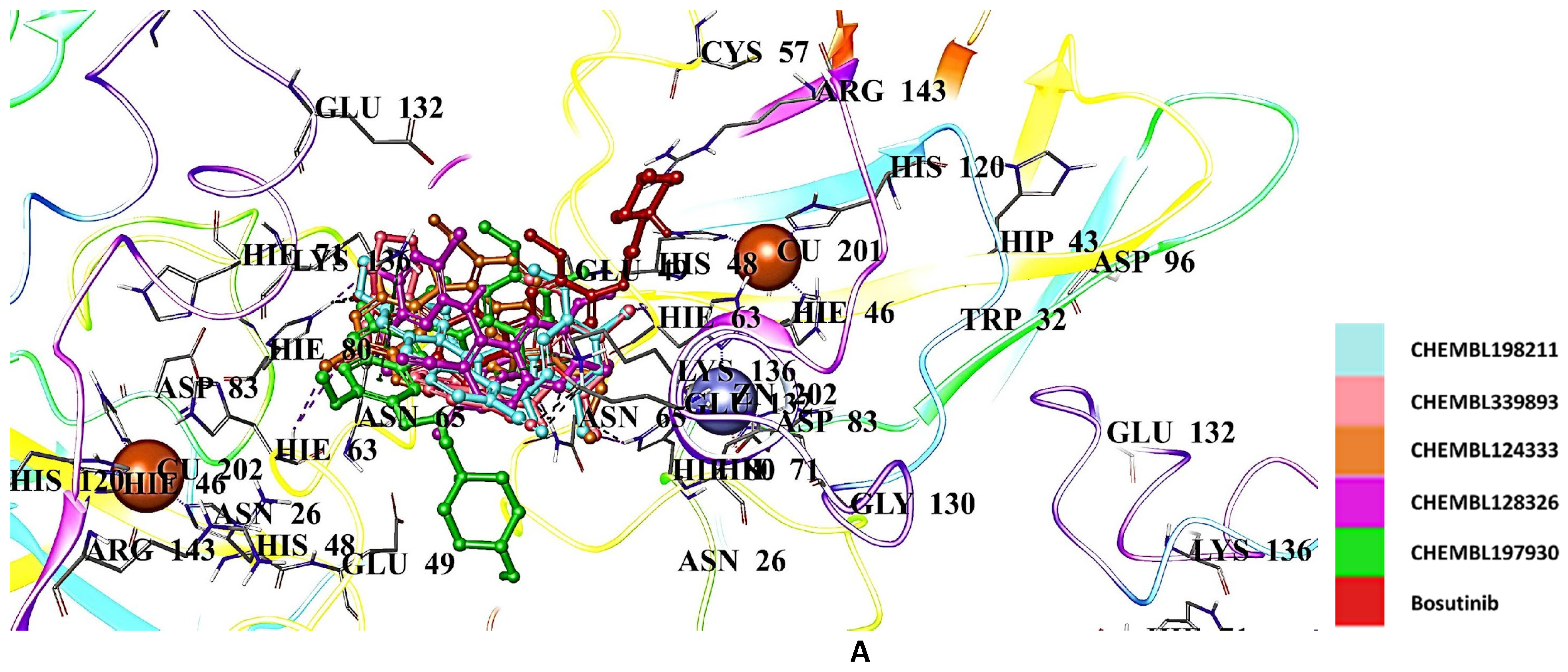
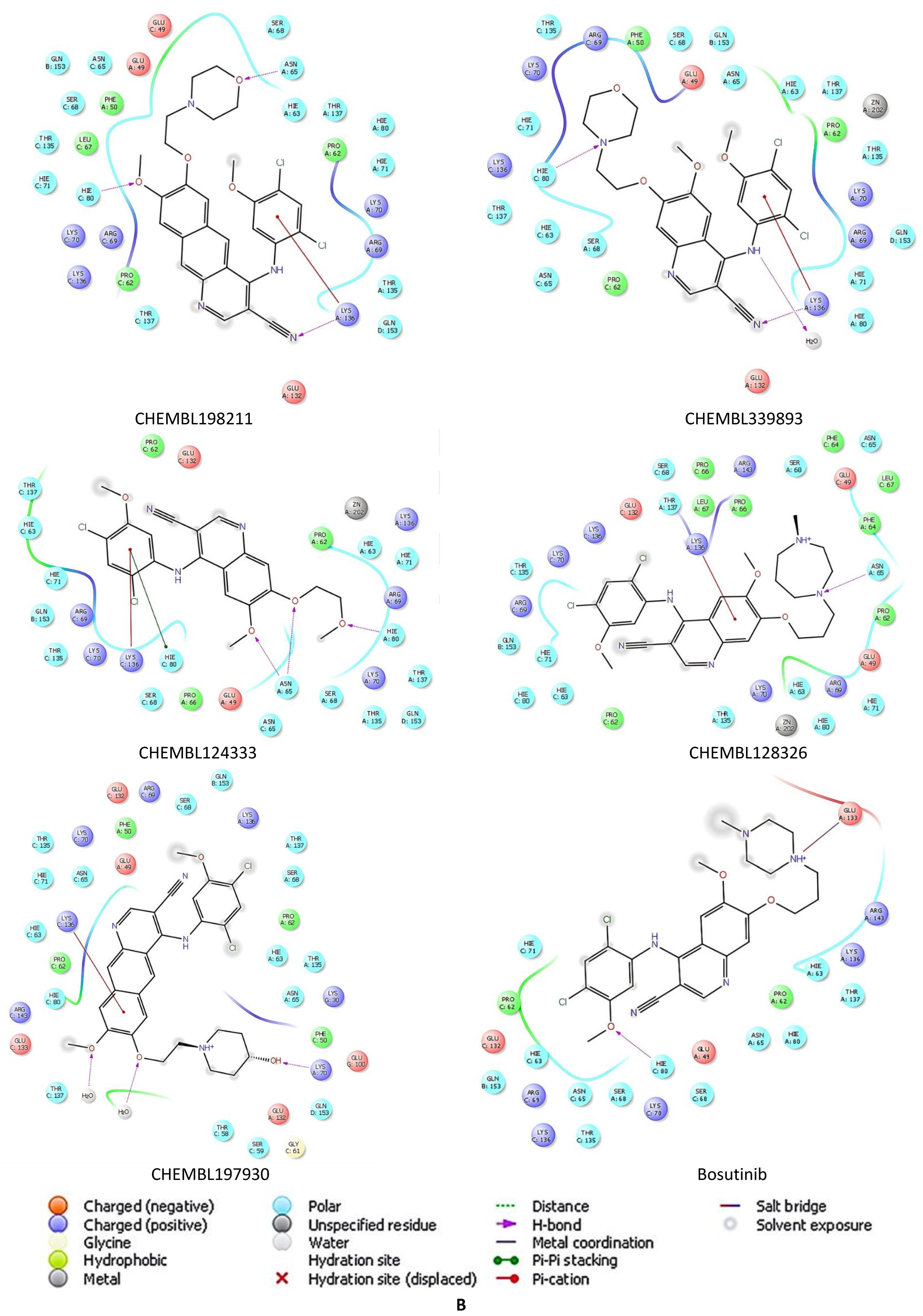
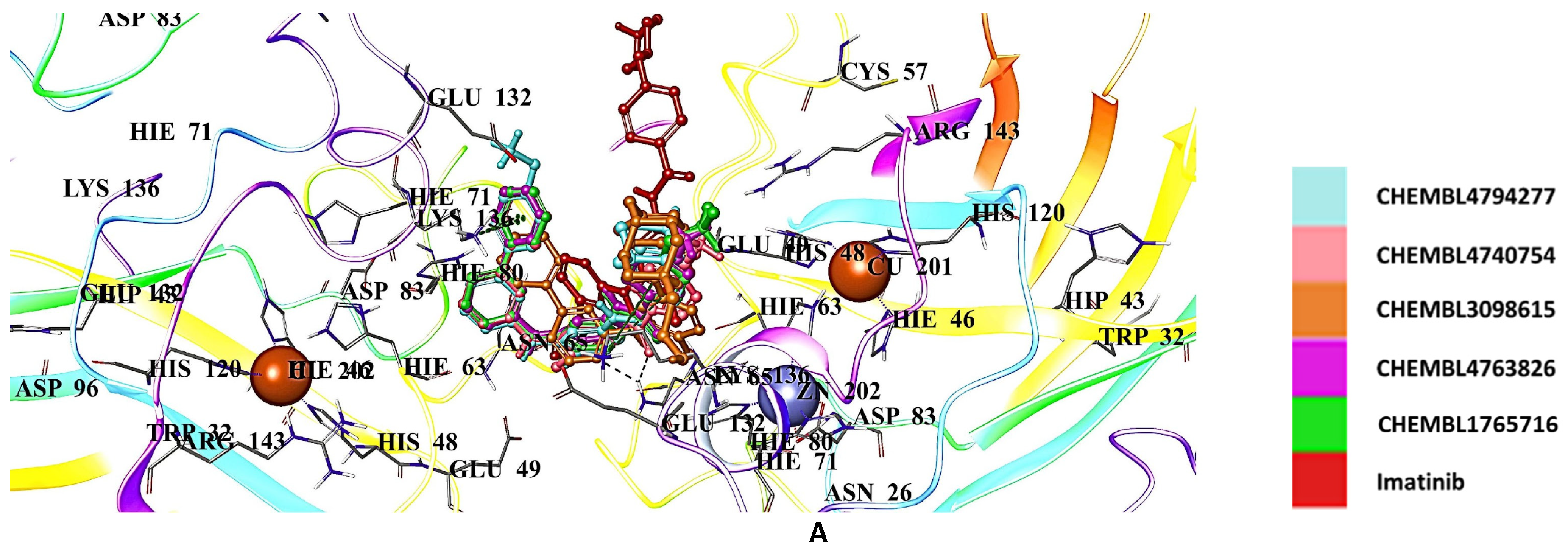
2.5. Docking Studies of 9IYK Structure
3. Discussion
4. Materials and Methods
4.1. Expression and Purification of hSOD1
4.2. Crystallization of hSOD1
4.3. Data Collection, Processing, and Structure Determination of hSOD1
4.4. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andrés, C.M.C.; Pérez de la Lastra, J.M.; Andrés Juan, C.; Plou, F.J.; Pérez-Lebeña, E. Superoxide anion chemistry—Its role at the core of the innate immunity. Int. J. Mol. Sci. 2023, 24, 1841. [Google Scholar] [CrossRef]
- Fridovich, I. Superoxide dismutases. Annu. Rev. Biochem. 1975, 44, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Trist, B.G.; Hilton, J.B.; Hare, D.J.; Crouch, P.J.; Double, K.L. Superoxide dismutase 1 in health and disease: How a frontline antioxidant becomes neurotoxic. Angew. Chem. (Int. Ed. Engl.) 2020, 60, 9215–9246. [Google Scholar] [CrossRef]
- Deng, H.-X.; Hentati, A.; Tainer, J.A.; Iqbal, Z.; Cayabyab, A.; Hung, W.-Y.; Getzoff, E.D.; Hu, P.; Herzfeldt, B.; Roos, R.P.; et al. Amyotrophic lateral ssclerosis and structural defects in Cu,Zn superoxide dismutase. Science 1993, 261, 1047–1051. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, S.; Ono, K.; Takeuchi, H. Uv resonance raman scattering from metal-coordinating histidine residues in Cu,Zn-superoxide dismutase. J. Raman Spectrosc. 1998, 29, 969–975. [Google Scholar] [CrossRef]
- Rakhit, R.; Chakrabartty, A. Structure, folding, and misfolding of Cu,Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2006, 1762, 1025–1037. [Google Scholar] [CrossRef]
- Hart, P.J.; Balbirnie, M.M.; Ogihara, N.L.; Nersissian, A.M.; Weiss, M.S.; Valentine, J.S.; Eisenberg, D. A structure-based mechanism for copper−zinc superoxide dismutase. Biochemistry 1999, 38, 2167–2178. [Google Scholar] [CrossRef]
- Klug, D.; Rabani, J.; Fridovich, I. A direct demonstration of the catalytic action of superoxide dismutase through the use of pulse radiolysis. J. Biol. Chem. 1972, 247, 4839–4842. [Google Scholar] [CrossRef]
- McCord, J.M.; Fridovich, I. The reduction of cytochrome c by milk xanthine oxidase. J. Biol. Chem. 1968, 243, 5753–5760. [Google Scholar] [CrossRef]
- Tainer, J.A.; Getzoff, E.D.; Richardson, J.S.; Richardson, D.C. Structure and mechanism of copper, zinc superoxide dismutase. Nature 1983, 306, 284–287. [Google Scholar] [CrossRef]
- Ferraroni, M.; Rypniewski, W.R.; Bruni, B.; Orioli, P.; Mangani, S.; Ferraroni, M.; Rypniewski, W.R.; Bruni, B.; Orioli, P.; Mangani, S. Crystallographic determination of reduced bovine superoxide dismutase at pH 5. 0 and of anion binding to its active site. JBIC J. Biol. Inorg. Chem. 1998, 3, 411–422. [Google Scholar] [CrossRef]
- Hough, M.A.; Hasnain, S.S. Structure of fully reduced bovine copper zinc superoxide dismutase at 1.15 Å. Structure 2003, 11, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.S.A.; Antonyuk, S.V.; Hasnain, S.S. The biophysics of superoxide dismutase-1 and amyotrophic lateral sclerosis. Q. Rev. Biophys. 2019, 52, e12. [Google Scholar] [CrossRef] [PubMed]
- Djinović-Carugo, K.; Polticelli, F.; Desideri, A.; Rotilio, G.; Wilson, K.S.; Bolognesi, M. Crystallographic study of azide-inhibited bovine Cu,Zn superoxide dismutase. J. Mol. Biol. 1994, 240, 179–183. [Google Scholar] [CrossRef]
- Getzoff, E.D.; Tainer, J.A.; Weiner, P.K.; Kollman, P.A.; Richardson, J.S.; Richardson, D.C. Electrostatic recognition between superoxide and copper, zinc superoxide dismutase. Nature 1983, 306, 287–290. [Google Scholar] [CrossRef]
- Hough, M.A.; Hasnain, S. Crystallographic structures of bovine copper-zinc superoxide dismutase reveal asymmetry in two subunits: Functionally important three and five coordinate copper sites captured in the same crystal. J. Mol. Biol. 1999, 287, 579–592. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Borsari, M.; Viezzoli, M.S.; Hallewell, R.A. Mutation of the metal-bridging proton-donor his63 residue in human Cu, Zn superoxide dismutase. Eur. J. Biochem. 1995, 232, 220–225. [Google Scholar] [CrossRef]
- Shin, D.S.; DiDonato, M.; Barondeau, D.P.; Hura, G.L.; Hitomi, C.; Berglund, J.A.; Getzoff, E.D.; Cary, S.C.; Tainer, J.A. Superoxide dismutase from the eukaryotic thermophile alvinella pompejana: Structures, stability, mechanism, and insights into amyotrophic lateral sclerosis. J. Mol. Biol. 2009, 385, 1534–1555. [Google Scholar] [CrossRef]
- Healy, E.F.; Flores, R.; Lynch, V.M.; Toledo, S. Protein dynamics of [Cu-Zn] superoxide dismutase (SOD1): How protein motions at the global and local levels impact the reactivity of SOD1. J. Inorg. Biochem. 2020, 210, 111161. [Google Scholar] [CrossRef]
- Sever, B.; Ciftci, H.; DeMirci, H.; Sever, H.; Ocak, F.; Yulug, B.; Tateishi, H.; Tateishi, T.; Otsuka, M.; Fujita, M.; et al. Comprehensive research on past and future therapeutic strategies devoted to treatment of amyotrophic lateral sclerosis. Int. J. Mol. Sci. 2022, 23, 2400. [Google Scholar] [CrossRef]
- Rosen, D.R.; Siddique, T.; Patterson, D.; Figlewicz, D.A.; Sapp, P.; Hentati, A.; Donaldson, D.; Goto, J.; O’Regan, J.P.; Deng, H.-X.; et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993, 362, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Abel, O.; Powell, J.F.; Andersen, P.M.; Al-Chalabi, A. ALSoD: A user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum. Mutat. 2012, 33, 1345–1351. [Google Scholar] [CrossRef]
- Müller, K.; Oh, K.-W.; Nordin, A.; Panthi, S.; Kim, S.H.; Nordin, F.; Freischmidt, A.; Ludolph, A.C.; Ki, C.S.; Forsberg, K.; et al. De novo mutations in SOD1 are a cause of ALS. J. Neurol. Neurosurg. Psychiatry 2022, 93, 201–206. [Google Scholar] [CrossRef]
- Grad, L.I.; Rouleau, G.A.; Ravits, J.; Cashman, N.R. Clinical spectrum of amyotrophic lateral sclerosis (ALS). Cold Spring Harb. Perspect. Med. 2017, 7, a024117. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, M.K. Riluzole and edaravone: A tale of two amyotrophic lateral sclerosis drugs. Med. Res. Rev. 2019, 39, 733–748. [Google Scholar] [CrossRef]
- Miller, T.M.; Cudkowicz, M.E.; Genge, A.; Shaw, P.J.; Sobue, G.; Bucelli, R.C.; Chiò, A.; Van Damme, P.; Ludolph, A.C.; Glass, J.D.; et al. Trial of antisense oligonucleotide tofersen for SOD1 ALS. N. Engl. J. Med. 2022, 387, 1099–1110. [Google Scholar] [CrossRef]
- Wiesenfarth, M.; Dorst, J.; Brenner, D.; Elmas, Z.; Parlak, Ö.; Uzelac, Z.; Kandler, K.; Mayer, K.; Weiland, U.; Herrmann, C.; et al. Effects of tofersen treatment in patients with SOD1-ALS in a “real-world” setting—A 12-month multicenter cohort study from the german early access program. EClinicalMedicine 2024, 69, 102495. [Google Scholar] [CrossRef]
- Platzer, M.; Kiese, S.; Tybussek, T.; Herfellner, T.; Schneider, F.; Schweiggert-Weisz, U.; Eisner, P. Radical scavenging mechanisms of phenolic compounds: A quantitative structure-property relationship (QSPR) study. Front. Nutr. 2022, 9, 882458. [Google Scholar] [CrossRef]
- Bruce, R.M.; Santodonato, J.; Neal, M.W. Summary review of the health effects associated with phenol. Toxicol. Ind. Health 1987, 3, 535–568. [Google Scholar] [CrossRef]
- Watanabe, K.; Tanaka, M.; Yuki, S.; Hirai, M.; Yamamoto, Y. How is edaravone effective against acute ischemic stroke and amyotrophic lateral sclerosis? J. Clin. Biochem. Nutr. 2018, 62, 20–38. [Google Scholar] [CrossRef]
- Watanabe, K.; Morinaka, Y.; Iseki, K.; Watanabe, T.; Yuki, S.; Nishi, H. Structure–activity relationship of 3-methyl-1-phenyl-2-pyrazolin-5-one (edaravone). Redox Rep. 2003, 8, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhong, S.; Yang, H.; Wang, X.; Lv, B.; Bian, Y.; Pei, Y.; Xu, C.; Zhao, Q.; Wu, Y.; et al. Current therapy in amyotrophic lateral sclerosis (ALS): A review on past and future therapeutic strategies. Eur. J. Med. Chem. 2024, 272, 116496. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Wate, R.; Zhang, J.; Ohnishi, S.; Kaneko, S.; Ito, H.; Nakano, S.; Kusaka, H. Treatment with edaravone, initiated at symptom onset, slows motor decline and decreases SOD1 deposition in ALS mice. Exp. Neurol. 2008, 213, 448–455. [Google Scholar] [CrossRef]
- Aoki, M.; Warita, H.; Mizuno, H.; Suzuki, N.; Yuki, S.; Itoyama, Y. Feasibility study for functional test battery of SOD transgenic rat (H46R) and evaluation of edaravone, a free radical scavenger. Brain Res. 2011, 1382, 321–325. [Google Scholar] [CrossRef]
- Badhani, B.; Sharma, N.; Kakkar, R. Gallic acid: A versatile antioxidant with promising therapeutic and industrial applications. RSC Adv. 2015, 5, 27540–27557. [Google Scholar] [CrossRef]
- Ortega-Arellano, H.F.; Jimenez-Del-Rio, M.; Velez-Pardo, C. Dmp53, basket and drICE gene knockdown and polyphenol gallic acid increase life span and locomotor activity in a drosophila parkinson’s disease model. Genet. Mol. Biol. 2013, 36, 608–615. [Google Scholar] [CrossRef]
- Parihar, P.; Jat, D.; Ghafourifar, P.; Parihar, M.S. Efficiency of mitochondrially targeted gallic acid in reducing brain mitochondrial oxidative damage. Cell. Mol. Biol. 2014, 60, 35–41. [Google Scholar] [CrossRef]
- Seo, E.-J.; Fischer, N.; Efferth, T. Phytochemicals as inhibitors of NF-κB for treatment of alzheimer’s disease. Pharmacol. Res. 2018, 129, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Rabiei, Z.; Solati, K.; Amini-Khoei, H. Phytotherapy in treatment of parkinson’s disease: A review. Pharm. Biol. 2019, 57, 355–362. [Google Scholar] [CrossRef]
- AL Zahrani, N.A.; El-Shishtawy, R.M.; Asiri, A.M. Recent developments of gallic acid derivatives and their hybrids in medicinal chemistry: A review. Eur. J. Med. Chem. 2020, 204, 112609. [Google Scholar] [CrossRef]
- Baek, Y.; Lee, S.-J.; Ryu, J.; Jeong, S.; Lee, J.-H.; Ha, N.-C. Gallic acid inhibits filament formation and promotes the disassembly of superoxide dismutase 1, a protein involved in the pathogenesis of amyotrophic lateral sclerosis. Food Biosci. 2023, 55, 102942. [Google Scholar] [CrossRef]
- Semchyshyn, H.M. Reactive carbonyl species in vivo: Generation and dual biological effects. Sci. World J. 2014, 2014, 417842. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, A.; Osawa, T.; Hitomi, K.; Uchida, K. 4-hydroxy-2-nonenal cytotoxicity in renal proximal tubular cells: Protein modification and redox alteration. Arch. Biochem. Biophys. 1996, 333, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Herbst, U.; Toborek, M.; Kaiser, S.; Mattson, M.P.; Hennig, B. 4-hydroxynonenal induces dysfunction and apoptosis of cultured endothelial cells. J. Cell. Physiol. 1999, 181, 295–303. [Google Scholar] [CrossRef]
- Dalleau, S.; Baradat, M.; Guéraud, F.; Huc, L. Cell death and diseases related to oxidative stress:4-hydroxynonenal (HNE) in the balance. Cell Death Differ. 2013, 20, 1615–1630. [Google Scholar] [CrossRef]
- Noguchi, K.; Ali, T.F.; Miyoshi, J.; Orito, K.; Negoto, T.; Biswas, T.; Taira, N.; Koga, R.; Okamoto, Y.; Fujita, M.; et al. Neuroprotective effects of a novel carnosine-hydrazide derivative on hippocampal ca1 damage after transient cerebral ischemia. Eur. J. Med. Chem. 2019, 163, 207–214. [Google Scholar] [CrossRef]
- Martin, L.J. Neuronal death in amyotrophic lateral sclerosis is apoptosis: Possible contribution of a programmed cell death mechanism. J. Neuropathol. Exp. Neurol. 1999, 58, 459–471. [Google Scholar] [CrossRef]
- Ranganathan, S.; Bowser, R. p53 and cell cycle proteins participate in spinal motor neuron cell death in ALS. Open Pathol. J. 2010, 4, 11–22. [Google Scholar] [CrossRef]
- Sathasivam, S.; Ince, P.G.; Shaw, P.J. Apoptosis in amyotrophic lateral sclerosis: A review of the evidence. Neuropathol. Appl. Neurobiol. 2001, 27, 257–274. [Google Scholar] [CrossRef]
- Katsumata, R.; Ishigaki, S.; Katsuno, M.; Kawai, K.; Sone, J.; Huang, Z.; Adachi, H.; Tanaka, F.; Urano, F.; Sobue, G. c-Abl inhibition delays motor neuron degeneration in the G93A mouse, an animal model of amyotrophic lateral sclerosis. PLoS ONE 2012, 7, e46185. [Google Scholar] [CrossRef]
- Guo, W.; Vandoorne, T.; Steyaert, J.; Staats, K.A.; Van Den Bosch, L. The multifaceted role of kinases in amyotrophic lateral sclerosis: Genetic, pathological and therapeutic implications. Brain 2020, 143, 1651–1673. [Google Scholar] [CrossRef] [PubMed]
- Karagiannis, P.; Inoue, H. ALS, a cellular whodunit on motor neuron degeneration. Mol. Cell. Neurosci. 2020, 107, 103524. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.W.; Jeong, Y.E.; Wong, M.; Martin, L.J. DNA damage accumulates and responses are engaged in human ALS brain and spinal motor neurons and DNA repair is activatable in IPSC-derived motor neurons with SOD1 mutations. Acta Neuropathol. Commun. 2020, 8, 7. [Google Scholar] [CrossRef]
- Palomo, V.; Nozal, V.; Rojas-Prats, E.; Gil, C.; Martinez, A. Protein kinase inhibitors for amyotrophic lateral sclerosis therapy. Br. J. Pharmacol. 2020, 178, 1316–1335. [Google Scholar] [CrossRef]
- Motaln, H.; Rogelj, B. The role of c-abl tyrosine kinase in brain and its pathologies. Cells 2023, 12, 2041. [Google Scholar] [CrossRef] [PubMed]
- Rojas, F.; Gonzalez, D.; Cortes, N.; Ampuero, E.; Hernã¡Ndez, D.E.; Fritz, E.; Abarzua, S.; Martinez, A.; Elorza, A.A.; Alvarez, A.; et al. Reactive oxygen species trigger motoneuron death in non-cell-autonomous models of ALS through activation of c-Abl signaling. Front. Cell. Neurosci. 2015, 9, 203. [Google Scholar] [CrossRef]
- Imamura, K.; Izumi, Y.; Watanabe, A.; Tsukita, K.; Woltjen, K.; Yamamoto, T.; Hotta, A.; Kondo, T.; Kitaoka, S.; Ohta, A.; et al. The Src/c-Abl pathway is a potential therapeutic target in amyotrophic lateral sclerosis. Sci. Transl. Med. 2017, 9, eaaf3962. [Google Scholar] [CrossRef]
- Kim, M.O.; Nichols, S.E.; Wang, Y.; McCammon, J.A. Effects of histidine protonation and rotameric states on virtual screening of M. tuberculosis RmlC. J. Comput. Mol. Des. 2013, 27, 235–246. [Google Scholar] [CrossRef]
- Ihara, K.; Fujiwara, N.; Yamaguchi, Y.; Torigoe, H.; Wakatsuki, S.; Taniguchi, N.; Suzuki, K. Structural switching of Cu,Zn-superoxide dismutases at loop VI: Insights from the crystal structure of 2-mercaptoethanol-modified enzyme. Biosci. Rep. 2012, 32, 539–548. [Google Scholar] [CrossRef]
- Forman, H.J.; Fridovich, I. On the stability of bovine superoxide dismutase: The effects of metals. J. Biol. Chem. 1973, 248, 2645–2649. [Google Scholar] [CrossRef]
- Lepock, J.R.; Arnold, L.D.; Torrie, B.H.; Andrews, B.; Kruuv, J. Structural analyses of various Cu2+, Zn2+-superoxide dismutases by differential scanning calorimetry and raman spectroscopy. Arch. Biochem. Biophys. 1985, 241, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Arnesano, F.; Banci, L.; Bertini, I.; Martinelli, M.; Furukawa, Y.; O’Halloran, T.V. The unusually stable quaternary structure of human Cu,Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. J. Biol. Chem. 2004, 279, 47998–48003. [Google Scholar] [CrossRef] [PubMed]
- Ferraroni, M.; Rypniewski, W.; Wilson, K.; Viezzoli, M.; Banci, L.; Bertini, I.; Mangani, S. The crystal structure of the monomeric human SOD mutant F50E/G51E/E133Q at atomic resolution. The enzyme mechanism revisited. J. Mol. Biol. 1999, 288, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Caruano-Yzermans, A.; Rodriguez, A.; Scheurmann, J.P.; Slunt, H.H.; Cao, X.; Gitlin, J.; Hart, P.J.; Borchelt, D.R. Disease-associated mutations at copper ligand histidine residues of superoxide dismutase 1 diminish the binding of copper and compromise dimer stability. J. Biol. Chem. 2007, 282, 345–352. [Google Scholar] [CrossRef]
- Cao, X.; Antonyuk, S.V.; Seetharaman, S.V.; Whitson, L.J.; Taylor, A.B.; Holloway, S.P.; Strange, R.W.; Doucette, P.A.; Valentine, J.S.; Tiwari, A.; et al. Structures of the G85R variant of SOD1 in familial amyotrophic lateral sclerosis. J. Biol. Chem. 2008, 283, 16169–16177. [Google Scholar] [CrossRef]
- Rakhit, R.; Cunningham, P.; Furtos-Matei, A.; Dahan, S.; Qi, X.-F.; Crow, J.P.; Cashman, N.R.; Kondejewski, L.H.; Chakrabartty, A. Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J. Biol. Chem. 2002, 277, 47551–47556. [Google Scholar] [CrossRef]
- Rakhit, R.; Crow, J.P.; Lepock, J.R.; Kondejewski, L.H.; Cashman, N.R.; Chakrabartty, A. Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J. Biol. Chem. 2004, 279, 15499–15504. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, H.; Eggers, D.K.; Nersissian, A.M.; Faull, K.F.; Goto, J.J.; Ai, J.; Sanders-Loehr, J.; Gralla, E.B.; Valentine, J.S. Copper(2+) binding to the surface residue cysteine 111 of His46Arg human copper−zinc superoxide dismutase, a familial amyotrophic lateral sclerosis mutant. Biochemistry 2000, 39, 8125–8132. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Ding, F.; Dokholyan, N.V. Probing protein aggregation using simplified models and discrete molecular dynamics. Front. Biosci. 2008, 13, 4795–4808. [Google Scholar] [CrossRef]
- Shi, Y.; Rhodes, N.R.; Abdolvahabi, A.; Kohn, T.; Cook, N.P.; Marti, A.A.; Shaw, B.F. Deamidation of asparagine to aspartate destabilizes Cu, Zn superoxide dismutase, accelerates fibrillization, and mirrors ALS-linked mutations. J. Am. Chem. Soc. 2013, 135, 15897–15908. [Google Scholar] [CrossRef]
- Trist, B.G.; Genoud, S.; Roudeau, S.; Rookyard, A.; Abdeen, A.; Cottam, V.; Hare, D.J.; White, M.; Altvater, J.; Fifita, J.A.; et al. Altered SOD1 maturation and post-translational modification in amyotrophic lateral sclerosis spinal cord. Brain 2022, 145, 3108–3130. [Google Scholar] [CrossRef]
- Xia, Y.; Chen, Z.; Xu, G.; Borchelt, D.R.; Ayers, J.I.; Giasson, B.I. Novel SOD1 monoclonal antibodies against the electrostatic loop preferentially detect misfolded SOD1 aggregates. Neurosci. Lett. 2021, 742, 135553. [Google Scholar] [CrossRef] [PubMed]
- Getzoff, E.D.; Cabelli, D.E.; Fisher, C.L.; Parge, H.E.; Viezzoli, M.S.; Banci, L.; Hallewell, R.A. Faster superoxide dismutase mutants designed by enhancing electrostatic guidance. Nature 1992, 358, 347–351. [Google Scholar] [CrossRef]
- Wang, L.-Q.; Ma, Y.; Yuan, H.-Y.; Zhao, K.; Zhang, M.-Y.; Wang, Q.; Huang, X.; Xu, W.-C.; Dai, B.; Chen, J.; et al. Cryo-EM structure of an amyloid fibril formed by full-length human SOD1 reveals its conformational conversion. Nat. Commun. 2022, 13, 3491. [Google Scholar] [CrossRef]
- Fisher, C.L.; Cabelli, D.E.; Hallewell, R.A.; Beroza, P.; Lo, T.P.; Getzoff, E.D.; Tainer, J.A. Computational, pulse-radiolytic, and structural investigations of lysine-136 and its role in the electrostatic triad of human Cu,Zn superoxide dismutase. Proteins Struct. Funct. Bioinform. 1994, 29, 103–112. [Google Scholar] [CrossRef]
- Abe, K.; Aoki, M.; Tsuji, S.; Itoyama, Y.; Sobue, G.; Togo, M. Safety and efficacy of edaravone in well defined patients with amyotrophic lateral sclerosis: A randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2017, 16, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Ertem, F.B.; Guven, O.; Buyukdag, C.; Gocenler, O.; Ayan, E.; Yuksel, B.; Gul, M.; Usta, G.; Cakılkaya, B.; Johnson, J.A.; et al. Protocol for structure determination of SARS-CoV-2 main protease at near-physiological-temperature by serial femtosecond crystallography. STAR Protoc. 2022, 3, 101158. [Google Scholar] [CrossRef]
- Atalay, N.; Akcan, E.K.; Gül, M.; Ayan, E.; Destan, E.; Kuzucu, F.B.E.; Tokay, N.; Çakilkaya, B.; Nergiz, Z.; Usta, G.K.; et al. Cryogenic X-ray crystallographic studies of biomacromolecules at turkish light source “Turkish DeLight”. Turk. J. Biol. 2023, 47, 2. [Google Scholar] [CrossRef]
- Winter, G. Xia2: An expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 2009, 43, 186–190. [Google Scholar] [CrossRef]
- Winter, G.; Waterman, D.G.; Parkhurst, J.M.; Brewster, A.S.; Gildea, R.J.; Gerstel, M.; Fuentes-Montero, L.; Vollmar, M.; Michels-Clark, T.; Young, I.D.; et al. DIALS: Implementation and evaluation of a new integration package. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 85–97. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.S.; Antonyuk, S.V.; Kershaw, N.M.; Strange, R.W.; Hasnain, S.S. Ligand binding and aggregation of pathogenic SOD1. Nat. Commun. 2013, 4, 1758. [Google Scholar] [CrossRef] [PubMed]
- Ciftci, H.; Sever, B.; Ayan, E.; Can, M.; DeMirci, H.; Otsuka, M.; TuYuN, A.F.; Tateishi, H.; Fujita, M. Identification of new L-heptanoylphosphatidyl inositol pentakisphosphate derivatives targeting the interaction with HIV-1 gag by molecular modelling studies. Pharmaceuticals 2022, 15, 1255. [Google Scholar] [CrossRef]
- Guven, O.; Sever, B.; Başoğlu-Ünal, F.; Ece, A.; Tateishi, H.; Koga, R.; Radwan, M.O.; Demir, N.; Can, M.; Aytemir, M.D.; et al. Structural characterization of TRAF6 N-terminal for therapeutic uses and computational studies on new derivatives. Pharmaceuticals 2023, 16, 1608. [Google Scholar] [CrossRef]




| PDB ID | 9IYK |
|---|---|
| Data collection | |
| Beamline | Diamond Beamline i03 |
| Space group | C 1 2 1 |
| Cell dimensions | |
| a, b, c (Å) | 178.19 138.25 112.93 |
| α, β, γ (°) | 90 129.2 90 |
| Resolution (Å)2 | 56.47–2.34 (2.38–2.34) |
| CC(1/2) | 0.94 (0.26) |
| I/σI | 9.0 (0.5) |
| Completeness (%) | 99.1 (97.6) |
| Redundancy | 7.1 (7.3) |
| Refinement | |
| Resolution (Å) | 48.92–2.34 (2.37–2.34) |
| No. reflections | 77,004 (77,004) |
| Rwork/Rfree | 0.20/0.26 (0.35/0.39) |
| No. non-hydrogen atoms | 14,379 |
| Protein | 13,815 |
| Ligand | 73 |
| Water | 503 |
| B-factors | |
| Protein | 70.15 |
| Ligand | 64.69 |
| Water | 53.82 |
| Coordinate errors | 0.41 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.634 |
| Ramachandran plot | |
| Favored (%) | 96.64 |
| Allowed (%) | 3.30 |
| Disallowed (%) | 0.06 |
| Compound | Docking Score | Compound | Docking Score |
|---|---|---|---|
| CHEMBL4205125 | −7.641 | CHEMBL3425716 | −7.244 |
| CHEMBL1490540 | −7.002 | CHEMBL4451230 | −7.231 |
| CHEMBL1390061 | −6.865 | CHEMBL4439020 | −7.164 |
| CHEMBL1518895 | −6.767 | CHEMBL4865214 | −6.972 |
| CHEMBL1872892 | −6.484 | CHEMBL5283559 | −6.889 |
| CHEMBL1256379 | −7.121 | CHEMBL198211 | −6.459 |
| CHEMBL339835 | −7.027 | CHEMBL339893 | −6.291 |
| CHEMBL2146905 | −6.794 | CHEMBL124333 | −6.166 |
| CHEMBL47027 | −6.778 | CHEMBL128326 | −6.156 |
| CHEMBL95308 | −6.761 | CHEMBL197930 | −6.142 |
| CHEMBL1075867 | −9.760 | CHEMBL4794277 | −6.527 |
| CHEMBL429344 | −8.935 | CHEMBL4740754 | −6.491 |
| CHEMBL1075871 | −8.858 | CHEMBL3098615 | −6.489 |
| CHEMBL1075979 | −8.841 | CHEMBL4763826 | −6.488 |
| CHEMBL1076109 | −8.757 | CHEMBL1765716 | −6.193 |
| Edaravone | −4.831 | Dasatinib | −5.751 |
| Gallic acid | −5.428 | Bosutinib | −4.382 |
| CNN | −5.769 | Imatinib | −4.969 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yapici, I.; Tokur, A.G.; Sever, B.; Ciftci, H.; Basak, A.N.; DeMirci, H. Structural Insights into the Dynamics of Water in SOD1 Catalysis and Drug Interactions. Int. J. Mol. Sci. 2025, 26, 4228. https://doi.org/10.3390/ijms26094228
Yapici I, Tokur AG, Sever B, Ciftci H, Basak AN, DeMirci H. Structural Insights into the Dynamics of Water in SOD1 Catalysis and Drug Interactions. International Journal of Molecular Sciences. 2025; 26(9):4228. https://doi.org/10.3390/ijms26094228
Chicago/Turabian StyleYapici, Ilkin, Arda Gorkem Tokur, Belgin Sever, Halilibrahim Ciftci, Ayse Nazli Basak, and Hasan DeMirci. 2025. "Structural Insights into the Dynamics of Water in SOD1 Catalysis and Drug Interactions" International Journal of Molecular Sciences 26, no. 9: 4228. https://doi.org/10.3390/ijms26094228
APA StyleYapici, I., Tokur, A. G., Sever, B., Ciftci, H., Basak, A. N., & DeMirci, H. (2025). Structural Insights into the Dynamics of Water in SOD1 Catalysis and Drug Interactions. International Journal of Molecular Sciences, 26(9), 4228. https://doi.org/10.3390/ijms26094228









