Discovery of Boronic Acids-Based β-Lactamase Inhibitors Through In Situ Click Chemistry
Abstract
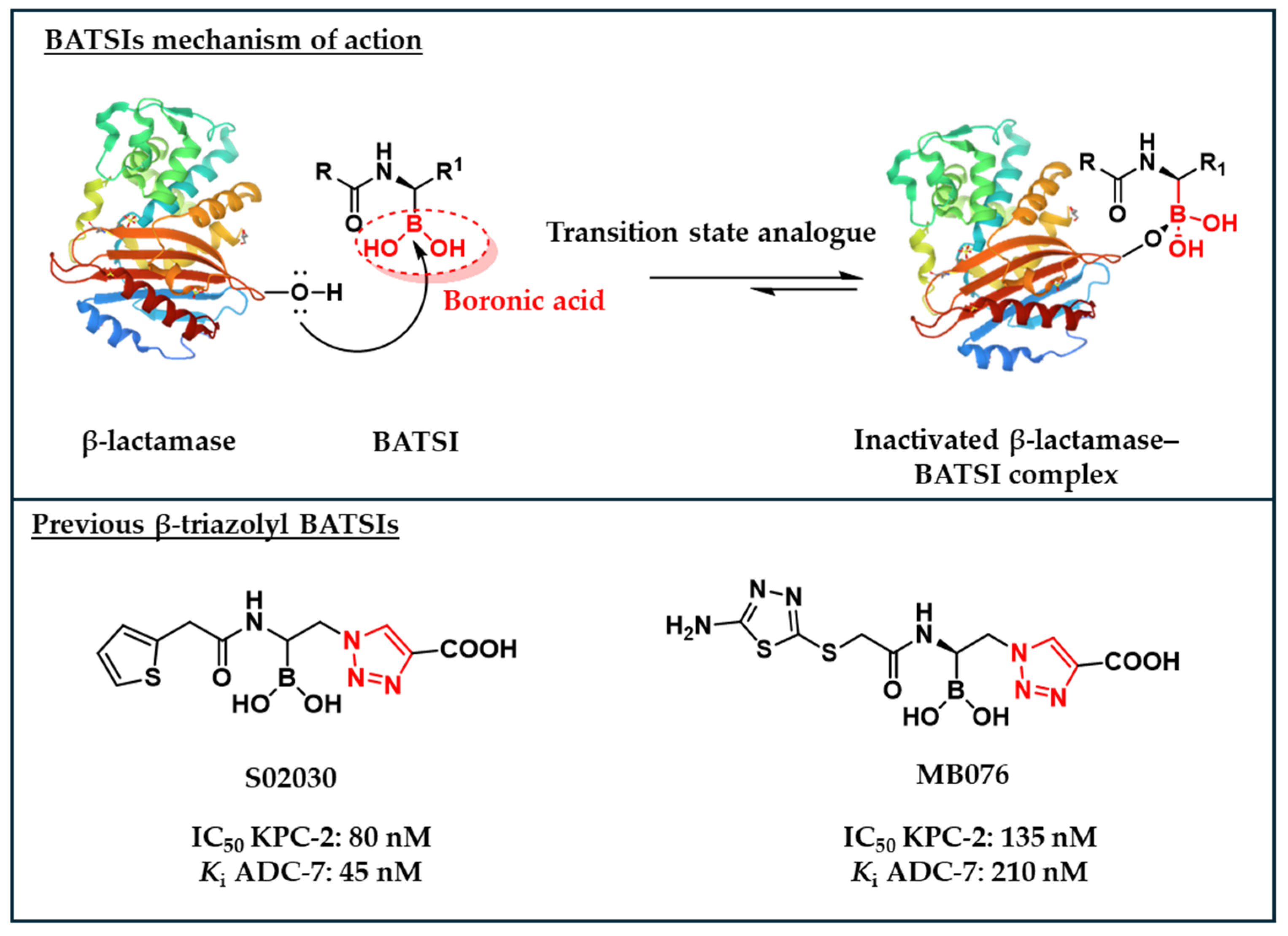
1. Introduction
2. Results
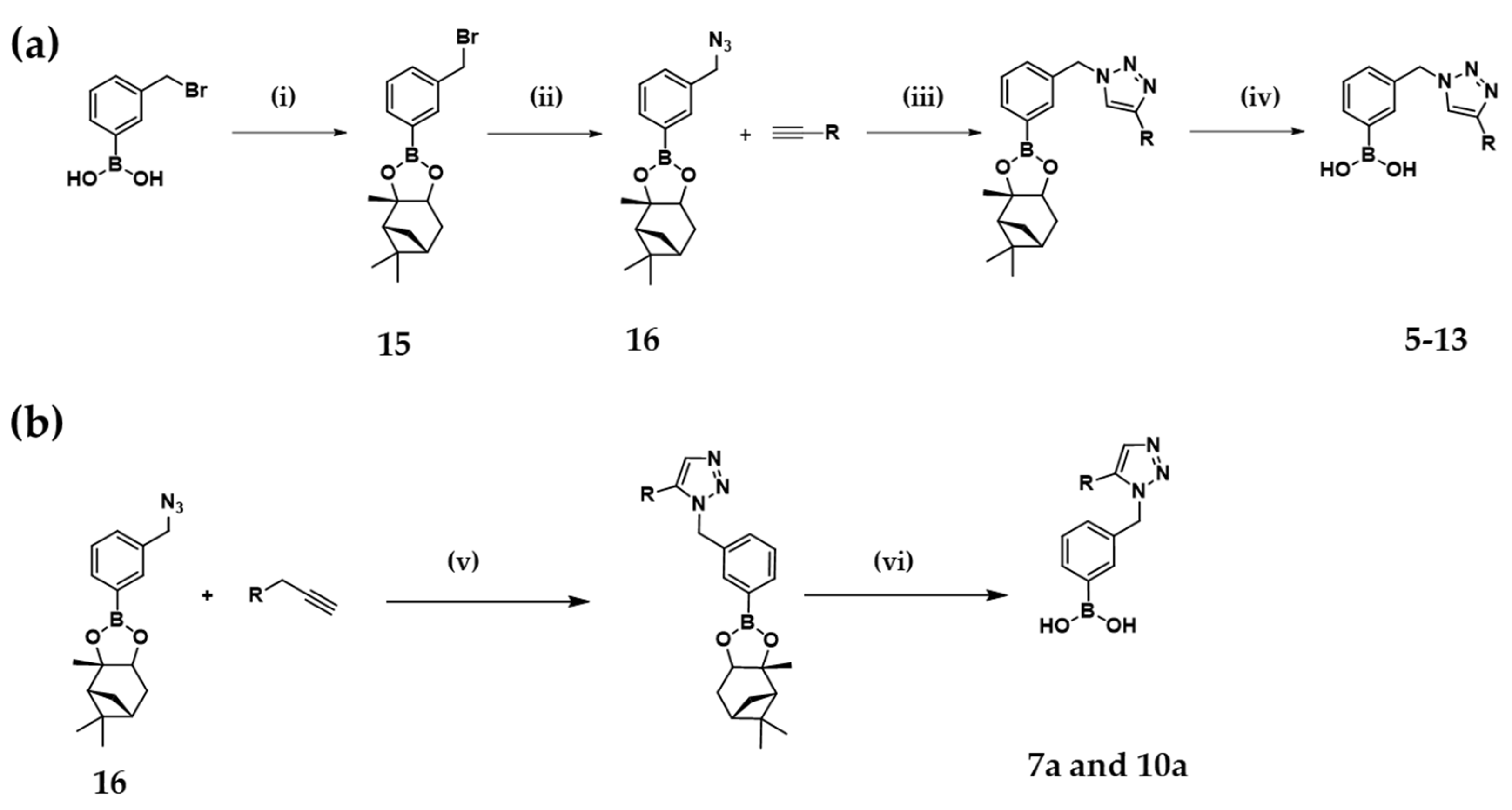
2.1. Design and Synthesis of the Warhead
2.2. Inhibition (%) of the Warheads on Representative BLs
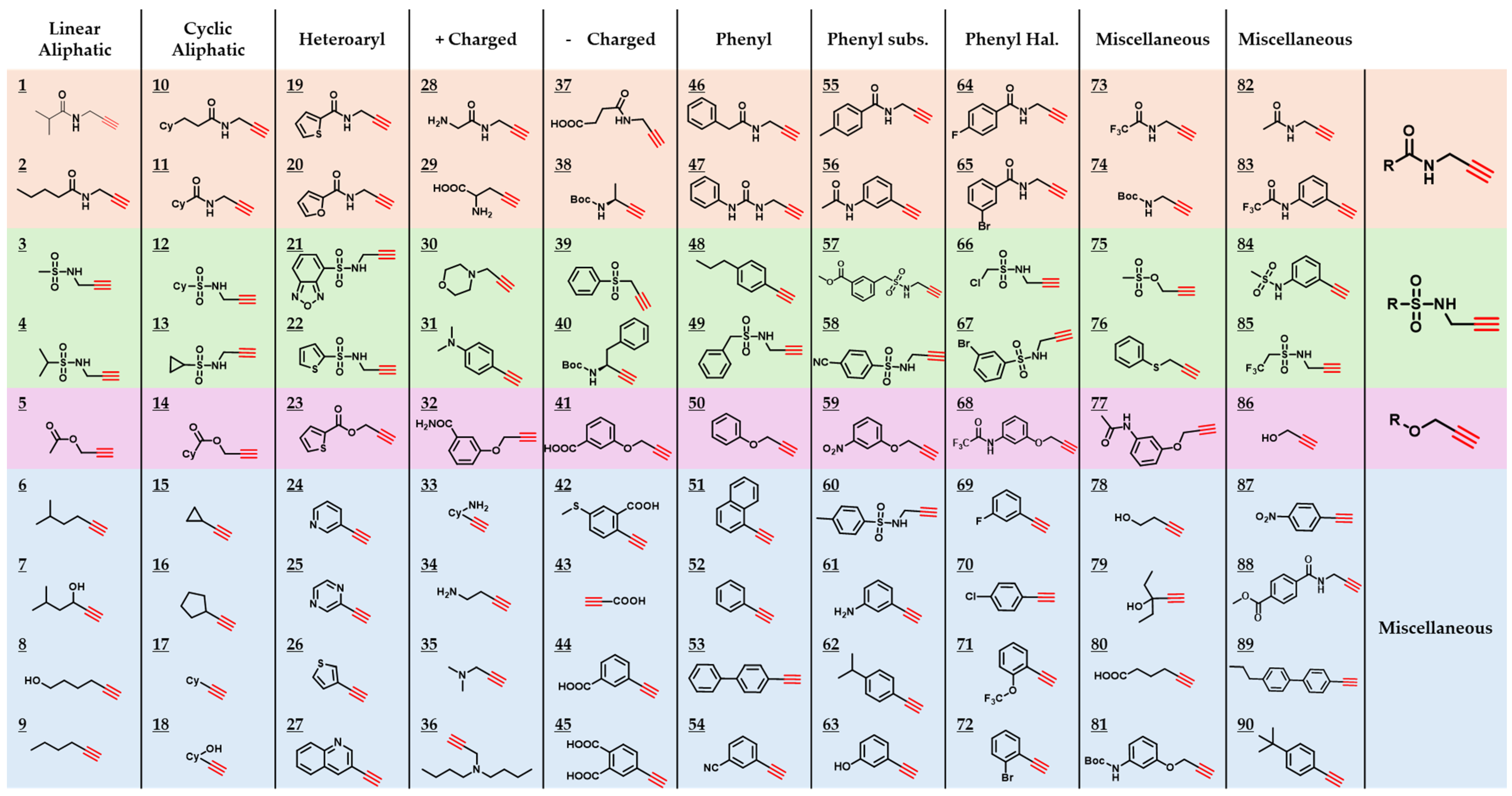
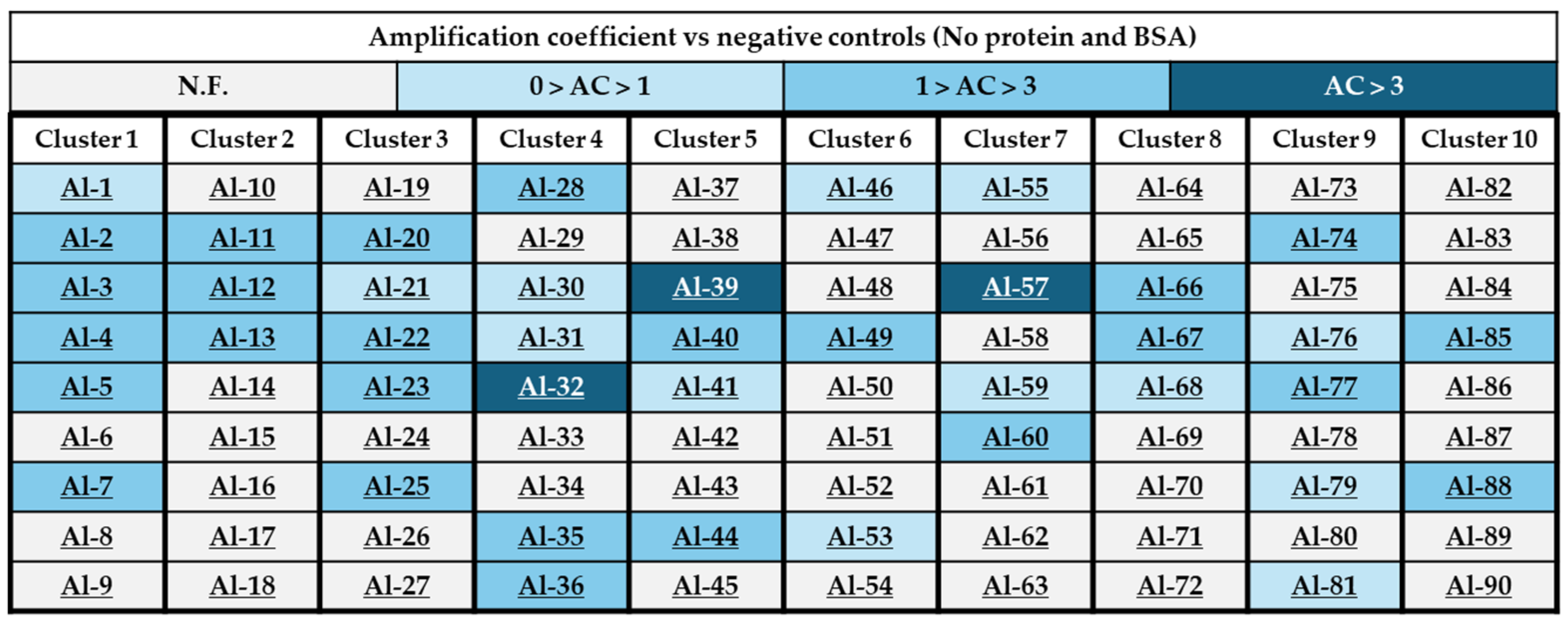
2.3. Generation of a 90-Component Alkyne Library
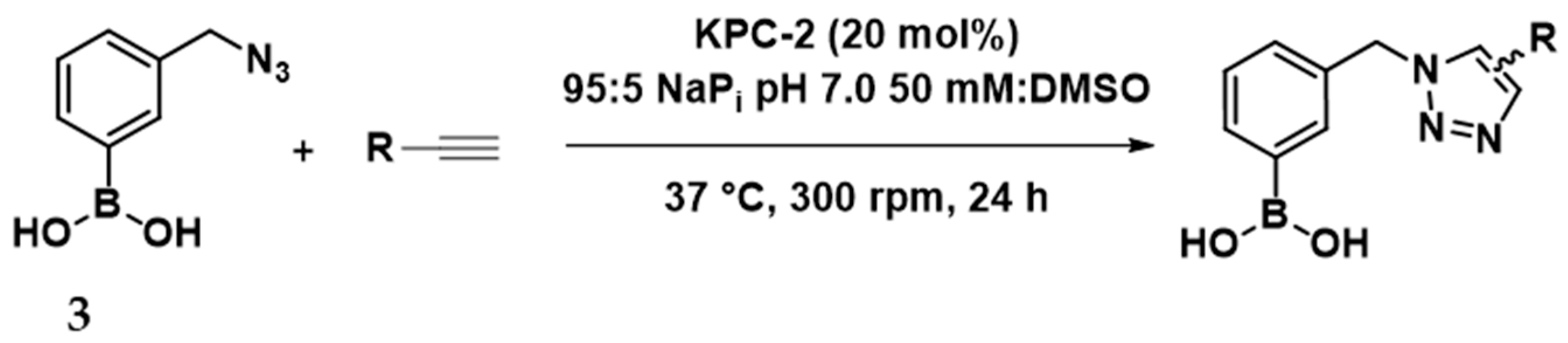
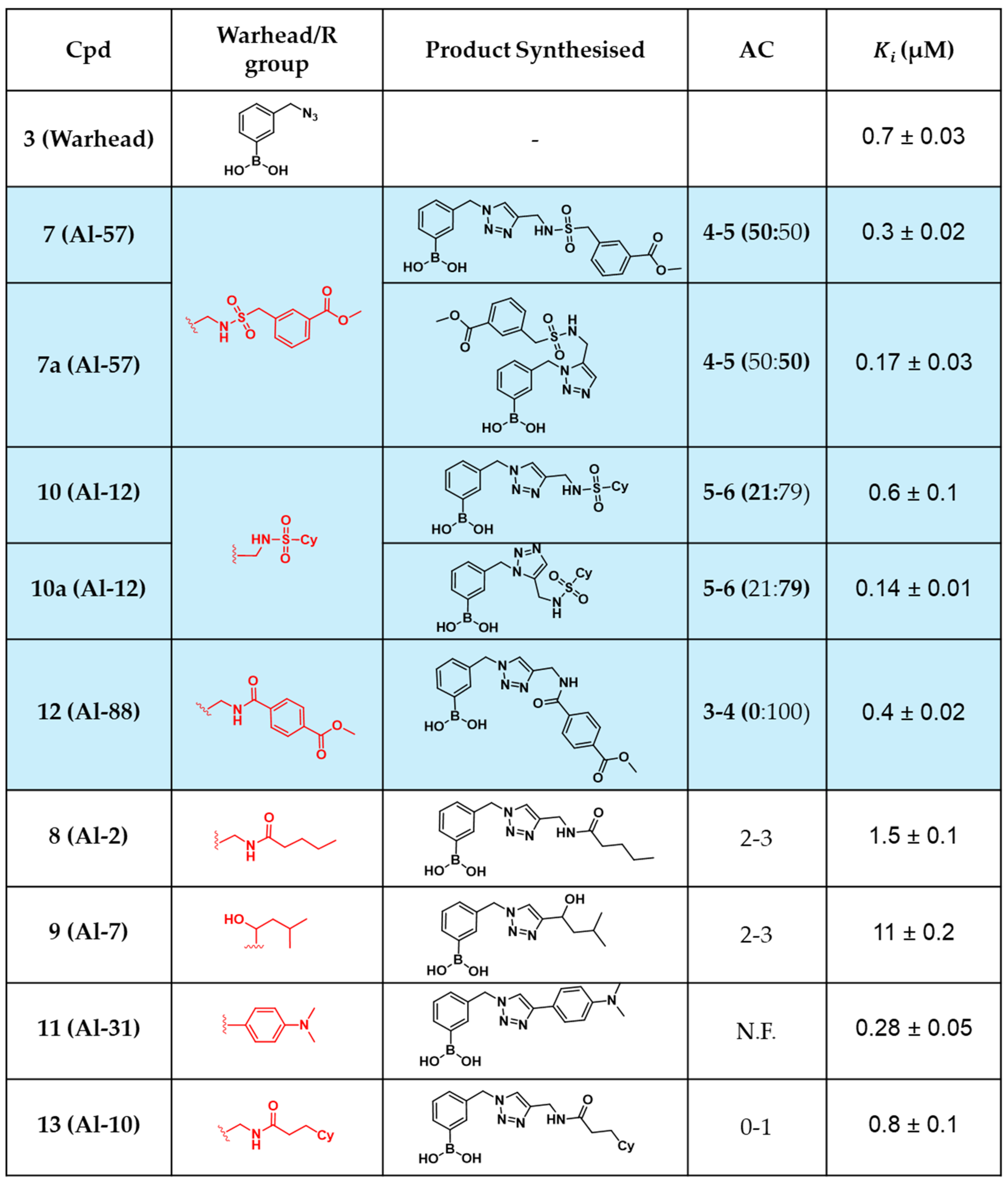
2.4. In Situ Click Chemistry with KPC-2
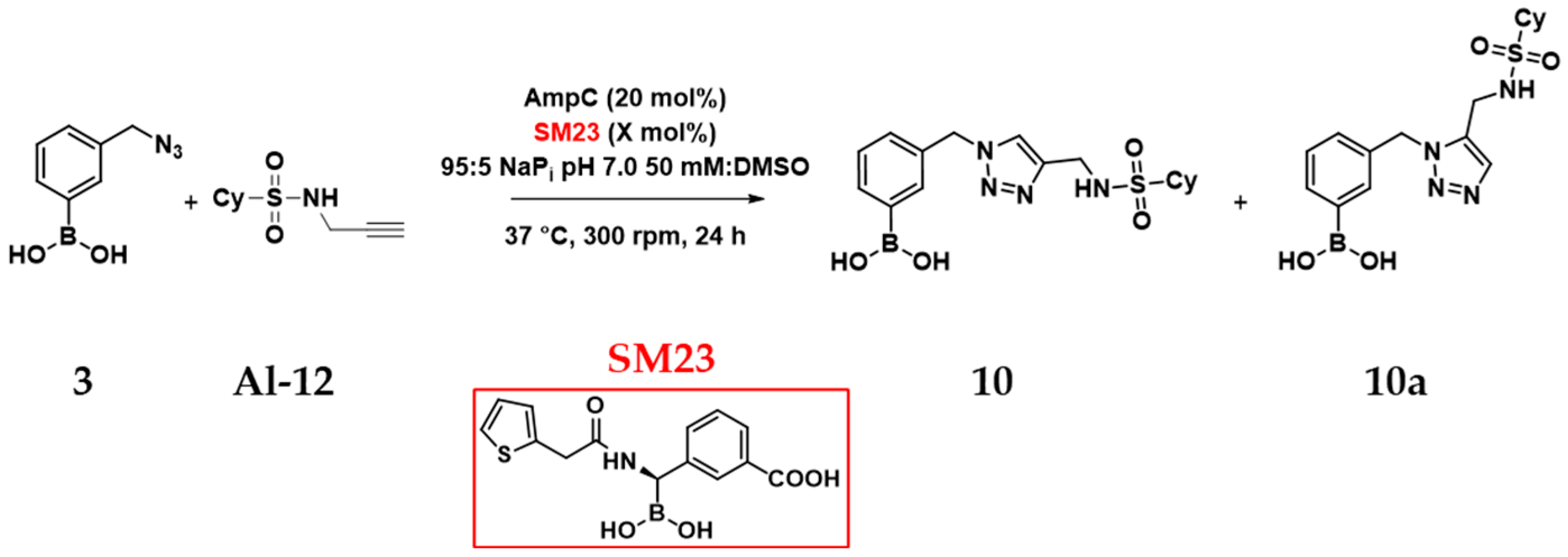
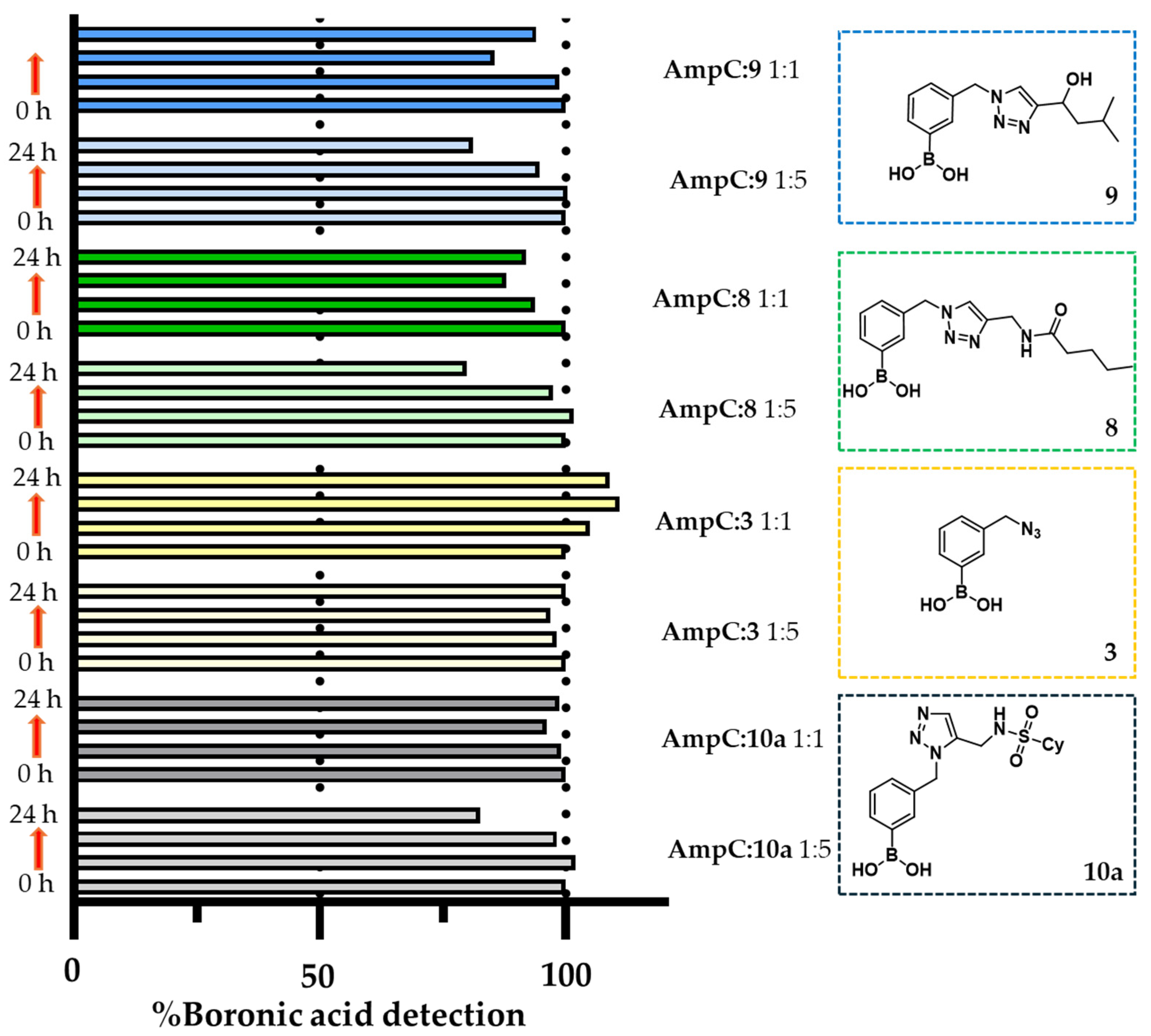
2.5. In Situ Click Chemistry with AmpC
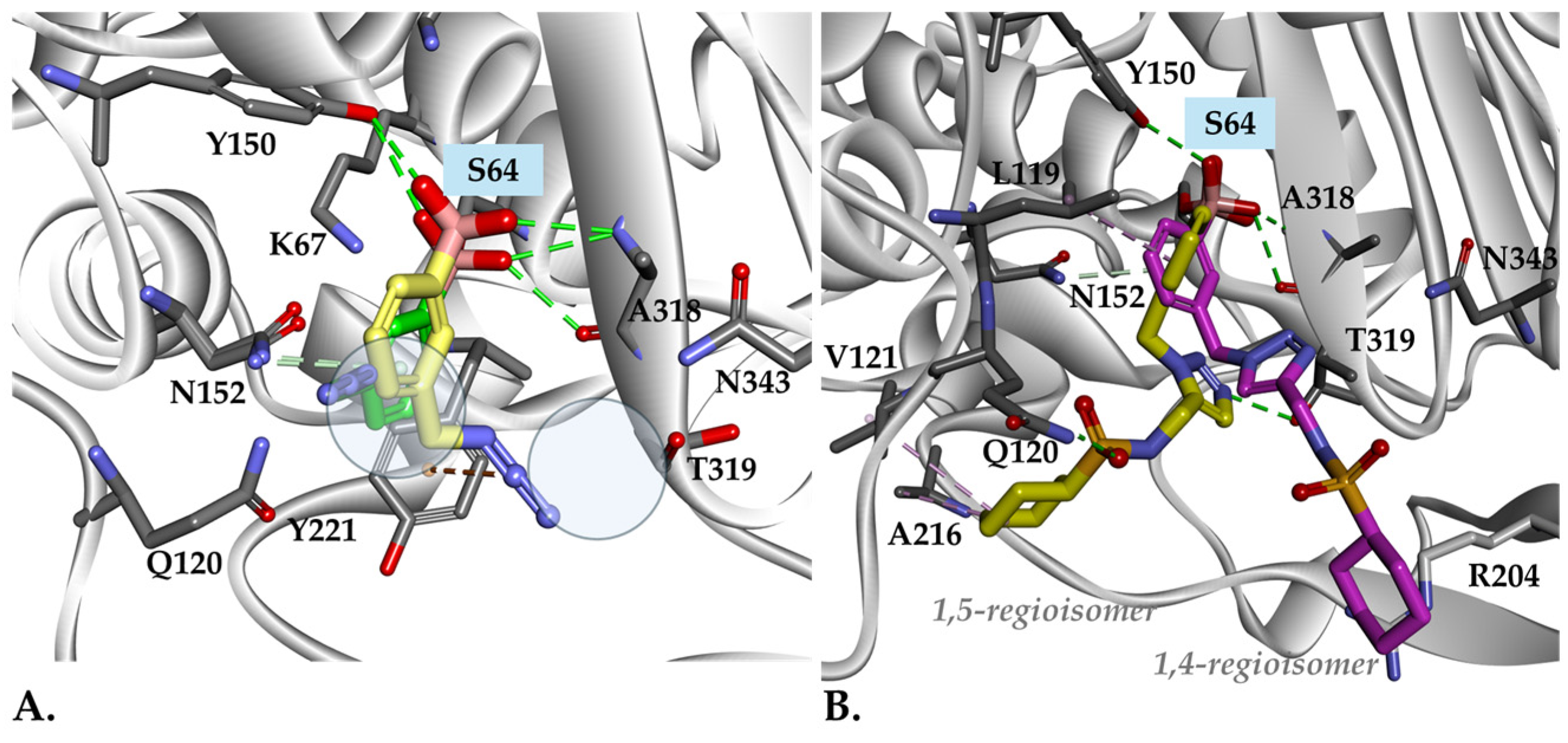
2.6. Molecular Docking Studies
3. Discussion
4. Materials and Methods
4.1. Chemistry
4.2. In Situ Click Chemistry
4.3. Microbiology and Determination of Ki
4.4. Docking Studies
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, S.S.; Apisarnthanarak, A.; Hsu, L.Y. Mechanisms of β-lactam antimicrobial resistance and epidemiology of major community- and healthcare-associated multidrug-resistant bacteria. Adv. Drug Deliv. Rev. 2014, 78, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.M.; Silva, B.N.M.d.; Barbosa, G.; Barreiro, E.J. β-lactam antibiotics: An overview from a medicinal chemistry perspective. Eur. J. Med. Chem. 2020, 208, 112829. [Google Scholar] [CrossRef] [PubMed]
- Docquier, J.-D.; Mangani, S. An update on β-lactamase inhibitor discovery and development. Drug Resist. Updates 2018, 36, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liao, X.; Ding, T.; Ahn, J. Role of β-Lactamase Inhibitors as Potentiators in Antimicrobial Chemotherapy Targeting Gram-Negative Bacteria. Antibiotics 2024, 13, 260. [Google Scholar] [CrossRef]
- Munita Jose, M.; Arias Cesar, A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 464–473. [Google Scholar] [CrossRef]
- Darby, E.M.; Trampari, E.; Siasat, P.; Gaya, M.S.; Alav, I.; Webber, M.A.; Blair, J.M.A. Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol. 2023, 21, 280–295. [Google Scholar] [CrossRef]
- Hall, B.G.; Barlow, M. Revised Ambler classification of β-lactamases. J. Antimicrob. Chemother. 2005, 55, 1050–1051. [Google Scholar] [CrossRef]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.A.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby George, A. Updated Functional Classification of β-Lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef]
- Hall, B.G.; Barlow, M. Evolution of the serine β-lactamases: Past, present and future. Drug Resist. Updates 2004, 7, 111–123. [Google Scholar] [CrossRef]
- Bradford Patricia, A. Extended-Spectrum β-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC-Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef]
- Hammoudi Halat, D.; Ayoub Moubareck, C. The Current Burden of Carbapenemases: Review of Significant Properties and Dissemination among Gram-Negative Bacteria. Antibiotics 2020, 9, 186. [Google Scholar] [CrossRef]
- Bedenic, B.; Plecko, V.; Sardelic, S.; Uzunovic, S.; Torkar, K.G. Carbapenemases in Gram-Negative Bacteria: Laboratory Detection and Clinical Significance. BioMed Res. Int. 2014, 2014, 841951. [Google Scholar] [CrossRef]
- Codjoe, F.S.; Donkor, E.S. Carbapenem Resistance: A Review. Med. Sci. 2018, 6, 1. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Interplay between β-lactamases and new β-lactamase inhibitors. Nat. Rev. Microbiol. 2019, 17, 295–306. [Google Scholar] [CrossRef]
- Caselli, E.; Romagnoli, C.; Vahabi, R.; Taracila, M.A.; Bonomo, R.A.; Prati, F. Click Chemistry in Lead Optimization of Boronic Acids as β-Lactamase Inhibitors. J. Med. Chem. 2015, 58, 5445–5458. [Google Scholar] [CrossRef]
- Caselli, E.; Romagnoli, C.; Powers, R.A.; Taracila, M.A.; Bouza, A.A.; Swanson, H.C.; Smolen, K.A.; Fini, F.; Wallar, B.J.; Bonomo, R.A.; et al. Inhibition of Acinetobacter-Derived Cephalosporinase: Exploring the Carboxylate Recognition Site Using Novel β-Lactamase Inhibitors. ACS Infect. Dis. 2018, 4, 337–348. [Google Scholar] [CrossRef]
- Ishikawa, T.; Furukawa, N.; Caselli, E.; Prati, F.; Taracila, M.A.; Bethel, C.R.; Ishii, Y.; Shimizu-Ibuka, A.; Bonomo, R.A. Insights Into the Inhibition of MOX-1 β-Lactamase by S02030, a Boronic Acid Transition State Inhibitor. Front. Microbiol. 2021, 12, 720036. [Google Scholar] [CrossRef]
- Rojas, L.J.; Taracila, M.A.; Papp-Wallace, K.M.; Bethel, C.R.; Caselli, E.; Romagnoli, C.; Winkler, M.L.; Spellberg, B.; Prati, F.; Bonomo, R.A. Boronic Acid Transition State Inhibitors Active against KPC and Other Class A β-Lactamases: Structure-Activity Relationships as a Guide to Inhibitor Design. Antimicrob. Agents Chemother. 2016, 60, 1751–1759. [Google Scholar] [CrossRef]
- Winkler, M.L.; Rodkey, E.A.; Taracila, M.A.; Drawz, S.M.; Bethel, C.R.; Papp-Wallace, K.M.; Smith, K.M.; Xu, Y.; Dwulit-Smith, J.R.; Romagnoli, C.; et al. Design and Exploration of Novel Boronic Acid Inhibitors Reveals Important Interactions with a Clavulanic Acid-Resistant Sulfhydryl-Variable (SHV) β-Lactamase. J. Med. Chem. 2013, 56, 1084–1097. [Google Scholar] [CrossRef] [PubMed]
- Hamrick Jodie, C.; Docquier, J.-D.; Uehara, T.; Myers Cullen, L.; Six David, A.; Chatwin Cassandra, L.; John Kaitlyn, J.; Vernacchio Salvador, F.; Cusick Susan, M.; Trout Robert, E.L.; et al. VNRX-5133 (Taniborbactam), a Broad-Spectrum Inhibitor of Serine- and Metallo-β-Lactamases, Restores Activity of Cefepime in Enterobacterales and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2020, 64, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Krajnc, A.; Brem, J.; Hinchliffe, P.; Calvopiña, K.; Panduwawala, T.D.; Lang, P.A.; Kamps, J.J.A.G.; Tyrrell, J.M.; Widlake, E.; Saward, B.G.; et al. Bicyclic Boronate VNRX-5133 Inhibits Metallo- and Serine-β-Lactamases. J. Med. Chem. 2019, 62, 8544–8556. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Trout, R.E.L.; Chu, G.-H.; McGarry, D.; Jackson, R.W.; Hamrick, J.C.; Daigle, D.M.; Cusick, S.M.; Pozzi, C.; De Luca, F.; et al. Discovery of Taniborbactam (VNRX-5133): A Broad-Spectrum Serine- and Metallo-β-lactamase Inhibitor for Carbapenem-Resistant Bacterial Infections. J. Med. Chem. 2020, 63, 2789–2801. [Google Scholar] [CrossRef]
- Wagenlehner Florian, M.; Gasink Leanne, B.; McGovern Paul, C.; Moeck, G.; McLeroth, P.; Dorr, M.; Dane, A.; Henkel, T. Cefepime–Taniborbactam in Complicated Urinary Tract Infection. N. Engl. J. Med. 2024, 390, 611–622. [Google Scholar] [CrossRef]
- Hecker, S.J.; Reddy, K.R.; Lomovskaya, O.; Griffith, D.C.; Rubio-Aparicio, D.; Nelson, K.; Tsivkovski, R.; Sun, D.; Sabet, M.; Tarazi, Z.; et al. Discovery of Cyclic Boronic Acid QPX7728, an Ultrabroad-Spectrum Inhibitor of Serine and Metallo-β-lactamases. J. Med. Chem. 2020, 63, 7491–7507. [Google Scholar] [CrossRef]
- Tsivkovski, R.; Totrov, M.; Lomovskaya, O. Biochemical Characterization of QPX7728, a New Ultrabroad-Spectrum Beta-Lactamase Inhibitor of Serine and Metallo-Beta-Lactamases. Antimicrob. Agents Chemother. 2020, 64, e00130-20. [Google Scholar] [CrossRef]
- Jacobs, L.M.C.; Consol, P.; Chen, Y. Drug Discovery in the Field of β-Lactams: An Academic Perspective. Antibiotics 2024, 13, 59. [Google Scholar] [CrossRef]
- Powers, R.A.; Caselli, E.; Focia, P.J.; Prati, F.; Shoichet, B.K. Structures of Ceftazidime and Its Transition-State Analogue in Complex with AmpC β-Lactamase: Implications for Resistance Mutations and Inhibitor Design. Biochemistry 2001, 40, 9207–9214. [Google Scholar] [CrossRef]
- Drawz, S.M.; Babic, M.; Bethel, C.R.; Taracila, M.; Distler, A.M.; Ori, C.; Caselli, E.; Prati, F.; Bonomo, R.A. Inhibition of the Class C β-Lactamase from Acinetobacter spp.: Insights into Effective Inhibitor Design. Biochemistry 2010, 49, 329–340. [Google Scholar] [CrossRef]
- Ke, W.; Sampson Jared, M.; Ori, C.; Prati, F.; Drawz Sarah, M.; Bethel Christopher, R.; Bonomo Robert, A.; van den Akker, F. Novel Insights into the Mode of Inhibition of Class A SHV-1 β-Lactamases Revealed by Boronic Acid Transition State Inhibitors. Antimicrob. Agents Chemother. 2011, 55, 174–183. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Eidam, O.; Romagnoli, C.; Caselli, E.; Babaoglu, K.; Pohlhaus, D.T.; Karpiak, J.; Bonnet, R.; Shoichet, B.K.; Prati, F. Design, Synthesis, Crystal Structures, and Antimicrobial Activity of Sulfonamide Boronic Acids as β-Lactamase Inhibitors. J. Med. Chem. 2010, 53, 7852–7863. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Caselli, E.; Fini, F.; Introvigne, M.L.; Stucchi, M.; Taracila, M.A.; Fish, E.R.; Smolen, K.A.; Rather, P.N.; Powers, R.A.; Wallar, B.J.; et al. 1,2,3-Triazolylmethaneboronate: A Structure Activity Relationship Study of a Class of β-Lactamase Inhibitors against Acinetobacter baumannii Cephalosporinase. ACS Infect. Dis. 2020, 6, 1965–1975. [Google Scholar] [CrossRef] [PubMed]
- Introvigne, M.L.; Taracila, M.A.; Prati, F.; Caselli, E.; Bonomo, R.A. α-Triazolylboronic Acids: A Promising Scaffold for Effective Inhibitors of KPCs. ChemMedChem 2020, 15, 1283–1288. [Google Scholar] [CrossRef]
- Powers, R.A.; June, C.M.; Fernando, M.C.; Fish, E.R.; Maurer, O.L.; Baumann, R.M.; Beardsley, T.J.; Taracila, M.A.; Rudin, S.D.; Hujer, K.M.; et al. Synthesis of a Novel Boronic Acid Transition State Inhibitor, MB076: A Heterocyclic Triazole Effectively Inhibits Acinetobacter-Derived Cephalosporinase Variants with an Expanded-Substrate Spectrum. J. Med. Chem. 2023, 66, 8510–8525. [Google Scholar] [CrossRef]
- Nguyen, N.Q.; Krishnan, N.P.; Rojas, L.J.; Prati, F.; Caselli, E.; Romagnoli, C.; Bonomo, R.A.; van den Akker, F. Crystal Structures of KPC-2 and SHV-1 β-Lactamases in Complex with the Boronic Acid Transition State Analog S02030. Antimicrob. Agents Chemother. 2016, 60, 1760–1766. [Google Scholar] [CrossRef]
- Neumann, S.; Biewend, M.; Rana, S.; Binder, W.H. The CuAAC: Principles, Homogeneous and Heterogeneous Catalysts, and Novel Developments and Applications. Macromol. Rapid Commun. 2020, 41, 1900359. [Google Scholar] [CrossRef]
- Zhao, R.; Zhu, J.; Jiang, X.; Bai, R. Click chemistry-aided drug discovery: A retrospective and prospective outlook. Eur. J. Med. Chem. 2024, 264, 116037. [Google Scholar] [CrossRef]
- Bosc, D.; Camberlein, V.; Gealageas, R.; Castillo-Aguilera, O.; Deprez, B.; Deprez-Poulain, R. Kinetic Target-Guided Synthesis: Reaching the Age of Maturity. J. Med. Chem. 2020, 63, 3817–3833. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Kaur, J.; Wuest, M.; Wuest, F. In situ click chemistry generation of cyclooxygenase-2 inhibitors. Nat. Commun. 2017, 8, 1. [Google Scholar] [CrossRef]
- Whiting, M.; Muldoon, J.; Lin, Y.-C.; Silverman, S.M.; Lindstrom, W.; Olson, A.J.; Kolb, H.C.; Finn, M.G.; Sharpless, K.B.; Elder, J.H.; et al. Inhibitors of HIV-1 Protease by Using In Situ Click Chemistry. Angew. Chem. Int. Ed. 2006, 45, 1435–1439. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Sunazuka, T.; Sugawara, A.; Endo, A.; Iguchi, K.; Yamamoto, T.; Ui, H.; Shiomi, K.; Watanabe, T.; Sharpless, K.B.; et al. Chitinase inhibitors: Extraction of the active framework from natural argifin and use of in situ click chemistry. J. Antibiot. 2009, 62, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Grimster, N.P.; Stump, B.; Fotsing, J.R.; Weide, T.; Talley, T.T.; Yamauchi, J.G.; Nemecz, Á.; Kim, C.; Ho, K.-Y.; Sharpless, K.B.; et al. Generation of Candidate Ligands for Nicotinic Acetylcholine Receptors via in situ Click Chemistry with a Soluble Acetylcholine Binding Protein Template. J. Am. Chem. Soc. 2012, 134, 6732–6740. [Google Scholar] [CrossRef] [PubMed]
- Tieu, W.; Soares da Costa, T.P.; Yap, M.Y.; Keeling, K.L.; Wilce, M.C.J.; Wallace, J.C.; Booker, G.W.; Polyak, S.W.; Abell, A.D. Optimising in situ click chemistry: The screening and identification of biotin protein ligase inhibitors. Chem. Sci. 2013, 4, 3533–3537. [Google Scholar] [CrossRef]
- Camberlein, V.; Fléau, C.; Sierocki, P.; Li, L.; Gealageas, R.; Bosc, D.; Guillaume, V.; Warenghem, S.; Leroux, F.; Rosell, M.; et al. Discovery of the First Selective Nanomolar Inhibitors of ERAP2 by Kinetic Target-Guided Synthesis. Angew. Chem. Int. Ed. Engl. 2022, 61, e202203560. [Google Scholar] [CrossRef]
- Parvatkar, P.T.; Wagner, A.; Manetsch, R. Biocompatible reactions: Advances in kinetic target-guided synthesis. Trends Chem. 2023, 5, 657–671. [Google Scholar] [CrossRef]
- Millward, S.W.; Agnew, H.D.; Lai, B.; Lee, S.S.; Lim, J.; Nag, A.; Pitram, S.; Rohde, R.; Heath, J.R. In situ click chemistry: From small molecule discovery to synthetic antibodies. Integr. Biol. 2013, 5, 87–95. [Google Scholar] [CrossRef]
- Oueis, E.; Sabot, C.; Renard, P.-Y. New insights into the kinetic target-guided synthesis of protein ligands. Chem. Commun. 2015, 51, 12158–12169. [Google Scholar] [CrossRef]
- Manetsch, R.; Krasiński, A.; Radić, Z.; Raushel, J.; Taylor, P.; Sharpless, K.B.; Kolb, H.C. In Situ Click Chemistry: Enzyme Inhibitors Made to Their Own Specifications. J. Am. Chem. Soc. 2004, 126, 12809–12818. [Google Scholar] [CrossRef]
- Krasiński, A.; Radić, Z.; Manetsch, R.; Raushel, J.; Taylor, P.; Sharpless, K.B.; Kolb, H.C. In Situ Selection of Lead Compounds by Click Chemistry: Target-Guided Optimization of Acetylcholinesterase Inhibitors. J. Am. Chem. Soc. 2005, 127, 6686–6692. [Google Scholar] [CrossRef]
- Lewis, W.G.; Green, L.G.; Grynszpan, F.; Radić, Z.; Carlier, P.R.; Taylor, P.; Finn, M.G.; Sharpless, K.B. Click Chemistry In Situ: Acetylcholinesterase as a Reaction Vessel for the Selective Assembly of a Femtomolar Inhibitor from an Array of Building Blocks. Angew. Chem. Int. Ed. 2002, 41, 1053–1057. [Google Scholar] [CrossRef]
- Unver, M.Y.; Gierse, R.M.; Ritchie, H.; Hirsch, A.K.H. Druggability Assessment of Targets Used in Kinetic Target-Guided Synthesis. J. Med. Chem. 2018, 61, 9395–9409. [Google Scholar] [CrossRef] [PubMed]
- Gladysz, R.; Vrijdag, J.; Van Rompaey, D.; Lambeir, A.-M.; Augustyns, K.; De Winter, H.; Van der Veken, P. Efforts towards an On-Target Version of the Groebke–Blackburn–Bienaymé (GBB) Reaction for Discovery of Druglike Urokinase (uPA) Inhibitors. Chem.–A Eur. J. 2019, 25, 12380–12393. [Google Scholar] [CrossRef] [PubMed]
- Nacheva, K.; Kulkarni, S.S.; Kassu, M.; Flanigan, D.; Monastyrskyi, A.; Iyamu, I.D.; Doi, K.; Barber, M.; Namelikonda, N.; Tipton, J.D.; et al. Going beyond Binary: Rapid Identification of Protein–Protein Interaction Modulators Using a Multifragment Kinetic Target-Guided Synthesis Approach. J. Med. Chem. 2023, 66, 5196–5207. [Google Scholar] [CrossRef]
- Lossouarn, A.; Renard, P.-Y.; Sabot, C. Tailored Bioorthogonal and Bioconjugate Chemistry: A Source of Inspiration for Developing Kinetic Target-Guided Synthesis Strategies. Bioconjugate Chem. 2021, 32, 63–72. [Google Scholar] [CrossRef]
- Morandi, S.; Morandi, F.; Caselli, E.; Shoichet, B.K.; Prati, F. Structure-based optimization of cephalothin-analogue boronic acids as β-lactamase inhibitors. Bioorganic Med. Chem. 2008, 16, 1195–1205. [Google Scholar] [CrossRef]
- Zhou, J.; Stapleton, P.; Haider, S.; Healy, J. Boronic acid inhibitors of the class A β-lactamase KPC-2. Bioorganic Med. Chem. 2018, 26, 2921–2927. [Google Scholar] [CrossRef]
- Zhou, J.; Stapleton, P.; Xavier-Junior, F.H.; Schatzlein, A.; Haider, S.; Healy, J.; Wells, G. Triazole-substituted phenylboronic acids as tunable lead inhibitors of KPC-2 antibiotic resistance. Eur. J. Med. Chem. 2022, 240, 114571. [Google Scholar] [CrossRef]
- Deprez-Poulain, R.; Hennuyer, N.; Bosc, D.; Liang, W.G.; Enée, E.; Marechal, X.; Charton, J.; Totobenazara, J.; Berte, G.; Jahklal, J.; et al. Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice. Nat. Commun. 2015, 6, 8250. [Google Scholar] [CrossRef]
- Kassu, M.; Parvatkar, P.T.; Milanes, J.; Monaghan, N.P.; Kim, C.; Dowgiallo, M.; Zhao, Y.; Asakawa, A.H.; Huang, L.; Wagner, A.; et al. Shotgun Kinetic Target-Guided Synthesis Approach Enables the Discovery of Small-Molecule Inhibitors against Pathogenic Free-Living Amoeba Glucokinases. ACS Infect. Dis. 2023, 9, 2190–2201. [Google Scholar] [CrossRef]
- Johansson, J.R.; Beke-Somfai, T.; Said Stålsmeden, A.; Kann, N. Ruthenium-Catalyzed Azide Alkyne Cycloaddition Reaction: Scope, Mechanism, and Applications. Chem. Rev. 2016, 116, 14726–14768. [Google Scholar] [CrossRef]
- Boren, B.C.; Narayan, S.; Rasmussen, L.K.; Zhang, L.; Zhao, H.; Lin, Z.; Jia, G.; Fokin, V.V. Ruthenium-Catalyzed Azide−Alkyne Cycloaddition: Scope and Mechanism. J. Am. Chem. Soc. 2008, 130, 8923–8930. [Google Scholar] [CrossRef] [PubMed]
- Morandi, F.; Caselli, E.; Morandi, S.; Focia, P.J.; Blázquez, J.; Shoichet, B.K.; Prati, F. Nanomolar Inhibitors of AmpC β-Lactamase. J. Am. Chem. Soc. 2003, 125, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Krajnc, A.; Lang, P.A.; Panduwawala, T.D.; Brem, J.; Schofield, C.J. Will morphing boron-based inhibitors beat the β-lactamases? Curr. Opin. Chem. Biol. 2019, 50, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Diller, D.J.; Merz Jr, K.M. High throughput docking for library design and library prioritization. Proteins Struct. Funct. Bioinform. 2001, 43, 113–124. [Google Scholar] [CrossRef]
- Rao, S.N.; Head, M.S.; Kulkarni, A.; LaLonde, J.M. Validation Studies of the Site-Directed Docking Program LibDock. J. Chem. Inf. Model. 2007, 47, 2159–2171. [Google Scholar] [CrossRef]
- Tirado-Rives, J.; Jorgensen, W.L. Contribution of Conformer Focusing to the Uncertainty in Predicting Free Energies for Protein−Ligand Binding. J. Med. Chem. 2006, 49, 5880–5884. [Google Scholar] [CrossRef]
- Sharpless, K.B.; Manetsch, R. In situ click chemistry: A powerful means for lead discovery. Expert Opin. Drug Discov. 2006, 1, 525–538. [Google Scholar] [CrossRef]
- Copeland, R.A. Enzymes. A Pratical Introduction to Structure, Mechanism and Data Analysis, 2nd ed.; John Wiley & Sons: New York, NY, USA, 2000. [Google Scholar]
- De Meester, F.; Joris, B.; Reckinger, G.; Bellefroid-Bourguignon, C.; Frère, J.-M.; Waley, S.G. Automated analysis of enzyme inactivation phenomena: Application to β-lactamases and DD-peptidases. Biochem. Pharmacol. 1987, 36, 2393–2403. [Google Scholar] [CrossRef]

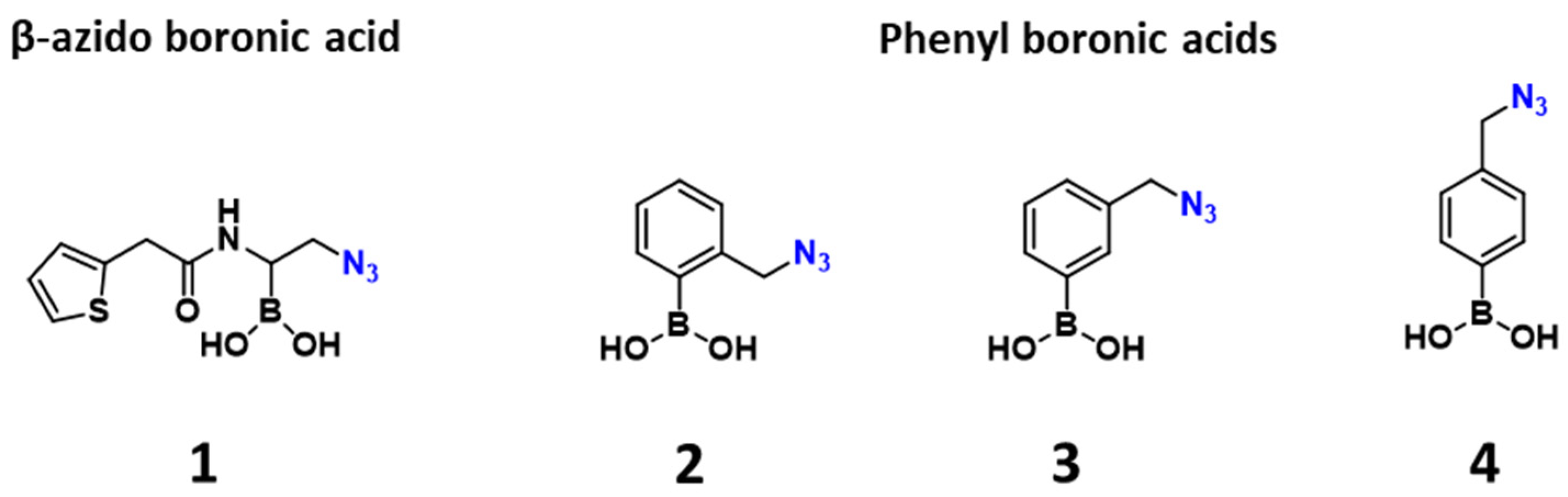


| Entry | β-lactamase 1 | Class | 2 | 3 | 4 |
|---|---|---|---|---|---|
| 1 | KPC-2 2 | A | 35 | 76 | 72 |
| 2 | CTX-M-15 | 0 | 42 | 22 | |
| 3 | KPC-53 | 54 | 65 | 55 | |
| 4 | SHV-12 | 14 | 38 | 48 | |
| 5 | NDM-1 | B | 23 | 20 | 24 |
| 6 | VIM-1 | 23 | 24 | 33 | |
| 7 | IMP-1 | 12 | 11 | 22 | |
| 8 | AmpC 2 | C | 57 | 100 | 81 |
| 9 | ADC-25 | 27 | 67 | 46 | |
| 10 | CMY-2 | 19 | 79 | 67 | |
| 11 | OXA-24 | D | 20 | 24 | 26 |
| 12 | OXA-48 | <1 | 2 | <1 |
| Entry * | Enzyme | %SM23 | Regioselectivity (10:10a) | AC 10/10a |
|---|---|---|---|---|
| 1 | None | - | 50:50 | - |
| 2 | BSA (20 mol%) | - | 60:40 | - |
| 3 | AmpC (20 mol%) | - | 21:79 | 5–6 |
| 4 | AmpC (20 mol%) | 1 mol% | 0:100 | 1–2 |
| 5 | AmpC (20 mol%) | 20 mol% | 50:50 | 0–1 |
| β-lactamase | Buffer | Substrate | KM substrate | [Substrate] | [β-lactamase] |
|---|---|---|---|---|---|
| KPC-2 | NaPi 1 | NCF 3 | 10 ± 1 µM | 50 µM | 1 nM |
| CTX-M-15 | NaPi | NCF | 35 ± 1 µM | 25 µM | 2.5 nM |
| KPC-53 | NaPi | NCF | 106 ± 2 µM | 100 µM | 30 nM |
| SHV-12 | NaPi | NCF | 50 ± 3 µM | 25 µM | 7 nM |
| NDM-1 | HEPES 2 | MPM 4 | 80 ± 1 µM | 100 µM | 4.5 nM |
| VIM-1 | HEPES | MPM | 130 ± 4 µM | 150 µM | 22 nM |
| IMP-1 | HEPES | MPM | 30 ± 1 µM | 80 µM | 13 nM |
| AmpC | NaPi | NCF | 118 ± 2 µM | 142 µM | 14 nM |
| ADC-25 | NaPi | NCF | 120 ± 3 µM | 24 µM | 3 nM |
| CMY-2 | NaPi | NCF | 8 ± 1 µM | 24 µM | 2.5 nM |
| OXA-24 | NaPi | NCF | 29 ± 1 µM | 142 µM | 4 nM |
| OXA-48 | NaPi | IMI 5 | 13 ± 1 µM | 50 µM | 75 nM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santi, N.; Piccirilli, A.; Corsini, F.; Taracila, M.A.; Perilli, M.; Bonomo, R.A.; Fini, F.; Prati, F.; Caselli, E. Discovery of Boronic Acids-Based β-Lactamase Inhibitors Through In Situ Click Chemistry. Int. J. Mol. Sci. 2025, 26, 4182. https://doi.org/10.3390/ijms26094182
Santi N, Piccirilli A, Corsini F, Taracila MA, Perilli M, Bonomo RA, Fini F, Prati F, Caselli E. Discovery of Boronic Acids-Based β-Lactamase Inhibitors Through In Situ Click Chemistry. International Journal of Molecular Sciences. 2025; 26(9):4182. https://doi.org/10.3390/ijms26094182
Chicago/Turabian StyleSanti, Nicolò, Alessandra Piccirilli, Federico Corsini, Magdalena A. Taracila, Mariagrazia Perilli, Robert A. Bonomo, Francesco Fini, Fabio Prati, and Emilia Caselli. 2025. "Discovery of Boronic Acids-Based β-Lactamase Inhibitors Through In Situ Click Chemistry" International Journal of Molecular Sciences 26, no. 9: 4182. https://doi.org/10.3390/ijms26094182
APA StyleSanti, N., Piccirilli, A., Corsini, F., Taracila, M. A., Perilli, M., Bonomo, R. A., Fini, F., Prati, F., & Caselli, E. (2025). Discovery of Boronic Acids-Based β-Lactamase Inhibitors Through In Situ Click Chemistry. International Journal of Molecular Sciences, 26(9), 4182. https://doi.org/10.3390/ijms26094182











