Comparative Transcriptome Analysis Reveals Key Genes Related to Erythritol Production in Yarrowia lipolytica and the Optimization of Culture Conditions
Abstract
1. Introduction
2. Results and Discussion
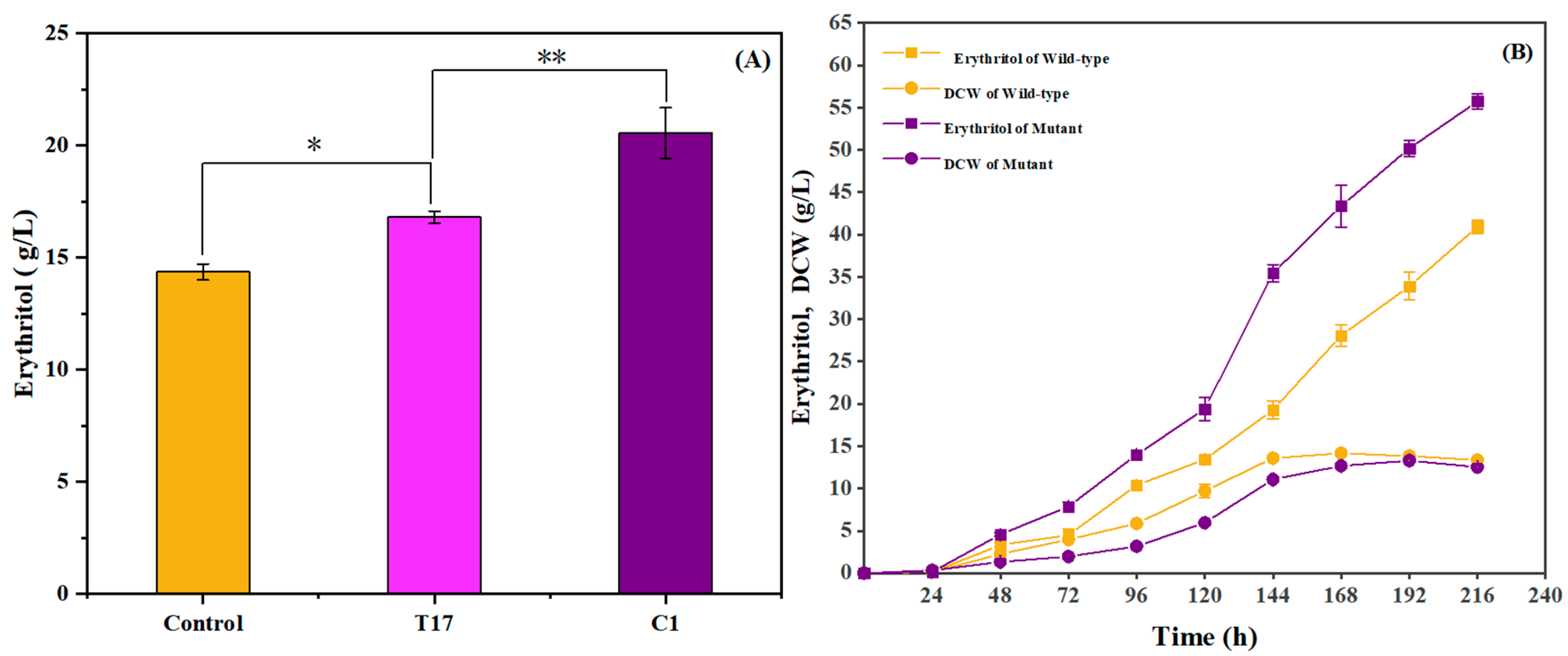
2.1. Mutation of Y. lipolytica for Improving Erythritol Production
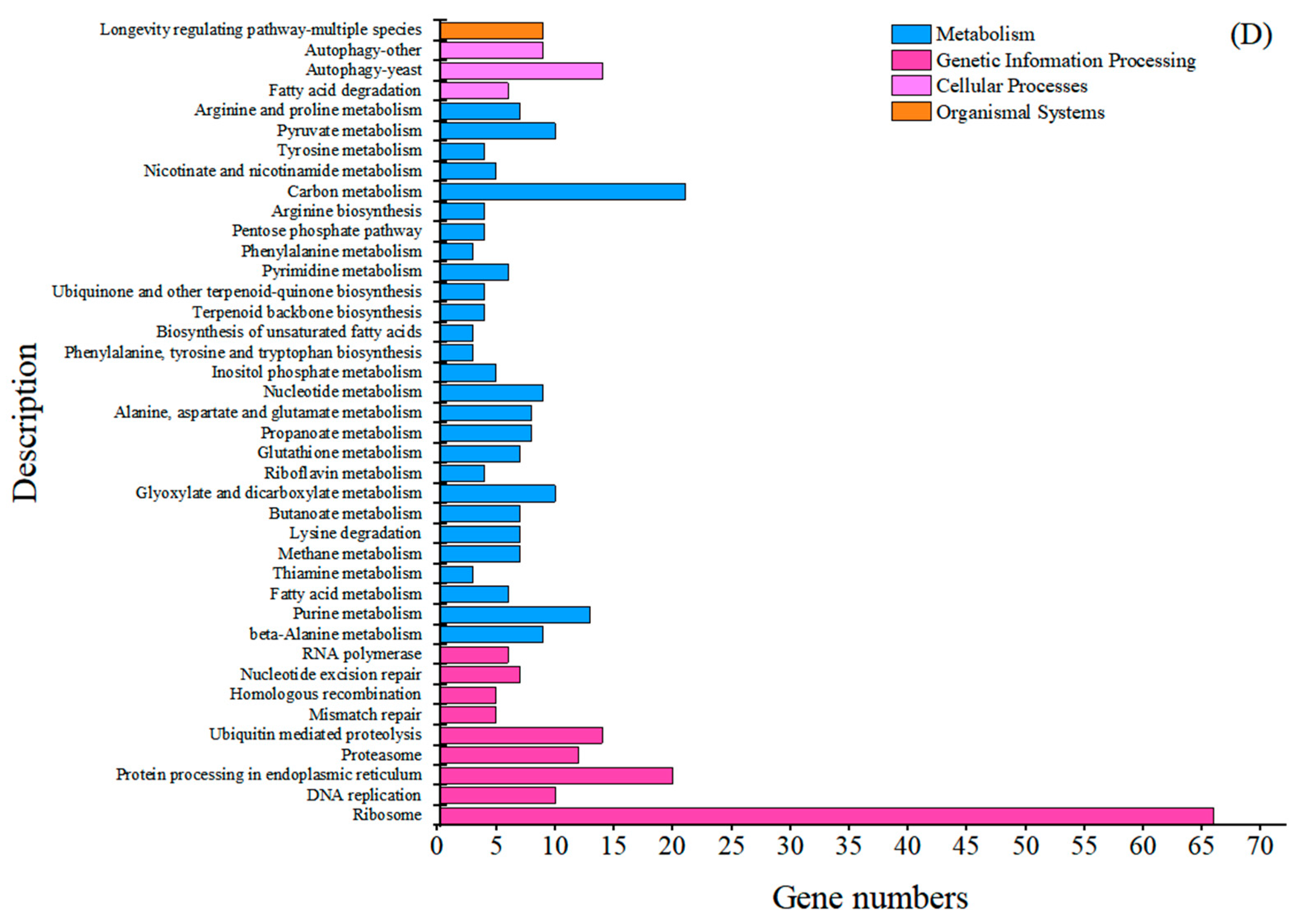
2.2. Comparative Transcriptome Analysis
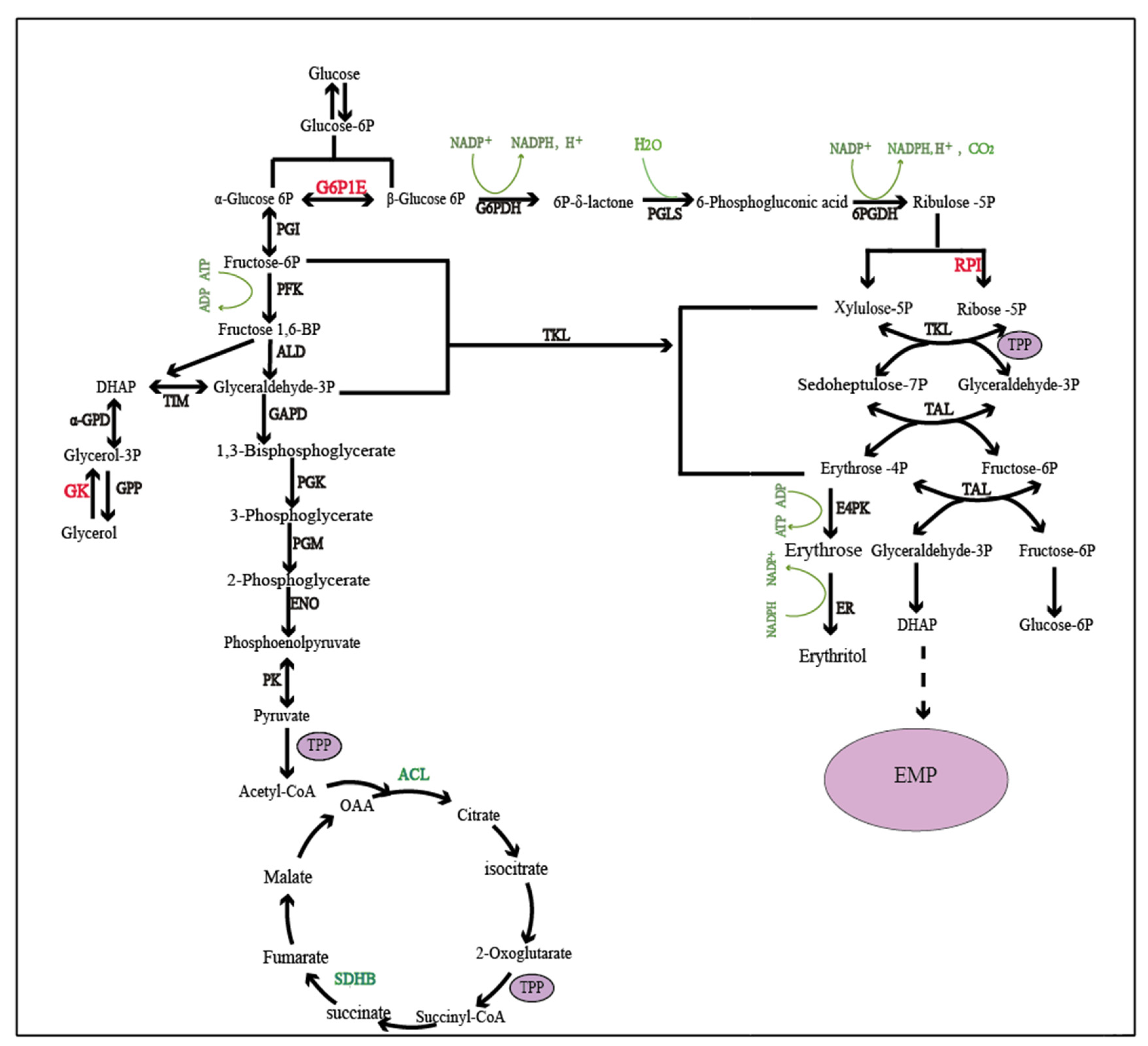
2.3. Key Genes Affecting Erythritol Production in Oxidative Synthesis
2.4. Key Genes Affecting Erythritol Production in Non-Oxidative Synthesis
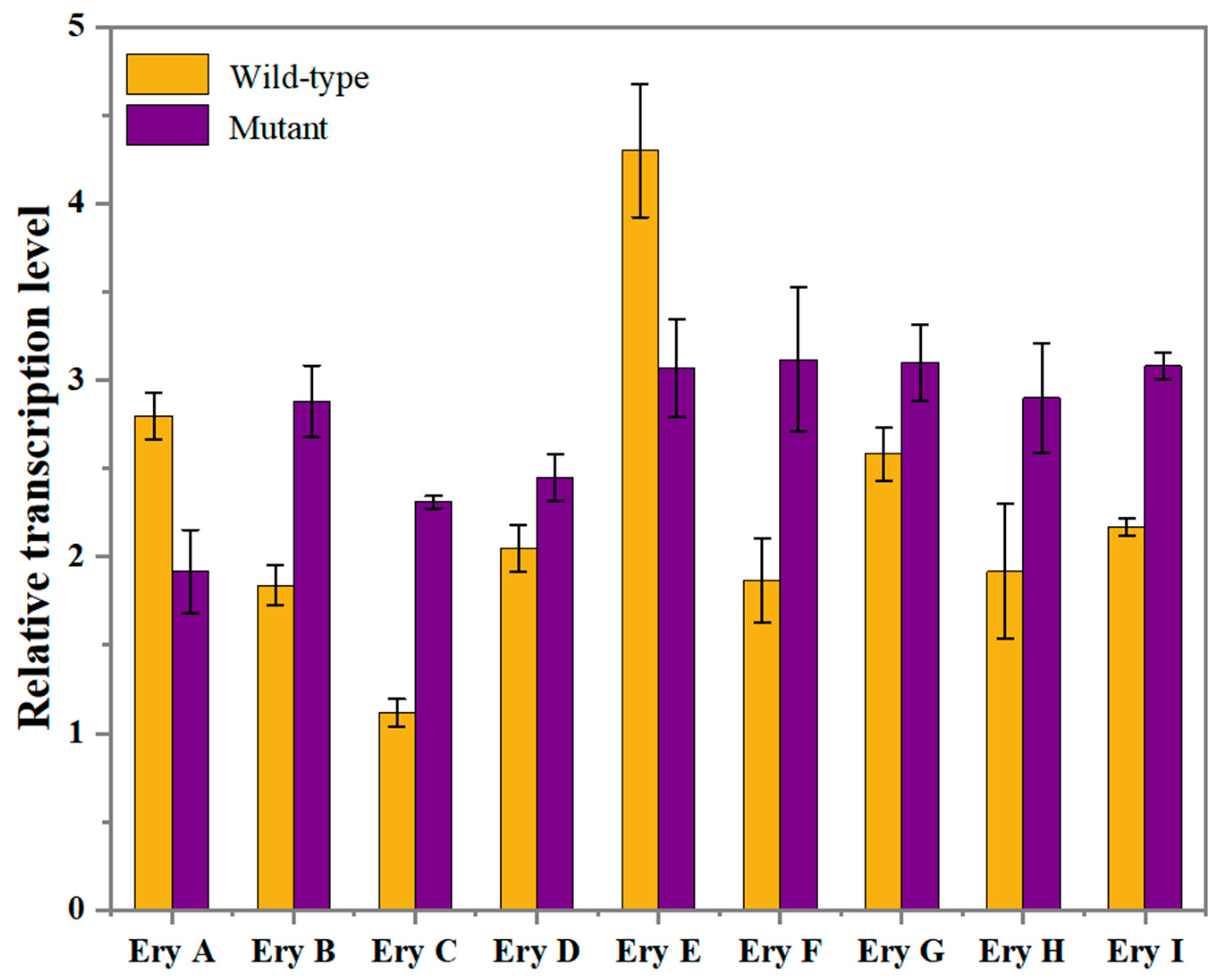
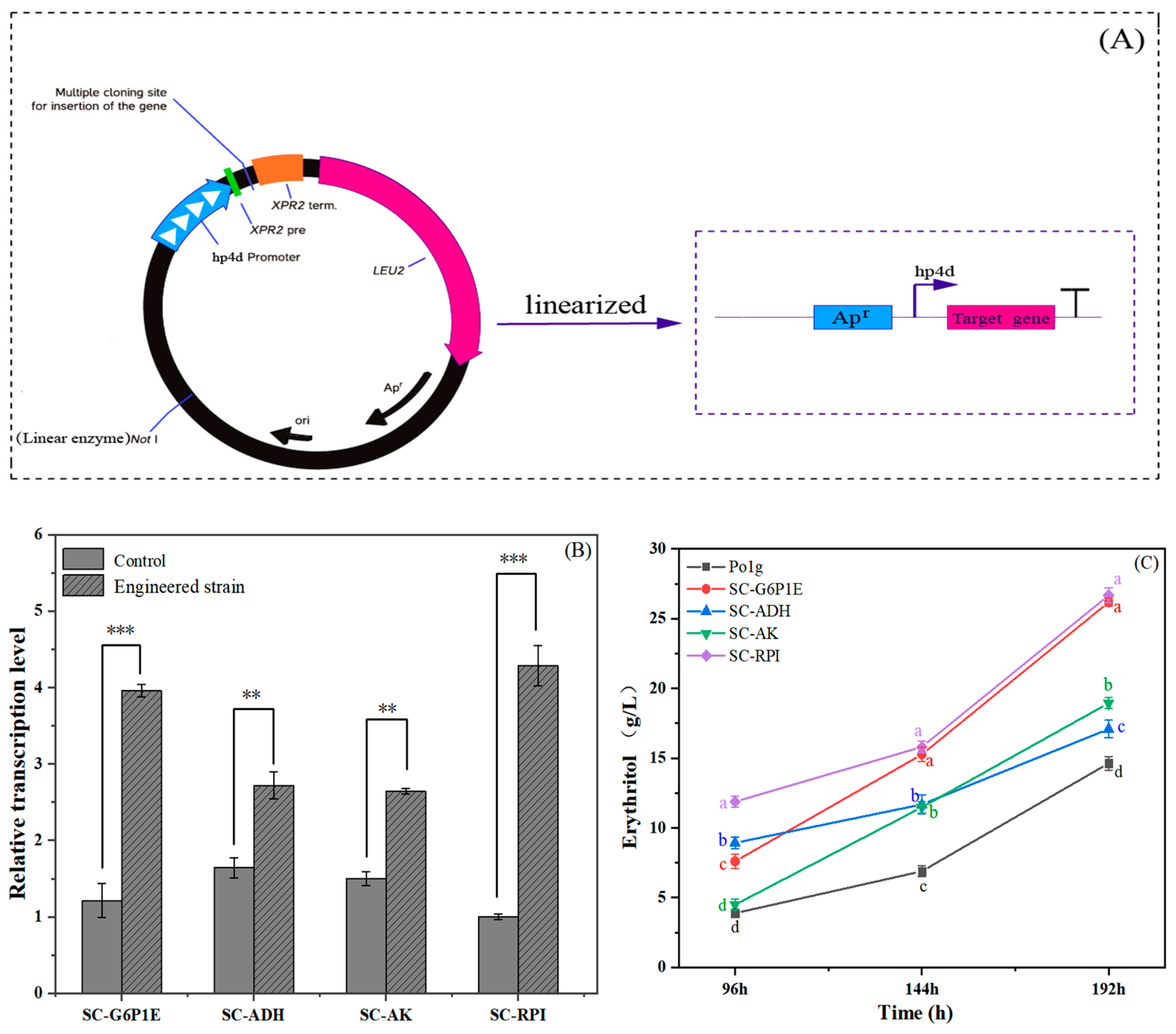
2.5. Validation of Key Genes Affecting Erythritol Production by RT-qPCR Expression Analysis
2.6. Rational Metabolic Engineering for Improving Erythritol Production
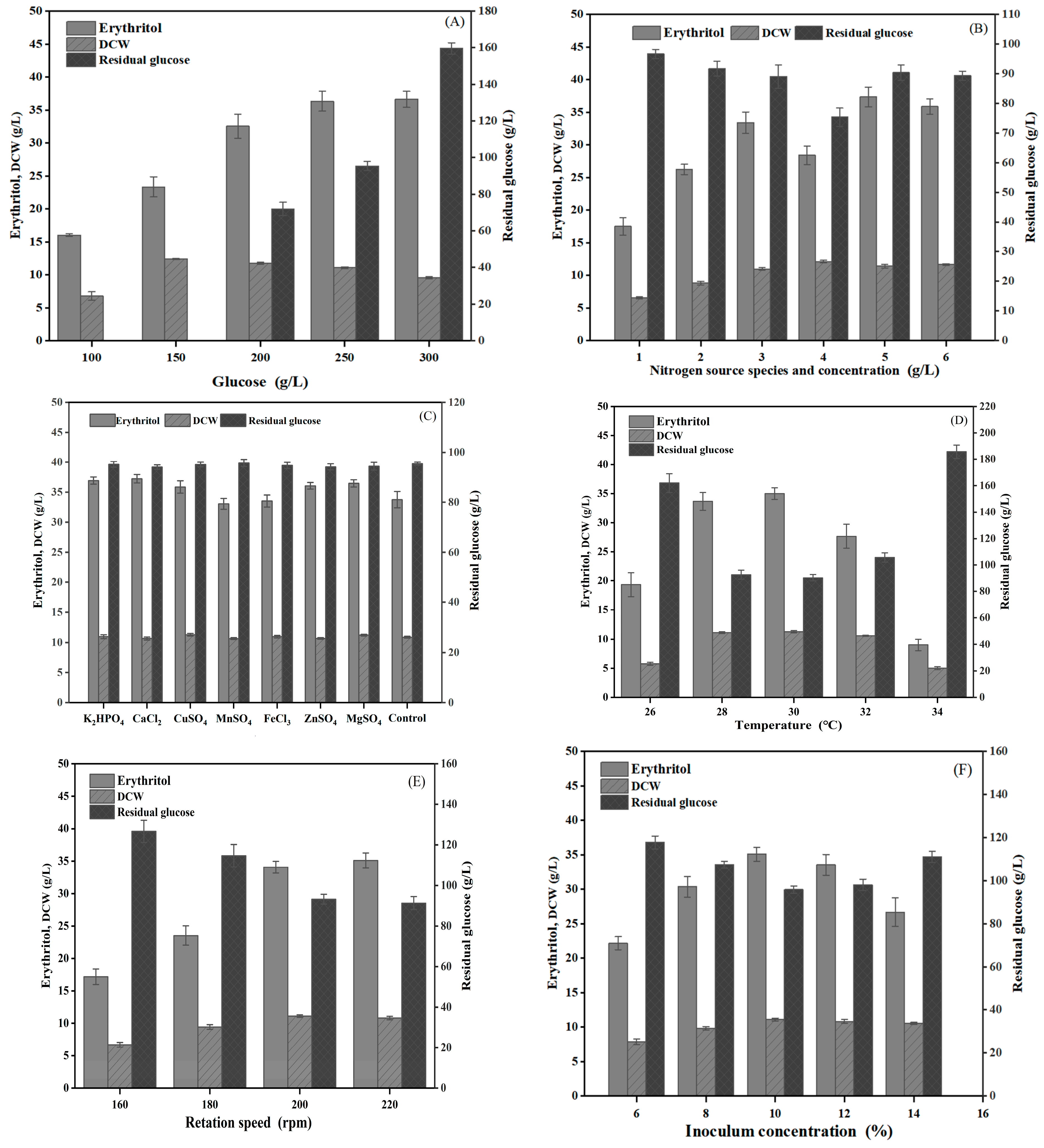
2.7. Influence of Culture Conditions on Erythritol Production
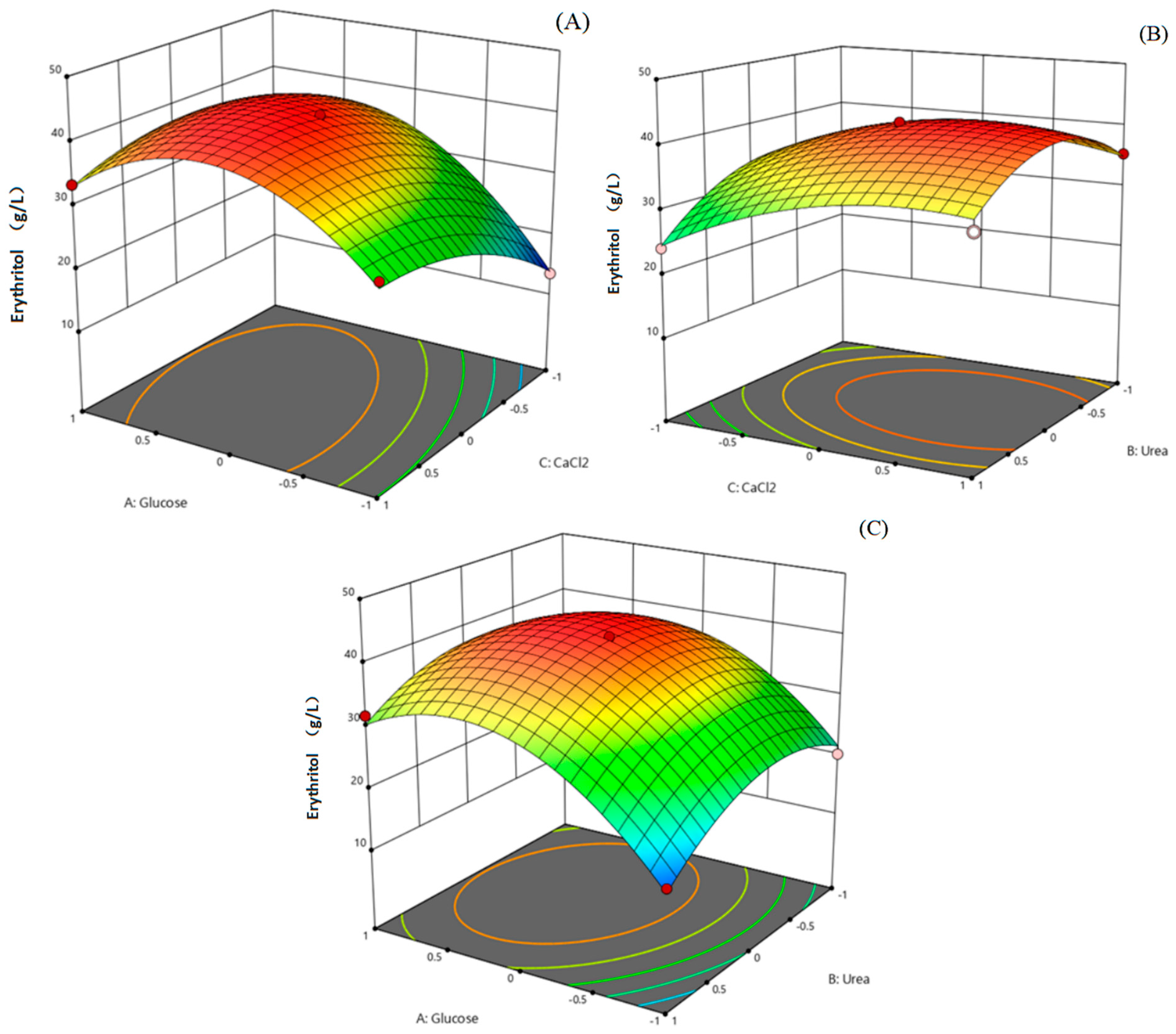
2.8. Optimization of the Culture Medium Using Response Surface Methodology
2.9. Evaluation of Erythritol Production by Incubating Mutant C1 in a 10 L Fermenter
3. Materials and Methods
3.1. Strain and Culture Conditions
3.2. Mutagenesis and Screening
3.3. Bioassay for Biomass Quantification
3.4. Transcriptome Analysis
3.5. Real-Time Quantitative PCR Analysis
3.6. Rational Metabolic Engineering of Y. lipolytica Po1g
3.7. Optimization of Culture Conditions
3.8. Batch Fermentation in a 10 L Fermenter
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moon, H.J.; Jeya, M.; Kim, I.W.; Lee, J.K. Biotechnological production of erythritol and its applications. Appl. Microbiol. Biotechnol. 2010, 86, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Mandar, S.D.; Pranav, P.K.; Pramod, S.K.; Anand, R.G. Erythritol production from sugar based feedstocks by Moniliella pollinis using lysate of recycled cells as nutrients source. Process Biochem. 2021, 112, 45–52. [Google Scholar]
- Martău, G.A.; Coman, V.; Vodnar, D.C. Recent advances in the biotechnological production of erythritol and mannitol. Crit. Rev. Biotechnol. 2020, 40, 608–622. [Google Scholar] [CrossRef]
- Mazi, T.A.; Stanhope, K.L. Erythritol: An In-Depth Discussion of Its Potential to Be a Beneficial Dietary Component. Nutrients 2023, 15, 204. [Google Scholar] [CrossRef]
- Lee, J.K.; Koo, B.S.; Kim, S.Y. Fumarate-Mediated Inhibition of Erythrose Reductase, a Key Enzyme for Erythritol Production by Torula corallina. Appl. Environ. Microbiol. 2002, 68, 4534–4538. [Google Scholar] [CrossRef]
- Oku, T.; Nakamura, S. Digestion, absorption, fermentation, and metabolism of functional sugar substitutes and their available energy. Pure Appl. Chem. 2002, 74, 1253–1261. [Google Scholar] [CrossRef]
- Rzechonek, D.A.; Dobrowolski, A.; Rymowicz, W.; Mirończuk, A.M. Recent advances in biological production of erythritol. Crit. Rev. Biotechnol. 2017, 38, 620–633. [Google Scholar] [CrossRef]
- Hootman, K.C.; Trezzi, J.P.; Kraemer, L.; Burwell, L.S.; Dong, X.; Guertin, K.A.; Cassano, P.A. Erythritol is a pentose-phosphate pathway metabolite and associated with adiposity gain in young adults (Medical Sciences). Proc. Natl. Acad. Sci. USA 2017, 114, 4233–4240. [Google Scholar] [CrossRef] [PubMed]
- Daza-Serna, L.; Serna-Loaiza, S.; Masi, A.; Mach, R.L.; Mach-Aigner, A.R.; Friedl, A. From the culture broth to the erythritol crystals: An opportunity for circular economy. Appl. Microbiol. Biotechnol. 2021, 105, 4467–4486. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Kasumi, T.; Ogihara, J.; Tamura, M.; Arai, T.; Tomishige, K. Erythritol: Another C4 Platform Chemical in Biomass Refinery. ACS Omega 2020, 5, 2520–2530. [Google Scholar] [CrossRef]
- Jeya, M.; Lee, K.M.; Tiwari, M.K.; Kim, J.S.; Gunasekaran, P.; Kim, S.Y.; Lee, J.K. Isolation of a novel high erythritol-producing Pseudozyma tsukubaensis and scale-up of erythritol fermentation to industrial level. Appl. Microbiol. Biotechnol. 2009, 83, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Khatape, A.B.; Rangaswamy, V.; Dastager, S.G. Strain improvement for enhanced erythritol production by Moniliella pollinis Mutant-58 using jaggery as a cost-effective substrate. Int. Microbiol. 2023, 27, 581–596. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.J.; Wen, C.Y.; Liau, J.C.; Chu, W.S. Screening and production of erythritol by newly isolated osmophilic yeast-like fungi. Process Biochem. 2001, 36, 1249–1258. [Google Scholar] [CrossRef]
- Rymowicz, W.; Fatykhova, A.R.; Kamzolova, S.V.; Rywińska, A.; Morgunov, I.G. Citric acid production from glycerol-containing waste of biodiesel industry by Yarrowia lipolytica in batch, repeated batch, and cell recycle regimes. Appl. Microbiol. Biotechnol. 2010, 87, 971. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.G.; Xiao, B.W.; Wang, J.W.; Li, W.; Zhang, W.H.; Zhou, J.P.; Cai, X.; Zhang, B.; Liu, Z.Q.; Zheng, Y.G. Advances in efficient biosynthesis of erythritol by metabolic engineering of Yarrowia lipolytica. Chin. J. Biotechnol. 2024, 40, 665–686. [Google Scholar]
- Carly, F.; Fickers, P. Erythritol production by yeasts: A snapshot of current knowledge. Yeast 2018, 35, 455–463. [Google Scholar] [CrossRef]
- Ghezelbash, G.R.; Nahvi, I.; Emamzadeh, R. Improvement of Erythrose Reductase Activity, Deletion of By-products and Statistical Media Optimization for Enhanced Erythritol Production from Yarrowia lipolytica Mutant 49. Curr. Microbiol. 2014, 69, 149–157. [Google Scholar] [CrossRef]
- Beopoulos, A.; Cescut, J.; Haddouche, R.; Uribelarrea, J.L.; Molina-Jouve, C.; Nicaud, J.M. Yarrowia lipolytica as a model for bio-oil production. Prog. Lipid Res. 2009, 48, 375–387. [Google Scholar] [CrossRef]
- Tomaszewska, L.; Rymowicz, W.; Rywińska, A. Mineral supplementation increases erythrose reductase activity in erythritol biosynthesis from glycerol by Yarrowia lipolytica. Appl. Biochem. Biotechnol. 2014, 172, 3069–3078. [Google Scholar] [CrossRef]
- Tomaszewska, L.; Rywińska, A.; Gładkowski, W. Production of erythritol and mannitol by Yarrowia lipolytica yeast in media containing glycerol. J. Ind. Microbiol. Biotechnol. 2012, 39, 1333–1343. [Google Scholar] [CrossRef]
- Song, M.; He, W.; Cai, S.; Wang, F.; Xu, W.; Xu, W. Nysfungin Production Improvement by UV Mutagenesis in Streptomyces noursei D-3–14. Catalysts 2023, 13, 247. [Google Scholar] [CrossRef]
- Mirończuk, A.M.; Dobrowolski, A.; Rakicka, M.; Rywińska, A.; Rymowicz, W. Newly isolated mutant of Yarrowia lipolytica MK1 as a proper host for efficient erythritol biosynthesis from glycerol. Process Biochem. 2015, 50, 61–68. [Google Scholar] [CrossRef]
- Zhang, Q.; Miao, R.; Feng, R.; Yan, J.; Wang, T.; Gan, Y.; Gan, B. Application of Atmospheric and Room-Temperature Plasma (ARTP) to Microbial Breeding. Curr. Issues Mol. Biol. 2023, 45, 6466–6484. [Google Scholar] [CrossRef] [PubMed]
- Belov, O.V.; Chuluunbaatar, O.; Kapralov, M.I.; Sweilam, N.H. The role of the bacterial mismatch repair system in SOS-induced mutagenesis: A theoretical background. J. Theor. Biol. 2013, 332, 30–41. [Google Scholar] [CrossRef]
- Jiang, Y.; Shang, Y.P.; Li, H.; Zhang, C.; Pan, J.; Bai, Y.P.; Xu, J.H. Enhancing transglutaminase production of Streptomyces mobaraensis by iterative mutagenesis breeding with atmospheric and room-temperature plasma (ARTP). Bioresour. Bioprocess. 2017, 4, 37. [Google Scholar] [CrossRef]
- Kumar, A.K. UV mutagenesis treatment for improved production of endoglucanase and β-glucosidase from newly isolated thermotolerant actinomycetes, Streptomyces griseoaurantiacus. Bioresour. Bioprocess. 2015, 2, 22. [Google Scholar] [CrossRef]
- Ye, L.; Ye, R.; Hu, F.; Wang, G. Combination of atmospheric and room temperature plasma (ARTP) mutagenesis, genome shuffling and dimethyl sulfoxide (DMSO) feeding to improve FK506 production in Streptomyces tsukubaensis. Biotechnol. Lett. 2021, 43, 1809–1820. [Google Scholar] [CrossRef]
- Songnaka, N.; Lertcanawanichakul, M.; Hutapea, A.M.; Nisoa, M.; Krobthong, S.; Yingchutrakul, Y.; Atipairin, A. Atmospheric and Room Temperature Plasma (ARTP) Mutagenesis Improved the Anti-MRSA Activity of Brevibacillus sp. SPR20. Int. J. Mol. Sci. 2023, 24, 12016. [Google Scholar] [CrossRef]
- Nie, X.; Xing, Y.; Li, Q.; Gao, F.; Wang, S.; Liu, P.; Shi, H. ARTP mutagenesis promotes selenium accumulation in Saccharomyces boulardii. LWT—Food Sci. Technol. 2022, 168, 113916. [Google Scholar] [CrossRef]
- Feng, W.; Liang, J.; Wang, B.; Chen, J. Improvement of kojic acid production in Aspergillus oryzae AR-47 mutant strain by combined mutagenesis. Bioprocess. Biosyst. Eng. 2019, 42, 753–761. [Google Scholar] [CrossRef]
- Qiu, X.; Xu, P.; Zhao, X.; Du, G.; Zhang, J.; Li, J. Combining genetically-encoded biosensors with high throughput strain screening to maximize erythritol production in Yarrowia lipolytica. Metab. Eng. 2020, 60, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Qian, Z.; Luo, M.; Fan, S.; Zhang, X.; Zhang, L. Identification of Salt Stress Responding Genes Using Transcriptome Analysis in Green Alga Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2018, 19, 3359. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Nazarbekov, A.; Zou, X.; Lee, S.Y. Recent advances in systems metabolic engineering. Curr. Opin. Biotechnol. 2023, 84, 103004. [Google Scholar] [CrossRef]
- Guo, Q.; Peng, Q.Q.; Chen, Y.Y.; Song, P.; Ji, X.J.; Huang, H.; Shi, T.Q. High-yield α-humulene production in Yarrowia lipolytica from waste cooking oil based on transcriptome analysis and metabolic engineering. Microb. Cell Factories 2022, 21, 271. [Google Scholar] [CrossRef]
- Min, X.; Guo, L.; Li, L.; Yang, R.; Zhao, W.; Lyu, X. Comparative transcriptome analysis reveals the underlying mechanism for over-accumulation of menaquinone-7 in Bacillus subtilis natto mutant. Biochem. Eng. J. 2021, 174, 108097. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, Y.; Fu, J.; Yang, Q.; Feng, L. High-throughput screening of lycopene-overproducing mutants of Blakeslea trispora by combining ARTP mutation with microtiter plate cultivation and transcriptional changes revealed by RNA-seq. Biochem. Eng. J. 2020, 161, 107664. [Google Scholar] [CrossRef]
- Jiang, D.; Yang, M.; Chen, K.; Jiang, W.; Zhang, L.; Ji, X.J.; Lu, L. Exploiting synthetic biology platforms for enhanced biosynthesis of natural products in Yarrowia lipolytica. Bioresour. Technol. 2024, 4, 37. [Google Scholar] [CrossRef]
- Szczepańczyk, M.; Rzechonek, D.A.; Dobrowolski, A.; Mirończuk, A.M. The Overexpression of YALI0B07117g Results in Enhanced Erythritol Synthesis from Glycerol by the Yeast Yarrowia lipolytica. Molecules 2021, 26, 7549. [Google Scholar] [CrossRef]
- Zheng, S.; Jiang, B.; Zhang, T.; Chen, J. Combined mutagenesis and metabolic regulation to enhance D-arabitol production from Candida parapsilosis. J. Ind. Microbiol. Biotechnol. 2020, 23, 62–63. [Google Scholar] [CrossRef]
- Ng, I.S.; Ye, C.; Zhang, Z.; Lu, Y.; Jing, K. Daptomycin antibiotic production processes in fed-batch fermentation by Streptomyces roseosporus NRRL11379 with precursor effect and medium optimization. Bioprocess. Biosyst. Eng. 2013, 37, 415–423. [Google Scholar] [CrossRef]
- Song, X.; Zhang, Y.; Zhu, X.; Wang, Y.; Chu, J.; Zhuang, Y. Mutation breeding of high avermectin B 1a-producing strain by the combination of high energy carbon heavy ion irradiation and sodium nitrite mutagenesis based on high throughput screening. Biotechnol. Bioprocess. Eng. 2017, 22, 539–548. [Google Scholar] [CrossRef]
- Wang, P.; Yin, Y.; Wang, X.; Wen, J. Enhanced ascomycin production in Streptomyces hygroscopicus var. ascomyceticus by employing polyhydroxybutyrate as an intracellular carbon reservoir and optimizing carbon addition. Microb. Cell Factories 2021, 20, 70. [Google Scholar] [CrossRef] [PubMed]
- Yoon, V.; Nodwell, J.R. Activating secondary metabolism with stress and chemicals. J. Ind. Microbiol. Biotechnol. 2013, 41, 415–424. [Google Scholar] [CrossRef]
- Zhang, C.; Li, Y.; Zhang, T.; Zhao, H. Increasing chitosanase production in Bacillus cereus by a novel mutagenesis and screen method. Bioengineered 2021, 12, 266–277. [Google Scholar] [CrossRef]
- Xu, D.P.; Yao, Z.E.; Pan, J.B.; Feng, H.Y.; Guo, Z.Q.; Lu, X.L. Study on the multiple characteristics of M3 generation of pea mutants obtained by neutron irradiation. Nucl. Sci. Tech. 2020, 31, 67. [Google Scholar] [CrossRef]
- Noor, E.; Eden, E.; Milo, R.; Alon, U. Central Carbon Metabolism as a Minimal Biochemical Walk between Precursors for Biomass and Energy. Mol. Cell 2010, 39, 809–820. [Google Scholar] [CrossRef]
- Peng, Y.; Liu, H.; Liu, J.; Long, J. Post-translational modifications on mitochondrial metabolic enzymes in cancer. Free Radic. Biol. Med. 2021, 179, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Verschueren, K.H.G.; Blanchet, C.; Felix, J.; Dansercoer, A.; De Vos, D.; Bloch, Y.; Van Beeumen, J.; Svergun, D.; Gutsche, I.; Savvides, S.N. Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle. Nature 2019, 568, 571–575. [Google Scholar] [CrossRef]
- Zhang, G.; Zabed, H.M.; Zhang, Y.; Li, J.; Yun, J.; Qi, X. Random mutagenesis and transcriptomics-guided rational engineering in Zygosaccharomyces rouxii for elevating D-arabitol biosynthesis. Bioresour. Technol. 2024, 400, 130685. [Google Scholar] [CrossRef]
- Tyler, C.A.; Kopit, L.; Doyle, C.; Yu, A.O.; Hugenholtz, J.; Marco, M.L. Polyol production during heterofermentative growth of the plant isolate Lactobacillus florum 2F. J. Appl. Microbiol. 2016, 120, 1336–1345. [Google Scholar] [CrossRef]
- Wurster, B.; Hess, B. Glucose-6-phosphate-1-epimerase from baker’s yeast. A new enzyme. FEBS Lett. 1972, 23, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Chance, E.M.; Hess, B.; Plesser, T.; Wurster, B. Determination of the kinetic constants of glucose-6-phosphate 1-epimerase by non-linear optimization. Eur. J. Biochem. 1975, 50, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Graille, M.; Baltaze, J.P.; Leulliot, N.; Liger, D.; Quevillon-Cheruel, S.; van Tilbeurgh, H. Structure-based functional annotation: Yeast ymr099c codes for a D-hexose-6-phosphate mutarotase. J. Biol. Chem. 2006, 281, 30175–30185. [Google Scholar] [CrossRef]
- Zhang, L.; Nie, M.Y.; Liu, F.; Chen, J.; Wei, L.J.; Hua, Q. Multiple gene integration to promote erythritol production on glycerol in Yarrowia lipolytica. Biotechnol. Lett. 2021, 43, 1277–1287. [Google Scholar] [CrossRef]
- Suástegui, M.; Yu Ng, C.; Chowdhury, A.; Sun, W.; Cao, M.; House, E.; Shao, Z. Multilevel engineering of the upstream module of aromatic amino acid biosynthesis in Saccharomyces cerevisiae for high production of polymer and drug precursors. Metab. Eng. 2017, 43, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Adler, L.; Blomberg, A.; Nilsson, A. Glycerol metabolism and osmoregulation in the salt-tolerant yeast Debaryomyces hansenii. J. Bacteriol. 1985, 162, 300–306. [Google Scholar] [CrossRef]
- Grauslund, M.; Lopes, J.M.; Rønnow, B. Expression of GUT1, which encodes glycerol kinase in Saccharomyces cerevisiae, is controlled by the positive regulators Adr1p, Ino2p and Ino4p and the negative regulator Opi1p in a carbon source-dependent fashion. Nucleic Acids Res. 1999, 27, 4391–4398. [Google Scholar] [CrossRef]
- Carly, F.; Vandermies, M.; Telek, S.; Steels, S.; Thomas, S.; Nicaud, J.M.; Fickers, P. Enhancing erythritol productivity in Yarrowia lipolytica using metabolic engineering. Metab. Eng. 2017, 42, 19–24. [Google Scholar] [CrossRef]
- Subki, A.; Ho, C.L.; Ismail, N.F.N.; Abidin, A.A.Z.; Yusof, Z.N.B. Identification and characterisation of thiamine pyrophosphate (TPP) ribos itch in Elaeis guineensis. PLoS ONE 2020, 15, 20. [Google Scholar] [CrossRef]
- Walker, C.; Ryu, S.; Giannone, R.J.; Garcia, S.; Trinh, C.T. Understanding and eliminating the detrimental effect of thiamine deficiency on the Oleaginous yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 2019, 86, e02299-19. [Google Scholar] [CrossRef]
- Sayın Börekçi, B.; Kaya, M.; Kaban, G. Citric Acid Production by Yarrowia lipolytica NRRL Y-1094: Optimization of pH, Fermentation Time and Glucose Concentration Using Response Surface Methodology. Fermentation 2022, 8, 731. [Google Scholar] [CrossRef]
- Yaacob, N.; Mohamad Ali, M.S.; Salleh, A.B.; Abdul Rahman, N.A. Effects of glucose, ethanol and acetic acid on regulation of ADH2 gene from Lachancea fermentati. PeerJ 2016, 4, e1751. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-T.; Zhao, L.-X.; Wei, L.-J.; Chen, J.; Liu, Z.; Liu, F.; Hua, Q. Metabolic engineering of erythritol production from glycerol by Yarrowia lipolytica. Biotechnol. Bioprocess. Eng. 2024, 29, 119–127. [Google Scholar] [CrossRef]
- Hu, J.; Lin, S.; Zheng, B.; Cheung, P.C.K. Short-chain fatty acids in control of energy metabolism. Crit. Rev. Food Sci. Nutr. 2016, 58, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- Jagtap, S.S.; Bedekar, A.A.; Singh, V.; Jin, Y.S.; Rao, C.V. Metabolic engineering of the oleaginous yeast Yarrowia lipolytica PO1f for production of erythritol from glycerol. Biotechnol. Biofuels 2021, 14, 188. [Google Scholar] [CrossRef]
- Yang, S.; Pan, X.; Wang, Q.; Lv, Q.; Zhang, X.; Zhang, R.; Rao, Z. Enhancing erythritol production from crude glycerol in a wild-type Yarrowia lipolytica by metabolic engineering. Front. Microbiol. 2022, 13, 1054243. [Google Scholar] [CrossRef]
- Baptista, S.L.; Costa, C.E.; Cunha, J.T.; Soares, P.O.; Domingues, L. Metabolic engineering of Saccharomyces cerevisiae for the production of top value chemicals from biorefinery carbohydrates. Biotechnol. Adv. 2021, 312, 107697. [Google Scholar] [CrossRef]
- Li, X.; Huang, J.; Yun, J.; Zhang, G.; Zhang, Y.; Zhao, M.; Zabed, H.M.; Ravikumar, Y.; Qi, X. D-arabitol ameliorates obesity and metabolic disorders via the gut microbiota-SCFAs-WAT browning Axis. J. Agric. Food Chem. 2022, 71, 522–534. [Google Scholar] [CrossRef]
- Brabender, M.; Hussain, M.S.; Rodriguez, G.; Blenner, M.A. Urea and urine are a viable and cost-effective nitrogen source for Yarrowia lipolytica biomass and lipid accumulation. Appl. Microbiol. Biotechnol. 2018, 102, 2313–2322. [Google Scholar] [CrossRef]
- Lee, J.K.; Ha, S.J.; Kim, S.Y.; Oh, D.K. Increased erythritol production in Torula sp. by Mn2+ and Cu2+. Biotechnol. Lett. 2000, 22, 983–986. [Google Scholar] [CrossRef]
- Nambou, K.; Zhao, C.; Wei, L.; Chen, J.; Imanaka, T.; Hua, Q. Designing of a “cheap to run” fermentation platform for an enhanced production of single cell oil from Yarrowia lipolytica DSM3286 as a potential feedstock for biodiesel. Bioresour. Technol. 2014, 173, 324–333. [Google Scholar] [CrossRef]
- Lee, J.K.; Oh, D.K.; Song, H.Y.; Kim, I.W. Ca2+ and Cu2+ supplementation increases mannitol production by Candida magnoliae. Biotechnol. Lett. 2006, 29, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Vohra, A.; Satyanarayana, T. Statistical Optimization of the Medium Components by Response Surface Methodology to Enhance Phytase Production by Pichia anomala. Process Biochem. 2002, 37, 999–1004. [Google Scholar] [CrossRef]
- Bustos, G.; Moldes, A.B.; Alonso, J.L.; Vázquez, M. Optimization of D-Lactic Acid Production by Lactobacillus coryniformis Using Response Surface Methodology. Food Microbiol. 2004, 21, 143–148. [Google Scholar] [CrossRef]
- Savergave, L.S.; Gadre, R.V.; Vaidya, B.K.; Narayanan, K. Strain Improvement and Statistical Media Optimization for Enhanced Erythritol Production with Minimal By-Products from Candida magnoliae Mutant R23. Biochem. Eng. J. 2011, 55, 92–100. [Google Scholar] [CrossRef]
- Carly, F.; Steels, S.; Telek, S.; Vandermies, M.; Nicaud, J.M.; Fickers, P. Identification and characterization of EYD1, encoding an erythritol dehydrogenase in Yarrowia lipolytica and its application to bioconvert erythritol into erythrulose. Bioresour. Technol. 2017, 247, 963–969. [Google Scholar] [CrossRef] [PubMed]
- Rywińska, A.; Tomaszewska-Hetman, L.; Juszczyk, P.; Rakicka-Pustułka, M.; Bogusz, A.; Rymowicz, W. Enhanced Production of Erythritol from Glucose by the Newly Obtained UV Mutant Yarrowia lipolytica K1UV15. Molecules 2024, 29, 2187. [Google Scholar] [CrossRef]
- Qiu, X.; Gu, Y.; Du, G.; Zhang, J.; Xu, P.; Li, J. Conferring thermotolerant phenotype to wild-type Yarrowia lipolytica improves cell growth and erythritol production. Biotechnol. Bioeng. 2021, 118, 3117–3127. [Google Scholar] [CrossRef]
- Liu, J.M.; Hu, Y.H. The application of TLC in polyalcohol. Food Technol-Chic. 2007, 06, 224–226. [Google Scholar]
- Yao, Z.Y.; Gong, J.S.; Liu, Y.R.; Jiang, J.Y.; Zhang, Y.S.; Su, C.; Li, H.; Kang, C.L.; Liu, L.; Xu, Z.H.; et al. Genetic variation reveals the enhanced microbial hyaluronan biosynthesis via atmospheric and room temperature plasma. Carbohydr. Polym. 2023, 312, 120809. [Google Scholar] [CrossRef] [PubMed]
- Gong, M.; Zhang, H.N.; Wu, D.; Zhang, Z.; Zhang, J.S.; Bao, D.P.; Yang, Y. Key metabolism pathways and regulatory mechanisms of high polysaccharide yielding in Hericium erinaceus. BMC Genom. 2021, 22, 160. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, J.; Uehara, K.; Mogi, Y.; Suzuki, K.; Watanabe, T.; Yamazaki, T. Improved transformation of the halo-tolerant yeast Zygosaccharomyces rouxii by electroporation. Biosci. Biotechnol. Biochem. 2010, 74, 1092–1094. [Google Scholar] [CrossRef][Green Version]


| Levels (g/L) | |||
|---|---|---|---|
| Code | Type | −1 | +1 |
| X1 | Glucose | 125 | 250 |
| X2 | Urea | 1 | 1.5 |
| X3 | Glycine | 2 | 5 |
| X4 | KH2PO4 | 1 | 2 |
| X5 | MgSO4 | 1 | 1.25 |
| X6 | CaCl2 | 3 | 6 |
| X7 | CuSO4 | 0.1 | 0.125 |
| X8 | ZnSO4 | 0.1 | 0.125 |
| Source | Sum of Squares | df | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|
| Model | 759.76 | 8 | 94.97 | 13.99 | 0.0264 |
| X1(Glucose) | 532.97 | 1 | 532.97 | 78.53 | 0.0030 |
| X2(Urea) | 96.93 | 1 | 96.93 | 14.28 | 0.0325 |
| X3(Glycine) | 4.72 | 1 | 4.72 | 0.6957 | 0.4654 |
| X4(KH2PO4) | 38.73 | 1 | 38.73 | 5.71 | 0.0969 |
| X5(MgSO4) | 3.41 | 1 | 3.41 | 0.5030 | 0.5293 |
| X6(CaCl2) | 76.50 | 1 | 76.50 | 11.27 | 0.0438 |
| X7(CuSO4) | 0.0436 | 1 | 0.0436 | 0.0064 | 0.9411 |
| X8(ZnSO4) | 6.47 | 1 | 6.47 | 0.9532 | 0.4009 |
| Residual | 20.36 | 3 | 6.79 | ||
| Cor Total | 780.12 | 11 |
| Variables | Coding Levels | ||
|---|---|---|---|
| −1 | 0 | +1 | |
| A (Glucose) | 125 | 250 | 375 |
| B (Urea) | 0 | 1 | 2 |
| C (CaCl2) | 4 | 7 | 10 |
| Experimental Number | A (Glucose) | B (Urea) | C (CaCl2) | Predicted Value (g/L) | Experimental Value (g/L) |
|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 41.36 | 40.23 |
| 2 | −1 | 1 | 0 | 15.30 | 16.05 |
| 3 | 0 | 0 | 0 | 41.36 | 41.82 |
| 4 | 0 | −1 | −1 | 31.08 | 32.83 |
| 5 | 1 | 0 | −1 | 34.05 | 33.05 |
| 6 | 0 | −1 | 1 | 34.94 | 35.34 |
| 7 | −1 | 0 | −1 | 13.83 | 13.48 |
| 8 | 0 | 1 | −1 | 24.52 | 24.11 |
| 9 | −1 | 0 | 1 | 28.08 | 29.08 |
| 10 | 0 | 0 | 0 | 41.36 | 42.02 |
| 11 | 1 | 0 | 1 | 33.10 | 33.45 |
| 12 | −1 | −1 | 0 | 21.57 | 20.17 |
| 13 | 1 | 1 | 0 | 30.43 | 31.83 |
| 14 | 1 | −1 | 0 | 31.68 | 30.93 |
| 15 | 0 | 1 | 1 | 33.96 | 32.21 |
| Source | Sum of Squares | DF | Mean Square | F | Prob (P) > F | Significance |
|---|---|---|---|---|---|---|
| Model | 1036.74 | 9 | 115.19 | 36.66 | 0.0005 | *** |
| A (Glucose) | 318.53 | 1 | 318.53 | 101.38 | 0.0002 | |
| B (Urea) | 28.39 | 1 | 28.39 | 9.03 | 0.0299 | |
| C (CaCl2) | 88.51 | 1 | 88.51 | 28.17 | 0.0032 | |
| AB | 6.30 | 1 | 6.30 | 2.01 | 0.2159 | |
| AC | 57.76 | 1 | 57.76 | 18.38 | 0.0078 | |
| BC | 7.81 | 1 | 7.81 | 2.49 | 0.1757 | |
| A2 | 386.76 | 1 | 386.76 | 123.09 | 0.0001 | |
| B2 | 150.16 | 1 | 150.16 | 47.79 | 0.0010 | |
| C2 | 54.93 | 1 | 54.93 | 17.48 | 0.0086 | |
| Residual | 15.71 | 5 | 3.14 | |||
| Lack of Fit | 13.79 | 3 | 4.60 | 4.78 | 0.1780 | ns |
| Pure Error | 1.92 | 2 | 0.9620 | |||
| Cor Total | 1052.45 | 14 | ||||
| R2Adj | 0.9582 | |||||
| R2Pred | 0.7863 | |||||
| R2 | 0.9851 | |||||
| CV% | 5.82 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, W.; Xu, M.; Yang, F.; Li, X. Comparative Transcriptome Analysis Reveals Key Genes Related to Erythritol Production in Yarrowia lipolytica and the Optimization of Culture Conditions. Int. J. Mol. Sci. 2025, 26, 4180. https://doi.org/10.3390/ijms26094180
Fu W, Xu M, Yang F, Li X. Comparative Transcriptome Analysis Reveals Key Genes Related to Erythritol Production in Yarrowia lipolytica and the Optimization of Culture Conditions. International Journal of Molecular Sciences. 2025; 26(9):4180. https://doi.org/10.3390/ijms26094180
Chicago/Turabian StyleFu, Wei, Ming Xu, Fan Yang, and Xianzhen Li. 2025. "Comparative Transcriptome Analysis Reveals Key Genes Related to Erythritol Production in Yarrowia lipolytica and the Optimization of Culture Conditions" International Journal of Molecular Sciences 26, no. 9: 4180. https://doi.org/10.3390/ijms26094180
APA StyleFu, W., Xu, M., Yang, F., & Li, X. (2025). Comparative Transcriptome Analysis Reveals Key Genes Related to Erythritol Production in Yarrowia lipolytica and the Optimization of Culture Conditions. International Journal of Molecular Sciences, 26(9), 4180. https://doi.org/10.3390/ijms26094180






