B3 Superfamily in Cucumber (Cucumis sativus L.): Identification, Evolution, Expression Patterns, and Function in Glandular Trichome Development
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification and Phylogenetic Analysis of B3 Superfamily Genes in Cucumber
2.2. Gene Structural and Protein Motif Analysis
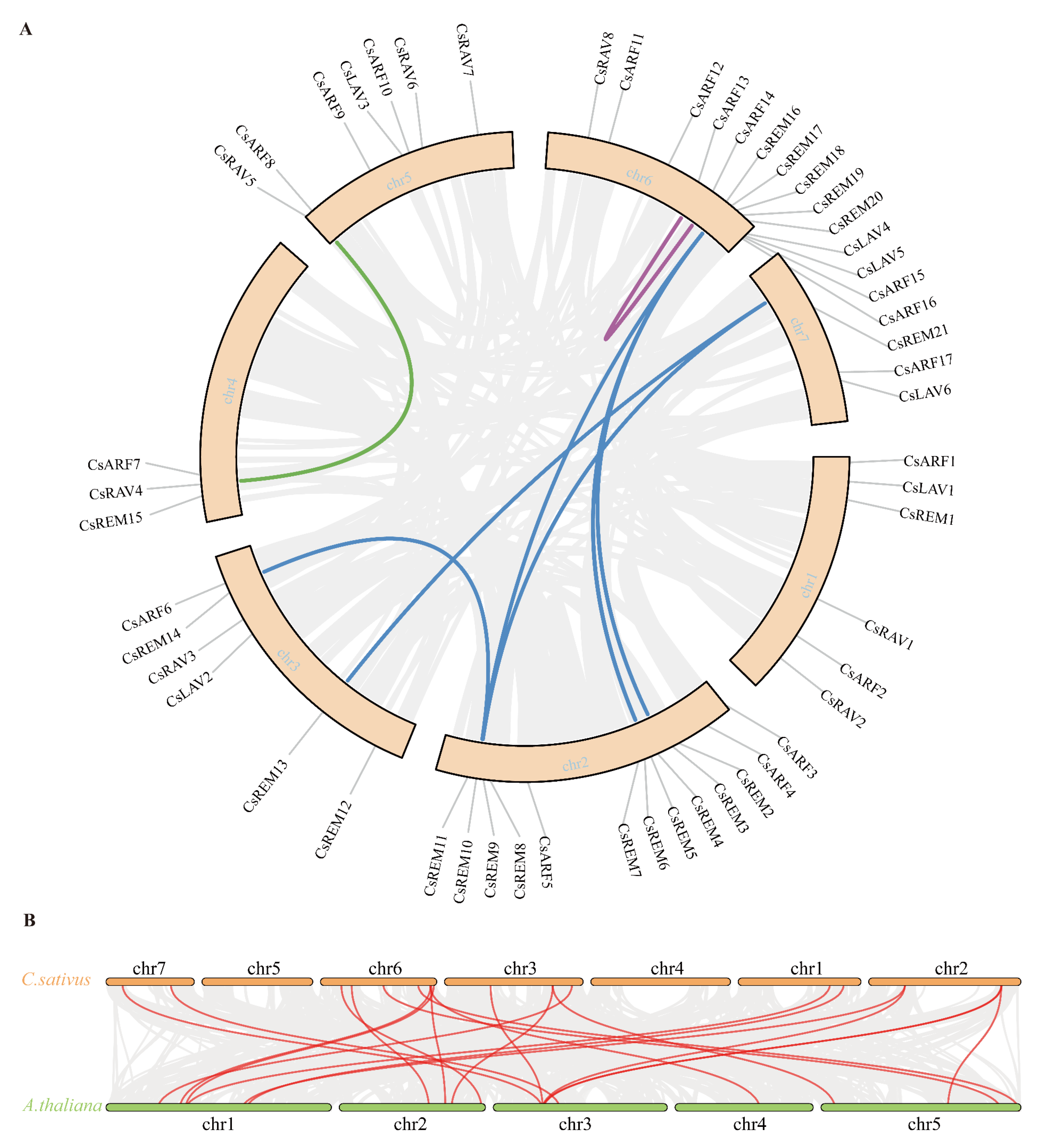
2.3. Collinearity Analysis of Cucumber B3 Genes
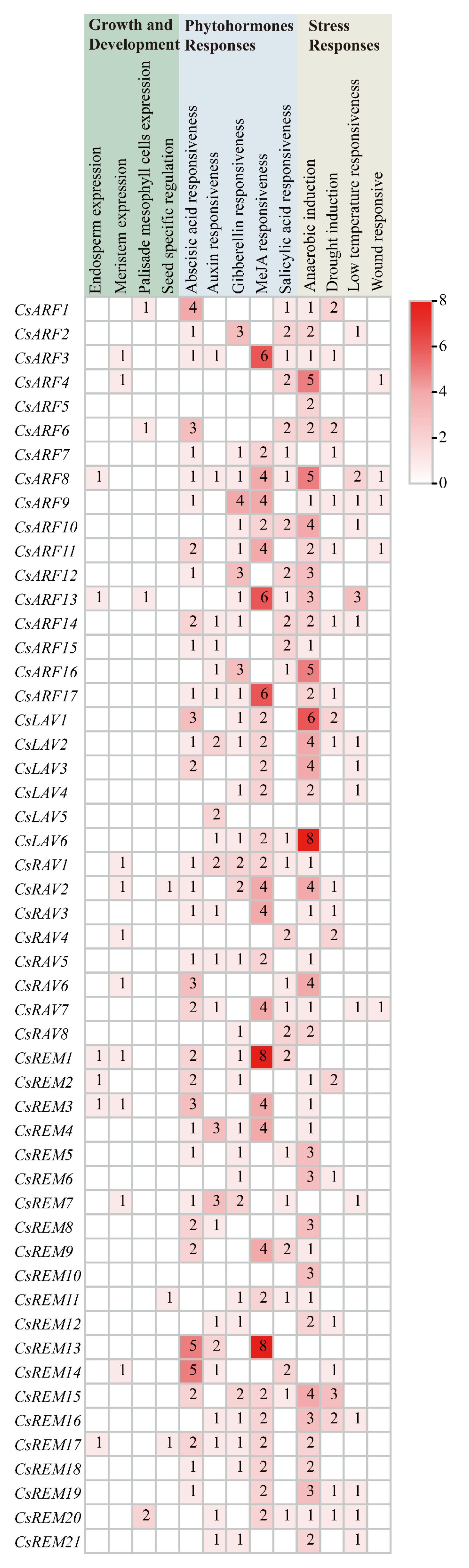
2.4. Analysis of Cis-Acting Elements in Cucumber B3 Gene Promoters
2.5. Expression Pattern of Cucumber B3 Genes
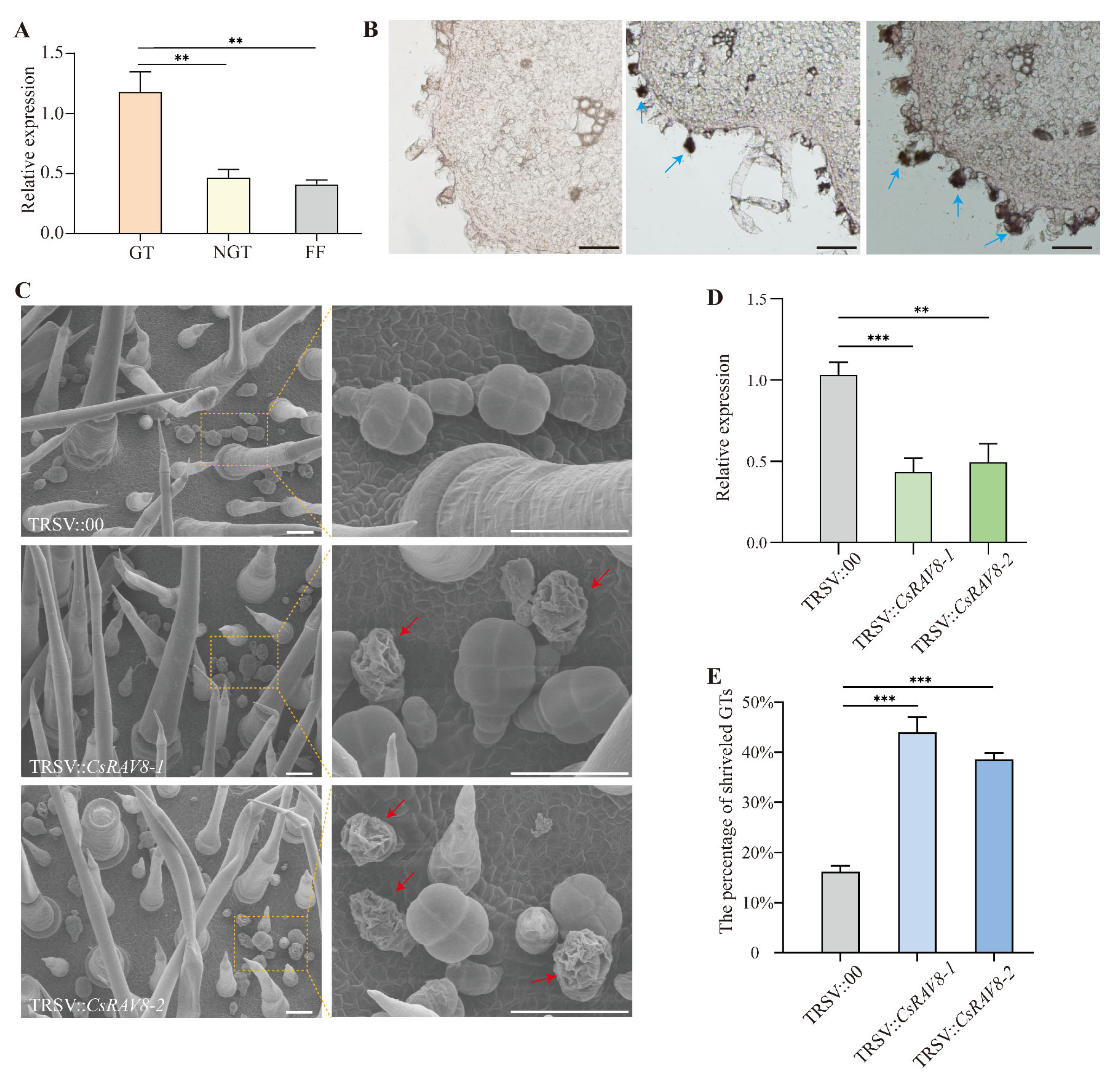
2.6. Preliminary Functional Validation of CsRAV8
3. Discussion
4. Materials and Methods
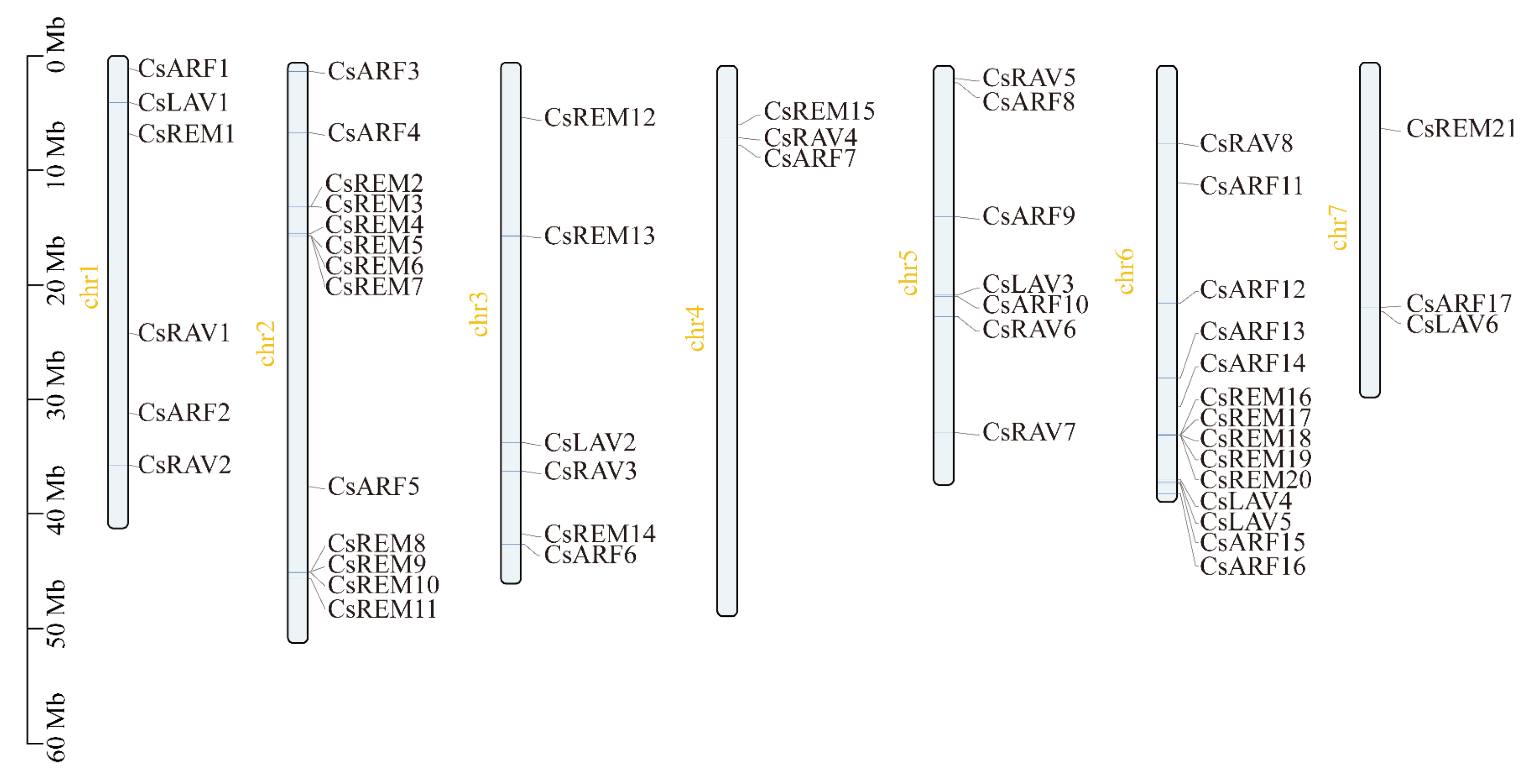
4.1. Gene Identification and Chromosomal Locations
4.2. Phylogenetic Analysis
4.3. Gene Structure Analysis
4.4. Synteny Analysis
4.5. Cis-Acting Element Analysis
4.6. Expression Profiling Analysis
4.7. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR) Analysis
4.8. In Situ Hybridization
4.9. VIGS Assay
4.10. Scanning Electron Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Swaminathan, K.; Peterson, K.; Jack, T. The plant B3 superfamily. Trends Plant Sci. 2008, 13, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.Y.; Weselake, R.J. Genome-wide identification and analysis of the B3 superfamily of transcription factors in Brassicaceae and major crop plants. Theor. Appl. Genet. 2013, 126, 1305–1319. [Google Scholar] [CrossRef] [PubMed]
- Tsukagoshi, H.; Saijo, T.; Shibata, D.; Morikami, A.; Nakamura, K. Analysis of a sugar response mutant of Arabidopsis identified a novel B3 domain protein that functions as an active transcriptional repressor. Plant Physiol. 2005, 138, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Kao, C.Y.; McCarty, D.R. The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 1997, 9, 799–807. [Google Scholar] [CrossRef]
- Ulmasov, T.; Murfett, J.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997, 9, 1963–1971. [Google Scholar] [CrossRef]
- Chandan, R.K.; Kumar, R.; Swain, D.M.; Ghosh, S.; Bhagat, P.K.; Patel, S.; Bagler, G.; Sinha, A.K.; Jha, G. RAV1 family members function as transcriptional regulators and play a positive role in plant disease resistance. Plant J. 2023, 114, 39–54. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Cubas, P.; Jarillo, J.A.; Fernández-Calvín, B.; Salinas, J.; Martínez-Zapater, J.M. AtREM1, a member of a new family of B3 domain-containing genes, is preferentially expressed in reproductive meristems. Plant Physiol. 2002, 128, 418–427. [Google Scholar] [CrossRef]
- Boulard, C.; Fatihi, A.; Lepiniec, L.; Dubreucq, B. Regulation and evolution of the interaction of the seed B3 transcription factors with NF-Y subunits. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2017, 1860, 1069–1078. [Google Scholar] [CrossRef]
- Chen, N.; Wang, H.; Abdelmageed, H.; Veerappan, V.; Tadege, M.; Allen, R.D. HSI2/VAL1 and HSL1/VAL2 function redundantly to repress DOG1 expression in Arabidopsis seeds and seedlings. New Phytol. 2020, 227, 840–856. [Google Scholar] [CrossRef]
- Chen, C.; Gong, X.; Li, Y.; Li, H.; Zhang, H.; Liu, L.; Liang, D.; Yuan, W. Interaction Analysis between the Arabidopsis Transcription Repressor VAL1 and Transcription Coregulators SIN3-LIKEs (SNLs). Int. J. Mol. Sci. 2022, 23, 6987. [Google Scholar] [CrossRef]
- Chandler, J.W. Auxin response factors. Plant Cell Environ. 2016, 39, 1014–1028. [Google Scholar] [CrossRef] [PubMed]
- Roosjen, M.; Paque, S.; Weijers, D. Auxin Response Factors: Output control in auxin biology. J. Exp. Bot. 2018, 69, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Han, S.; Qi, Y. Advances in structure and function of auxin response factor in plants. J. Integr. Plant Biol. 2023, 65, 617–632. [Google Scholar] [CrossRef] [PubMed]
- Pekker, I.; Alvarez, J.P.; Eshed, Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 2005, 17, 2899–2910. [Google Scholar] [CrossRef] [PubMed]
- Marzi, D.; Brunetti, P.; Saini, S.S.; Yadav, G.; Puglia, G.D.; Dello Ioio, R. Role of transcriptional regulation in auxin-mediated response to abiotic stresses. Front. Genet. 2024, 15, 1394091. [Google Scholar] [CrossRef]
- Matias-Hernandez, L.; Aguilar-Jaramillo, A.E.; Marin-Gonzalez, E.; Suarez-Lopez, P.; Pelaz, S. RAV genes: Regulation of floral induction and beyond. Ann. Bot. 2014, 114, 1459–1470. [Google Scholar] [CrossRef]
- Hu, H.; Tian, S.; Xie, G.; Liu, R.; Wang, N.; Li, S.; He, Y.; Du, J. TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2103895118. [Google Scholar] [CrossRef]
- Feng, C.Z.; Chen, Y.; Wang, C.; Kong, Y.H.; Wu, W.H.; Chen, Y.F. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 2014, 80, 654–668. [Google Scholar] [CrossRef]
- Trigueros, M.; Navarrete-Gomez, M.; Sato, S.; Christensen, S.K.; Pelaz, S.; Weigel, D.; Yanofsky, M.F.; Ferrandiz, C. The NGATHA genes direct style development in the Arabidopsis gynoecium. Plant Cell 2009, 21, 1394–1409. [Google Scholar] [CrossRef]
- Fourquin, C.; Ferrandiz, C. The essential role of NGATHA genes in style and stigma specification is widely conserved across eudicots. New Phytol. 2014, 202, 1001–1013. [Google Scholar] [CrossRef]
- Ballester, P.; Navarrete-Gomez, M.; Carbonero, P.; Onate-Sanchez, L.; Ferrandiz, C. Leaf expansion in Arabidopsis is controlled by a TCP-NGA regulatory module likely conserved in distantly related species. Physiol. Plant 2015, 155, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Mantegazza, O.; Gregis, V.; Mendes, M.A.; Morandini, P.; Alves-Ferreira, M.; Patreze, C.M.; Nardeli, S.M.; Kater, M.M.; Colombo, L. Analysis of the Arabidopsis REM gene family predicts functions during flower development. Ann. Bot. 2014, 114, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Caselli, F.; Beretta, V.M.; Mantegazza, O.; Petrella, R.; Leo, G.; Guazzotti, A.; Herrera-Ubaldo, H.; de Folter, S.; Mendes, M.A.; Kater, M.M.; et al. REM34 and REM35 Control Female and Male Gametophyte Development in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 1351. [Google Scholar] [CrossRef] [PubMed]
- Levy, Y.Y.; Mesnage, S.; Mylne, J.S.; Gendall, A.R.; Dean, C. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 2002, 297, 243–246. [Google Scholar] [CrossRef]
- Zhou, H.B.; Song, Z.H.; Zhong, S.; Zuo, L.Y.; Qi, Z.; Qu, L.J.; Lai, L.H. Mechanism of DNA-Induced Phase Separation for Transcriptional Repressor VRN1. Angew. Chem.-Int. Edit. 2019, 58, 4858–4862. [Google Scholar] [CrossRef]
- Feng, Z.; Bartholomew, E.S.; Liu, Z.; Cui, Y.; Dong, Y.; Li, S.; Wu, H.; Ren, H.; Liu, X. Glandular trichomes: New focus on horticultural crops. Hortic. Res. 2021, 8, 158. [Google Scholar] [CrossRef]
- Wang, W.-B.; Ao, T.; Zhang, Y.-Y.; Wu, D.; Xu, W.; Han, B.; Liu, A.-Z. Genome-wide analysis of the B3 transcription factors reveals that RcABI3/VP1 subfamily plays important roles in seed development and oil storage in castor bean (Ricinus communis). Plant Divers. 2022, 44, 201–212. [Google Scholar] [CrossRef]
- Gao, J.; Ma, G.; Chen, J.; Gichovi, B.; Cao, L.; Liu, Z.; Chen, L. The B3 gene family in Medicago truncatula: Genome-wide identification and the response to salt stress. Plant Physiol. Biochem. 2024, 206, 108260. [Google Scholar] [CrossRef]
- Ren, C.; Wang, H.; Zhou, Z.; Jia, J.; Zhang, Q.; Liang, C.; Li, W.; Zhang, Y.; Yu, G. Genome-wide identification of the B3 gene family in soybean and the response to melatonin under cold stress. Front. Plant Sci. 2022, 13, 1091907. [Google Scholar] [CrossRef]
- Ruan, C.C.; Chen, Z.; Hu, F.C.; Fan, W.; Wang, X.H.; Guo, L.J.; Fan, H.Y.; Luo, Z.W.; Zhang, Z.L. Genome-wide characterization and expression profiling of B3 superfamily during ethylene-induced flowering in pineapple (Ananas comosus L.). BMC Genom. 2021, 22, 561. [Google Scholar] [CrossRef]
- Park, Y.S.; Cho, H.J.; Kim, S. Identification and expression analyses of B3 genes reveal lineage-specific evolution and potential roles of REM genes in pepper. BMC Plant Biol. 2024, 24, 201. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Miao, H.; Zhang, Z.; Dong, S.; Zhou, Q.; Liu, X.; Beckles, D.M.; Gu, X.; Huang, S.; Zhang, S. A near-complete cucumber reference genome assembly and Cucumber-DB, a multi-omics database. Mol. Plant 2024, 17, 1178–1182. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.X.; Sun, L.; Dong, M.M.; Fan, S.S.; Shi, K.X.; Qu, Y.X.; Zhu, L.Y.; Shi, J.F.; Wang, W.J.; Liu, Y.H.; et al. Novel players in organogenesis and flavonoid biosynthesis in cucumber glandular trichomes. Plant Physiol. 2023, 192, 2723–2736. [Google Scholar] [CrossRef]
- Dong, M.; Xue, S.; Bartholomew, E.S.; Zhai, X.; Sun, L.; Xu, S.; Zhang, Y.; Yin, S.; Ma, W.; Chen, S.; et al. Transcriptomic and functional analysis provides molecular insights into multicellular trichome development. Plant Physiol. 2022, 189, 301–314. [Google Scholar] [CrossRef]
- Jiang, S.Y.; González, J.M.; Ramachandran, S. Comparative Genomic and Transcriptomic Analysis of Tandemly and Segmentally Duplicated Genes in Rice. PLoS ONE 2013, 8, e63551. [Google Scholar] [CrossRef]
- Zhou, D.; Song, R.Q.; Fang, Y.; Liu, R.; You, C.J.; Wang, Y.J.; Huang, L. Global identification and regulatory network analysis reveal the significant roles of lncRNAs during anther and pollen development in Arabidopsis. Plant Cell Rep. 2025, 44, 44. [Google Scholar] [CrossRef]
- Cai, X.X.; Zhang, H.; Mu, C.Q.; Chen, Y.J.; He, C.Z.; Liu, M.Y.; Laux, T.; Pi, L.M. A mobile miR160-triggered transcriptional axis controls root stem cell niche maintenance and regeneration in Arabidopsis. Dev. Cell 2025, 60, 459–471.e5. [Google Scholar] [CrossRef]
- Xue, S.; Dong, M.; Liu, X.; Xu, S.; Pang, J.; Zhang, W.; Weng, Y.; Ren, H. Classification of fruit trichomes in cucumber and effects of plant hormones on type II fruit trichome development. Planta 2019, 249, 407–416. [Google Scholar] [CrossRef]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L.; et al. The Pfam protein families database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J.; Paterson, A.H. MCScanX-transposed: Detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 2013, 29, 1458–1460. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Zhang, Y.; Shen, J.; Bartholomew, E.S.; Dong, M.; Chen, S.; Yin, S.; Zhai, X.; Feng, Z.; Ren, H.; Liu, X. TINY BRANCHED HAIR functions in multicellular trichome development through an ethylene pathway in Cucumis sativus L. Plant J. Cell Mol. Biol. 2021, 106, 753–765. [Google Scholar] [CrossRef]
- Fang, L.; Wei, X.Y.; Liu, L.Z.; Zhou, L.X.; Tian, Y.P.; Geng, C.; Li, X.D. A tobacco ringspot virus-based vector system for gene and microRNA function studies in cucurbits. Plant Physiol. 2021, 186, 853–864. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, M.; Sun, L.; Wang, W.; Wang, Y.; Shan, L.; Liu, X.; Ren, H. B3 Superfamily in Cucumber (Cucumis sativus L.): Identification, Evolution, Expression Patterns, and Function in Glandular Trichome Development. Int. J. Mol. Sci. 2025, 26, 4031. https://doi.org/10.3390/ijms26094031
Dong M, Sun L, Wang W, Wang Y, Shan L, Liu X, Ren H. B3 Superfamily in Cucumber (Cucumis sativus L.): Identification, Evolution, Expression Patterns, and Function in Glandular Trichome Development. International Journal of Molecular Sciences. 2025; 26(9):4031. https://doi.org/10.3390/ijms26094031
Chicago/Turabian StyleDong, Mingming, Lei Sun, Wujun Wang, Yaru Wang, Li Shan, Xingwang Liu, and Huazhong Ren. 2025. "B3 Superfamily in Cucumber (Cucumis sativus L.): Identification, Evolution, Expression Patterns, and Function in Glandular Trichome Development" International Journal of Molecular Sciences 26, no. 9: 4031. https://doi.org/10.3390/ijms26094031
APA StyleDong, M., Sun, L., Wang, W., Wang, Y., Shan, L., Liu, X., & Ren, H. (2025). B3 Superfamily in Cucumber (Cucumis sativus L.): Identification, Evolution, Expression Patterns, and Function in Glandular Trichome Development. International Journal of Molecular Sciences, 26(9), 4031. https://doi.org/10.3390/ijms26094031






