Identification of the MADS-Box Gene Family and the Key Role of BrAGL27 in the Regulation of Flowering in Chinese Cabbage (Brassica rapa L. ssp. pekinensis)
Abstract
1. Introduction
2. Results
2.1. Chromosomal Location Analysis of MADS-Box Genes
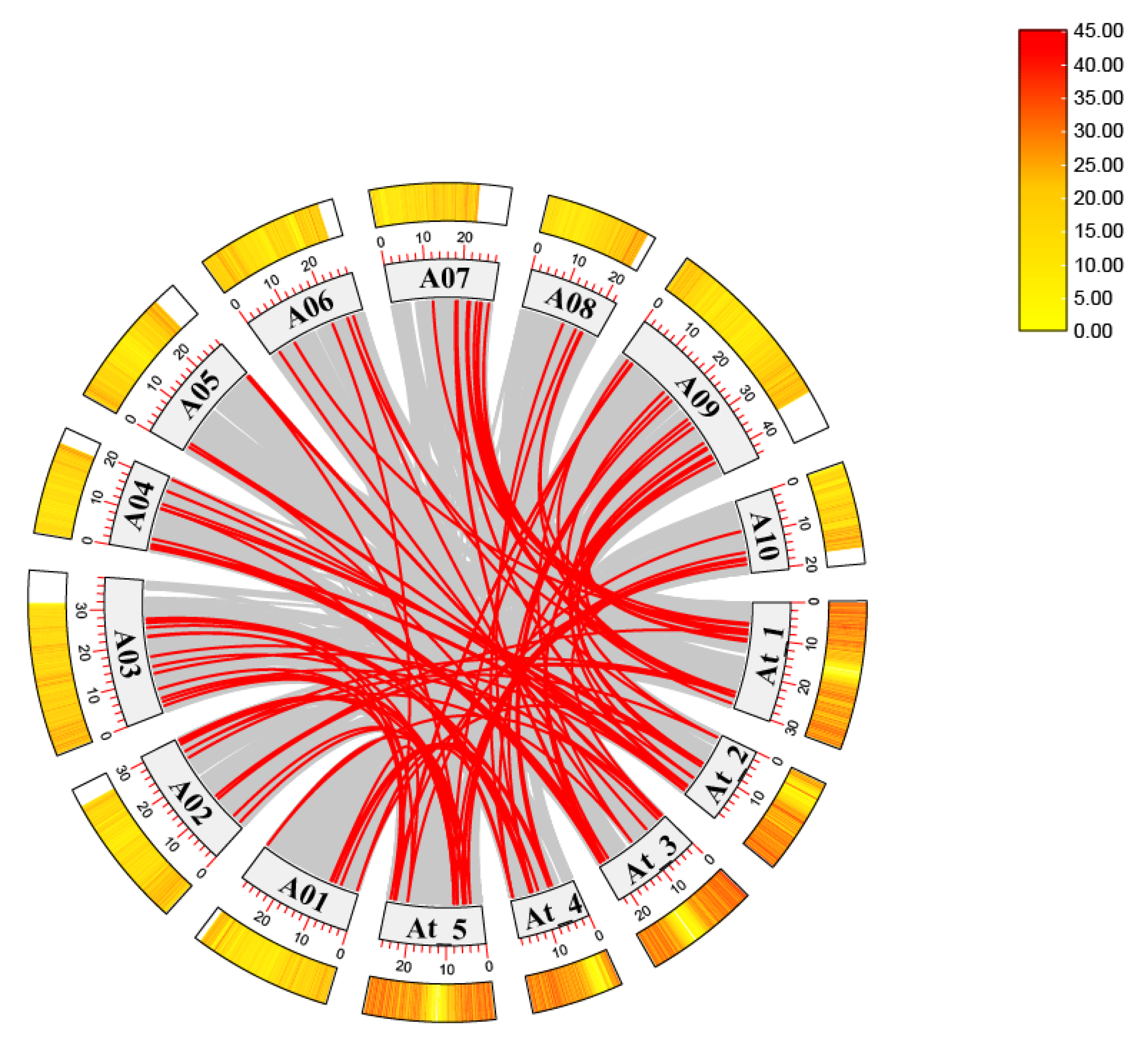
2.2. Gene Duplication and Synteny Analysis of the MADS-Box Family in Chinese Cabbage
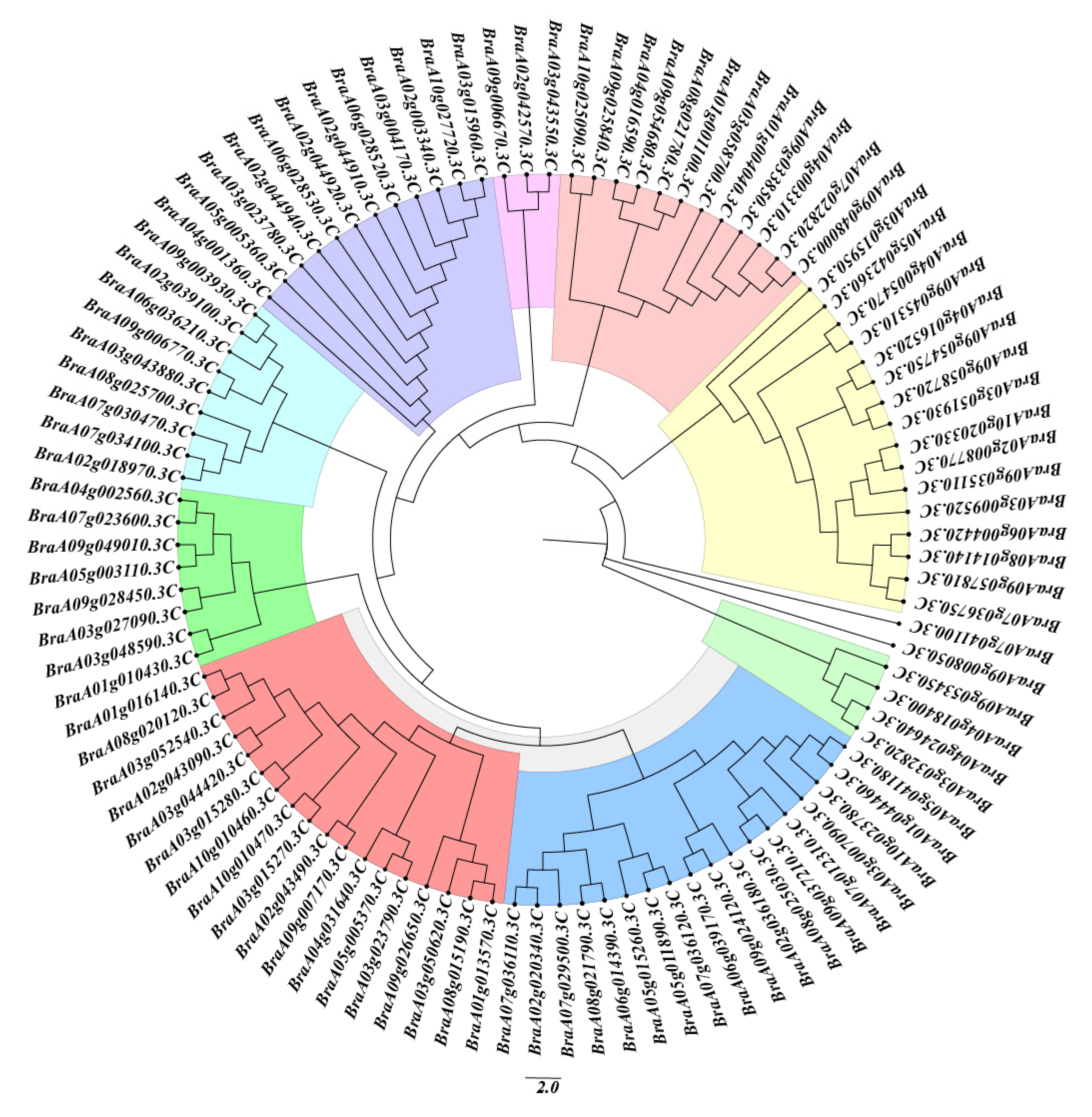
2.3. Phylogenetic Analysis of Chinese Cabbage MADS-Box Proteins
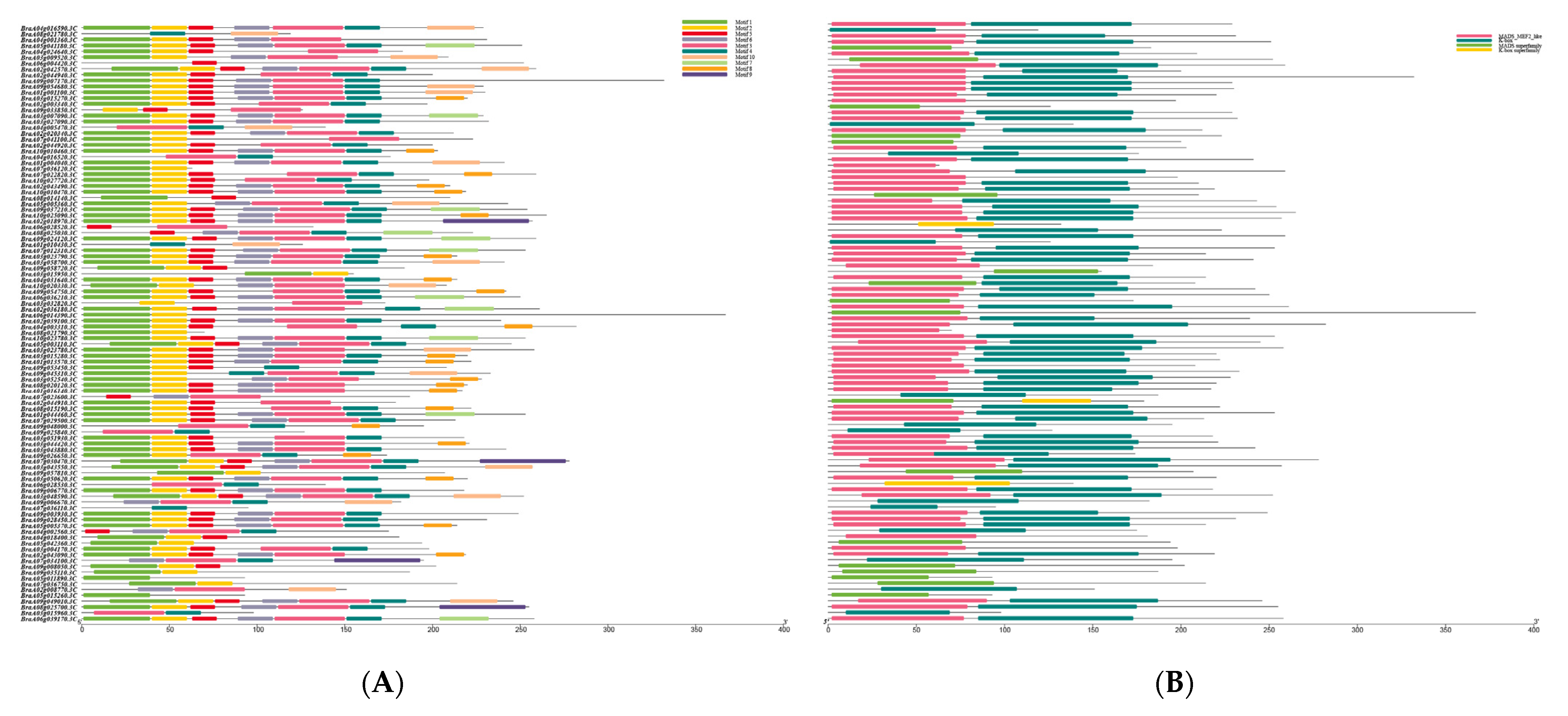
2.4. Analysis of Conserved Motifs, Structural Domains, and Gene Structure in Chinese Cabbage MADS-Box Genes
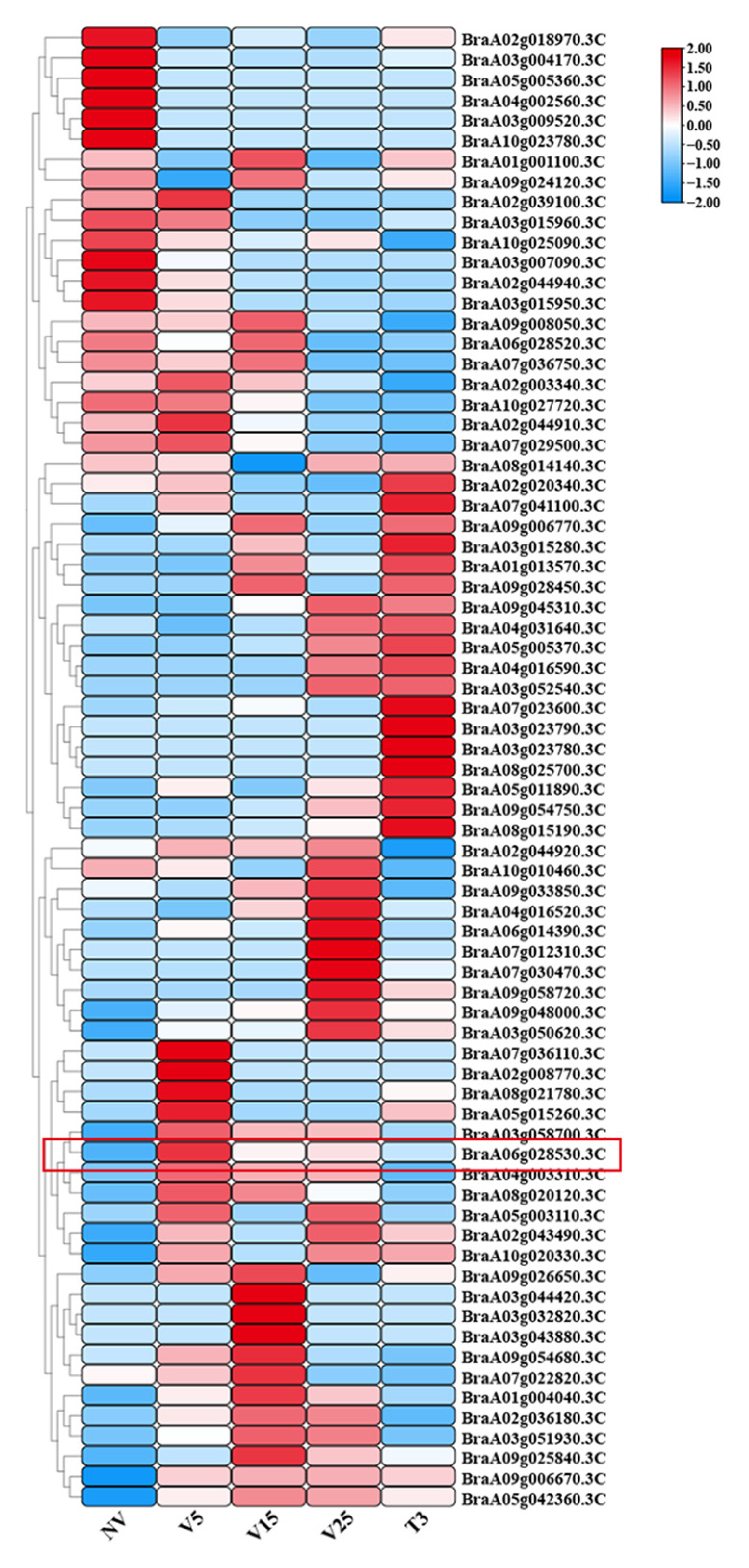
2.5. Heat Map of MADS-Box Gene Expression Under Different Vernalization Treatments
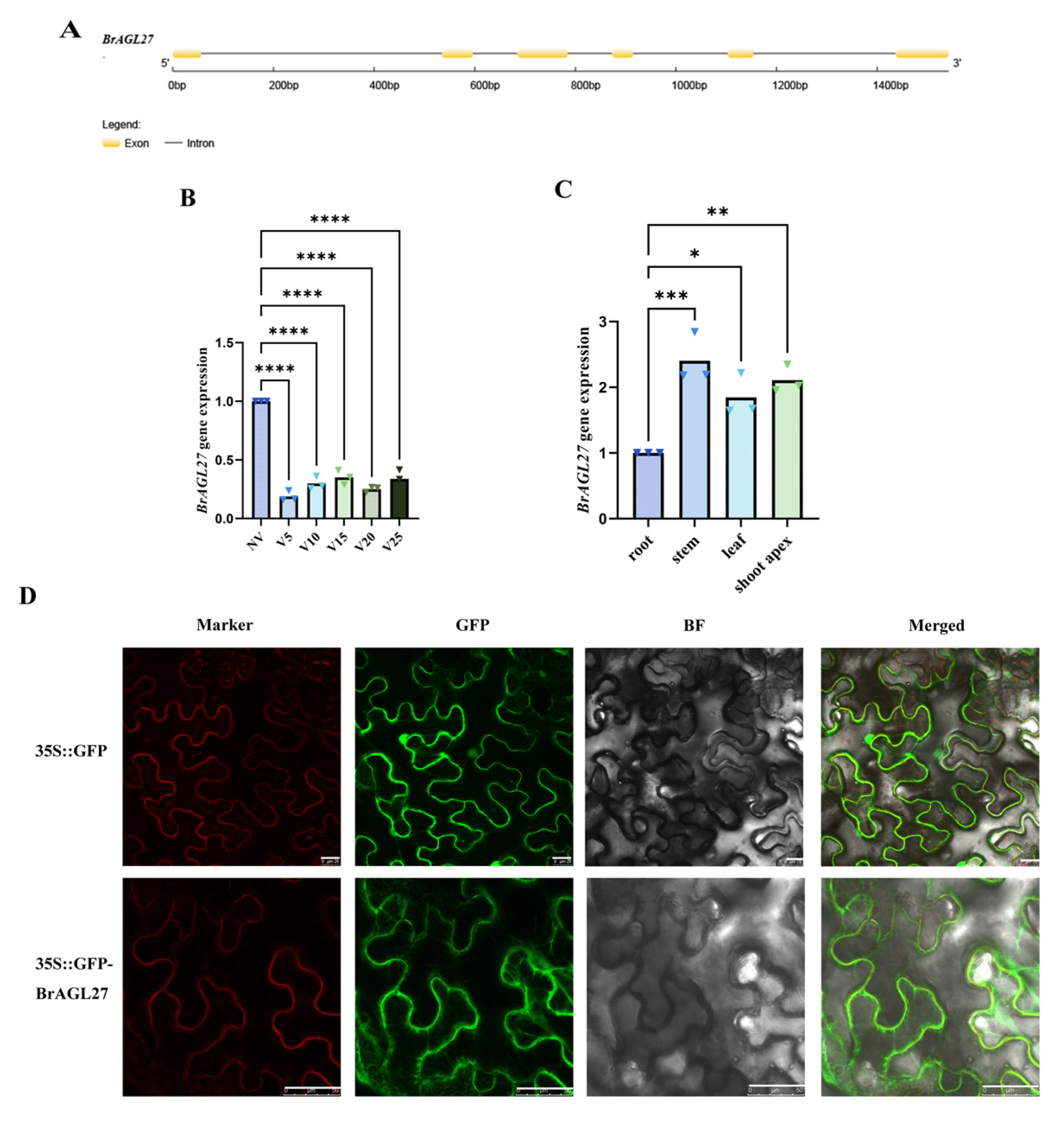
2.6. Expression Patterns of the BrAGL27 Gene in Response to Vernalization
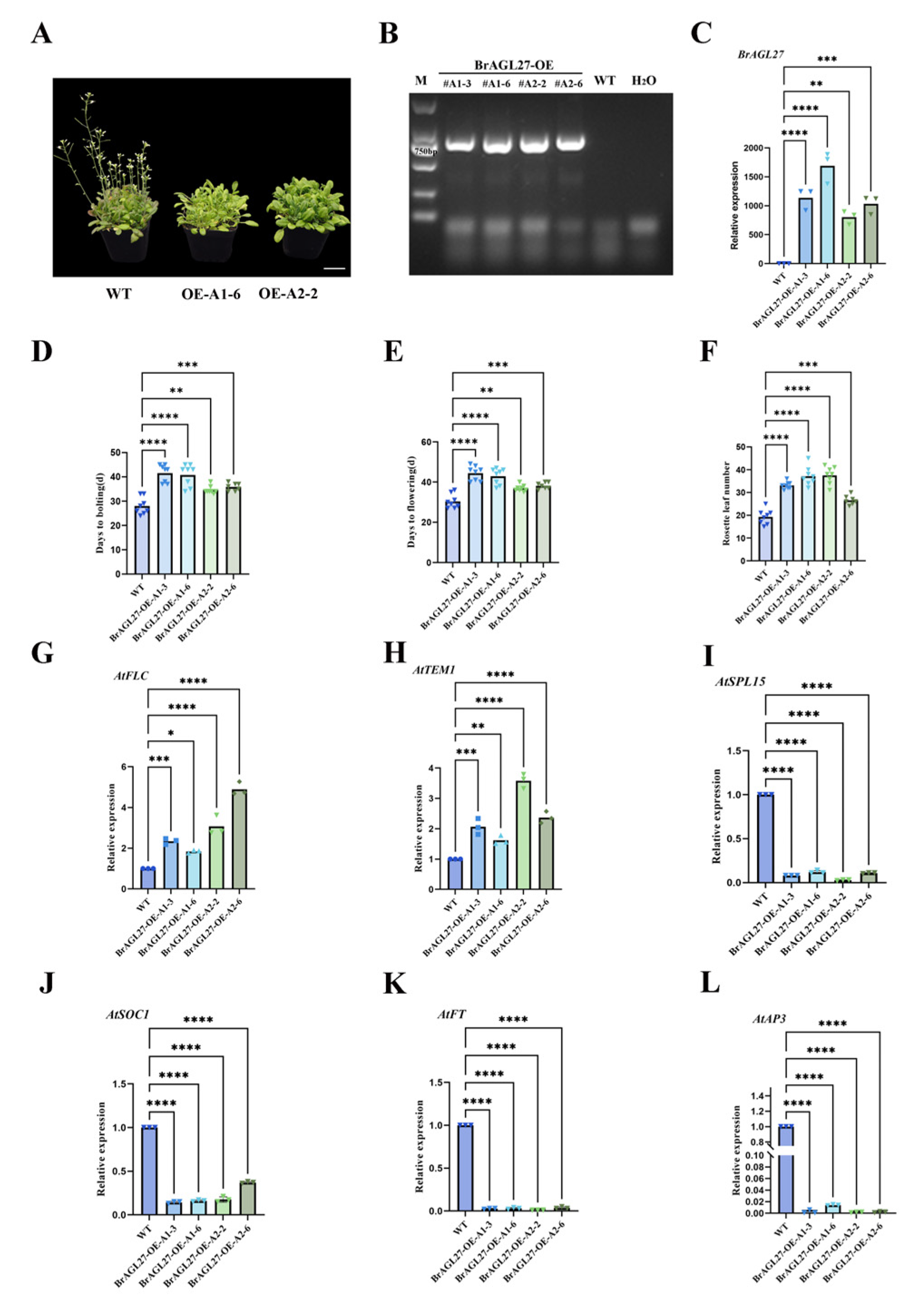
2.7. Overexpression of BrAGL27 Results in a Late-Flowering Phenotype in Plants
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Sources of Data and Plant Materials
4.3. Identification and Analysis of MADS-Box Genes in Chinese Cabbage
4.4. Chromosomal Localization and Gene Duplication Analysis of Chinese Cabbage MADS-Box Genes
4.5. Phylogenetic Analysis, Classification, and Structural and Motif Analysis of MADS-Box Genes
4.6. Gene Cloning and Vector Construction
4.7. Quantitative Real-Time PCR (qRT-PCR)
4.8. Subcellular Localization Analysis
4.9. Development of Overexpression Lines
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dai, Y.; Sun, X.; Wang, C.; Li, F.; Zhang, S.; Zhang, H.; Li, G.; Yuan, L.; Chen, G.; Sun, R.; et al. Gene co-expression network analysis reveals key pathways and hub genes in Chinese cabbage (Brassica Rapa L.) during vernalization. BMC Genom. 2021, 22, 236. [Google Scholar] [CrossRef] [PubMed]
- Laczi, E.; Apahidean, A.S.; Apahidean, A.I.; Varga, J. Agrobiological Particularities of Chinese Cabbage: Brassica rapa L. ssp. pekinensis (Lour) Hanelt and Brassica rapa L. ssp. chinensis (Lour) Hanelt. Bull. UASVM Hortic. 2010, 67, 1843–5254. [Google Scholar]
- Dai, Y.; Zhang, S.; Guan, J.; Wang, S.; Zhang, H.; Li, G.; Sun, R.; Li, F.; Zhang, S. Single-Cell Transcriptomic Analysis of Flowering Regulation and Vernalization in Chinese Cabbage Shoot Apex. Hortic. Res. 2024, 11, uhae214. [Google Scholar] [CrossRef]
- Kim, D.-H.; Sung, S. Genetic and Epigenetic Mechanisms Underlying Vernalization. Arab. Book 2014, 12, e0171. [Google Scholar] [CrossRef] [PubMed]
- Jack, T. Plant development going MADS. Plant Mol. Biol. 2001, 46, 515–520. [Google Scholar] [CrossRef]
- Norman, C.; Runswick, M.; Pollock, R.; Treisman, R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 1988, 55, 989–1003. [Google Scholar] [CrossRef]
- Passmore, S.; Maine, G.T.; Elble, R.; Christ, C.; Tye, B.-K. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MATα cells. J. Mol. Biol. 1988, 204, 593–606. [Google Scholar] [CrossRef]
- Sommer, H.; Beltrán, J.P.; Huijser, P.; Pape, H.; Lönnig, W.E.; Saedler, H.; Schwarz-Sommer, Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 1990, 9, 605–613. [Google Scholar] [CrossRef]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef]
- Ciannamea, S.; Kaufmann, K.; Frau, M.; Tonaco, I.A.N.; Petersen, K.; Nielsen, K.K.; Angenent, G.C.; Immink, R.G.H. Protein Interactions of MADS Box Transcription Factors Involved in Flowering in Lolium Perenne. J. Exp. Bot. 2006, 57, 3419–3431. [Google Scholar] [CrossRef]
- Yoo, S.K.; Wu, X.; Lee, J.S.; Ahn, J.H. AGAMOUS-LIKE 6 is a floral promoter that negatively regulates the FLC/MAF clade genes and positively regulates FT in Arabidopsis. Plant J. 2011, 65, 62–76. [Google Scholar] [CrossRef]
- Bowman, J.L.; Baum, S.F.; Eshed, Y.; Putterill, J.; Alvarez, J. Molecular genetics of gynoecium development in Arabidopsis. Curr. Top. Dev. Biol. 1999, 45, 155–205. [Google Scholar] [PubMed]
- Drews, G.N.; Bowman, J.L.; Meyerowitz, E.M. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 1991, 65, 991–1002. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Forde, B.G. An Arabidopsis MADS Box Gene That Controls Nutrient-Induced Changes in Root Architecture. Science 1998, 279, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Colombo, L.; Franken, J.; Koetje, E.; van Went, J.; Dons, H.J.; Angenent, G.C.; van Tunen, A.J. The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 1995, 7, 1859–1868. [Google Scholar]
- Ma, H.; Yanofsky, M.F.; Meyerowitz, E.M. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes. Dev. 1991, 5, 484–495. [Google Scholar] [CrossRef]
- Rounsley, S.D.; Ditta, G.S.; Yanofsky, M.F. Diverse roles for MADS box genes in Arabidopsis thaliana development. Plant Cell 1995, 7, 1259–1269. [Google Scholar]
- Parenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-Box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef]
- Grimplet, J.; Martínez-Zapater, J.M.; Carmona, M.J. Structural and functional annotation of the MADS-box transcription factor family in grapevine. BMC Genom. 2016, 17, 80. [Google Scholar] [CrossRef]
- Hu, L.; Liu, S. Genome-Wide Analysis of the MADS-Box Gene Family in Cucumber. Genome 2012, 55, 245–256. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, J.; Zhang, J.; Miao, H.; Wang, J.; Gao, P.; Hu, W.; Jia, C.; Wang, Z.; Xu, B. Genome-wide analysis of banana MADS-box family closely related to fruit development and ripening. Sci. Rep. 2017, 7, 3467. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Yang, Y.; Luo, W.; Yang, C.; Ding, P.; Liu, Y.; Qiao, L.; Chang, Z.; Geng, H.; Wang, P.; et al. Genome-wide identification and analysis of the MADS-Box gene family in bread wheat (Triticum aestivum L). PLoS ONE 2017, 12, e0181443. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Islam, T. Genome-wide analysis and expression profiling of glyoxalase gene families in soybean (Glycine max) indicate their development and abiotic stress specific response. BMC Plant Biol. 2016, 16, 87. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, J.; Feng, C.; Liu, M.; Wang, J.; Hu, Y. Genome-wide identification, characterization of the MADS-Box gene family in Chinese Jujube and their Involvement in flower development. Sci. Rep. 2017, 7, 1025. [Google Scholar] [CrossRef]
- Smaczniak, C.; Immink, R.G.H.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Chang, L.; Zhang, T.; Chen, H.; Zhang, L.; Lin, R.; Liang, J.; Wu, J.; Freeling, M.; Wang, X. Impacts of Allopolyploidization and Structural Variation on Intraspecific Diversification in Brassica Rapa. Genome Biol. 2021, 22, 166. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.-H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef]
- Kim, S.Y.; Park, B.S.; Kwon, S.J.; Kim, J.; Lim, M.H.; Park, Y.D.; Kim, D.Y.; Suh, S.C.; Jin, Y.M.; Ahn, J.H.; et al. Delayed Flowering Time in Arabidopsis and Brassica rapa by the overexpression of FLOWERING LOCUS C (FLC) homologs isolated from Chinese cabbage (Brassica rapa L.: ssp. pekinensis). Plant Cell Rep. 2007, 26, 327–336. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Krizek, B.A.; Meyerowitz, E.M. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 1996, 122, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef]
- Mizzotti, C.; Ezquer, I.; Paolo, D.; Rueda-Romero, P.; Guerra, R.F.; Battaglia, R.; Rogachev, I.; Aharoni, A.; Kater, M.M.; Caporali, E.; et al. SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat. PLOS Genet. 2014, 10, e1004856. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, K.A.; Goff, S.A. Chapter Four—The First Plant Genome Sequence—Arabidopsis Thaliana. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2014; Volume 69, pp. 91–117. [Google Scholar]
- Shen, L.; Zhang, Y.; Sawettalake, N. A Molecular switch for FLOWERING LOCUS C activation determines flowering time in Arabidopsis. Plant Cell 2022, 34, 818–833. [Google Scholar] [CrossRef]
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008, 67, 183. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010, 61, 2247–2254. [Google Scholar] [CrossRef]
- Huang, F.; Liu, T.; Tang, J.; Duan, W.; Hou, X. BcMAF2 activates BcTEM1 and represses flowering in Pak-choi (Brassica rapa ssp. chinensis). Plant Mol. Biol. 2019, 100, 19–32. [Google Scholar] [CrossRef]
- Dai, Y.; Li, G.; Gao, X.; Wang, S.; Li, Z.; Song, C.; Zhang, S.; Li, F.; Fang, Z.; Sun, R.; et al. Identification of long noncoding RNAs involved in plumule-vernalization of Chinese cabbage. Front. Plant Sci. 2023, 14, 1147494. [Google Scholar] [CrossRef]
- Zhang, L.; Cai, X.; Wu, J.; Liu, M.; Grob, S.; Cheng, F.; Liang, J.; Cai, C.; Liu, Z.; Liu, B.; et al. Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 2018, 5, 50. [Google Scholar] [CrossRef]
- Lu, J.-X.; Sun, J.-Y.; Wang, Z.; Ren, W.-C.; Xing, N.-N.; Liu, M.-Q.; Zhang, Z.-P.; Kong, L.-Y.; Su, X.-Y.; Liu, X.-B.; et al. In Silico Genome-Wide Analysis of B3 Transcription Factors in Cannabis Sativa L. Cannabis Cannabinoid Res. 2024, 9, 495–512. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A Simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.-S.; Niu, Q.-W.; Chua, N.-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Li, Y.; Dai, Y.; Li, X.; Huang, C.; Zhang, S.; Li, F.; Zhang, H.; Li, G.; Sun, R.; et al. Identification of the MADS-Box Gene Family and the Key Role of BrAGL27 in the Regulation of Flowering in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Int. J. Mol. Sci. 2025, 26, 2635. https://doi.org/10.3390/ijms26062635
Gao X, Li Y, Dai Y, Li X, Huang C, Zhang S, Li F, Zhang H, Li G, Sun R, et al. Identification of the MADS-Box Gene Family and the Key Role of BrAGL27 in the Regulation of Flowering in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). International Journal of Molecular Sciences. 2025; 26(6):2635. https://doi.org/10.3390/ijms26062635
Chicago/Turabian StyleGao, Xinyu, Yang Li, Yun Dai, Xiangqianchen Li, Can Huang, Shifan Zhang, Fei Li, Hui Zhang, Guoliang Li, Rifei Sun, and et al. 2025. "Identification of the MADS-Box Gene Family and the Key Role of BrAGL27 in the Regulation of Flowering in Chinese Cabbage (Brassica rapa L. ssp. pekinensis)" International Journal of Molecular Sciences 26, no. 6: 2635. https://doi.org/10.3390/ijms26062635
APA StyleGao, X., Li, Y., Dai, Y., Li, X., Huang, C., Zhang, S., Li, F., Zhang, H., Li, G., Sun, R., Song, H., Zhang, L., Chen, Z., & Zhang, S. (2025). Identification of the MADS-Box Gene Family and the Key Role of BrAGL27 in the Regulation of Flowering in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). International Journal of Molecular Sciences, 26(6), 2635. https://doi.org/10.3390/ijms26062635





