Isolation, Identification, and Genetic Evolution Analysis of VP1 Gene of Feline Calicivirus Strain ZZ202306
Abstract
1. Introduction
2. Results
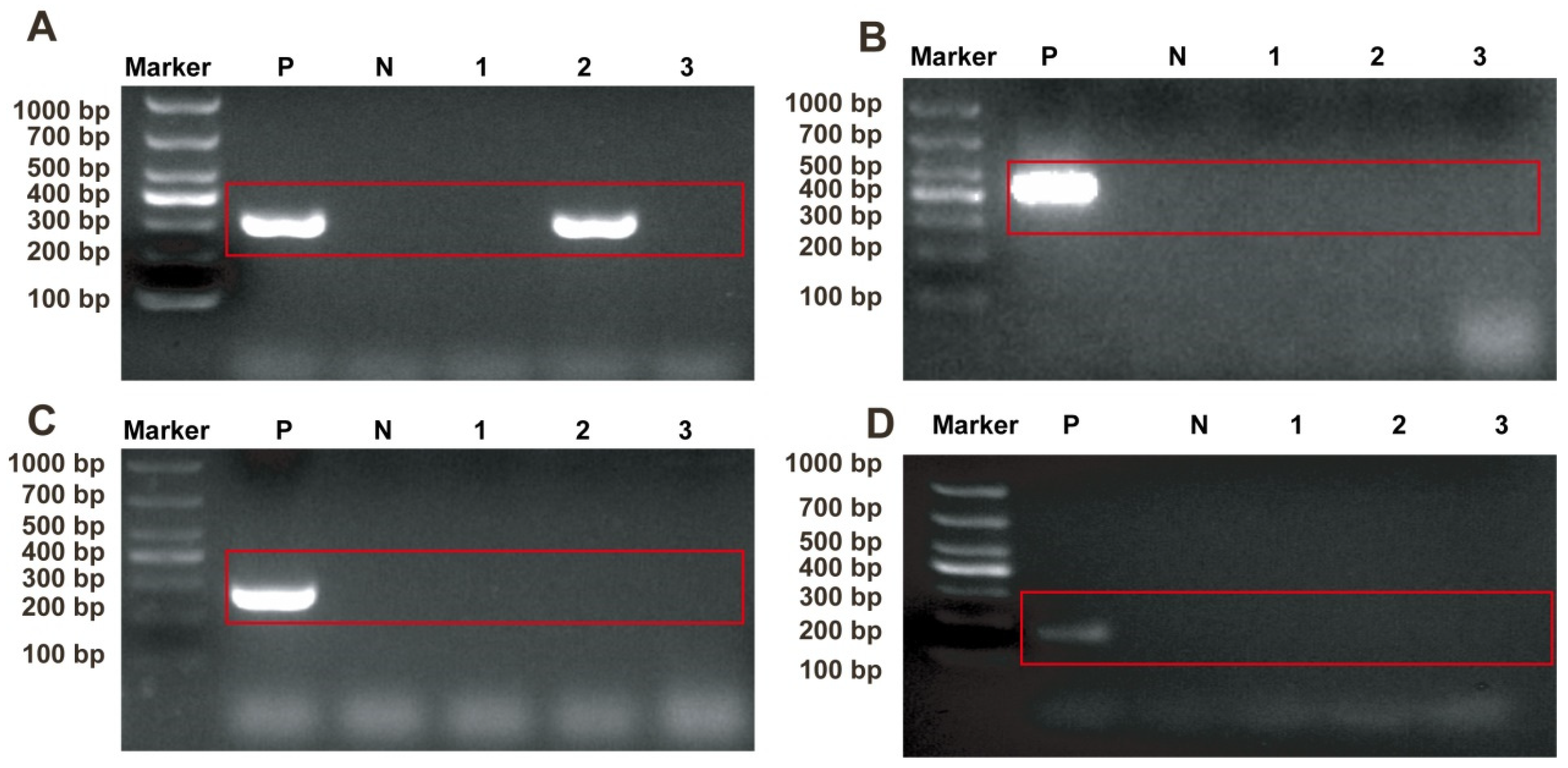
2.1. Identification of Pathogens in Sample
2.2. Isolation and Identification of FCV
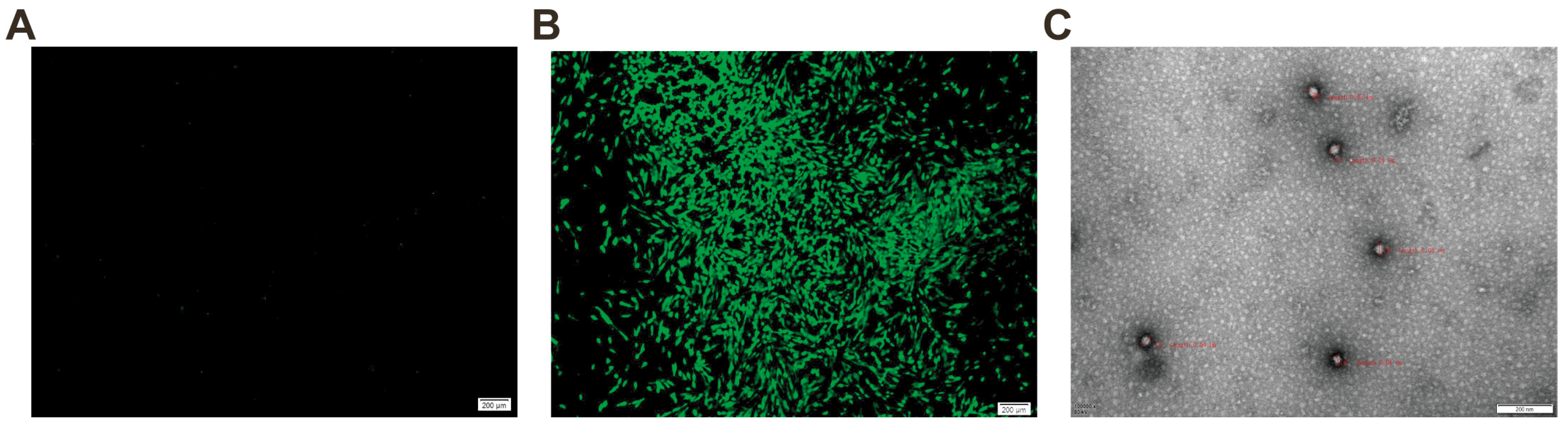
2.3. Detection of FCV Using Immunofluorescence Assay and Electron Microscopy
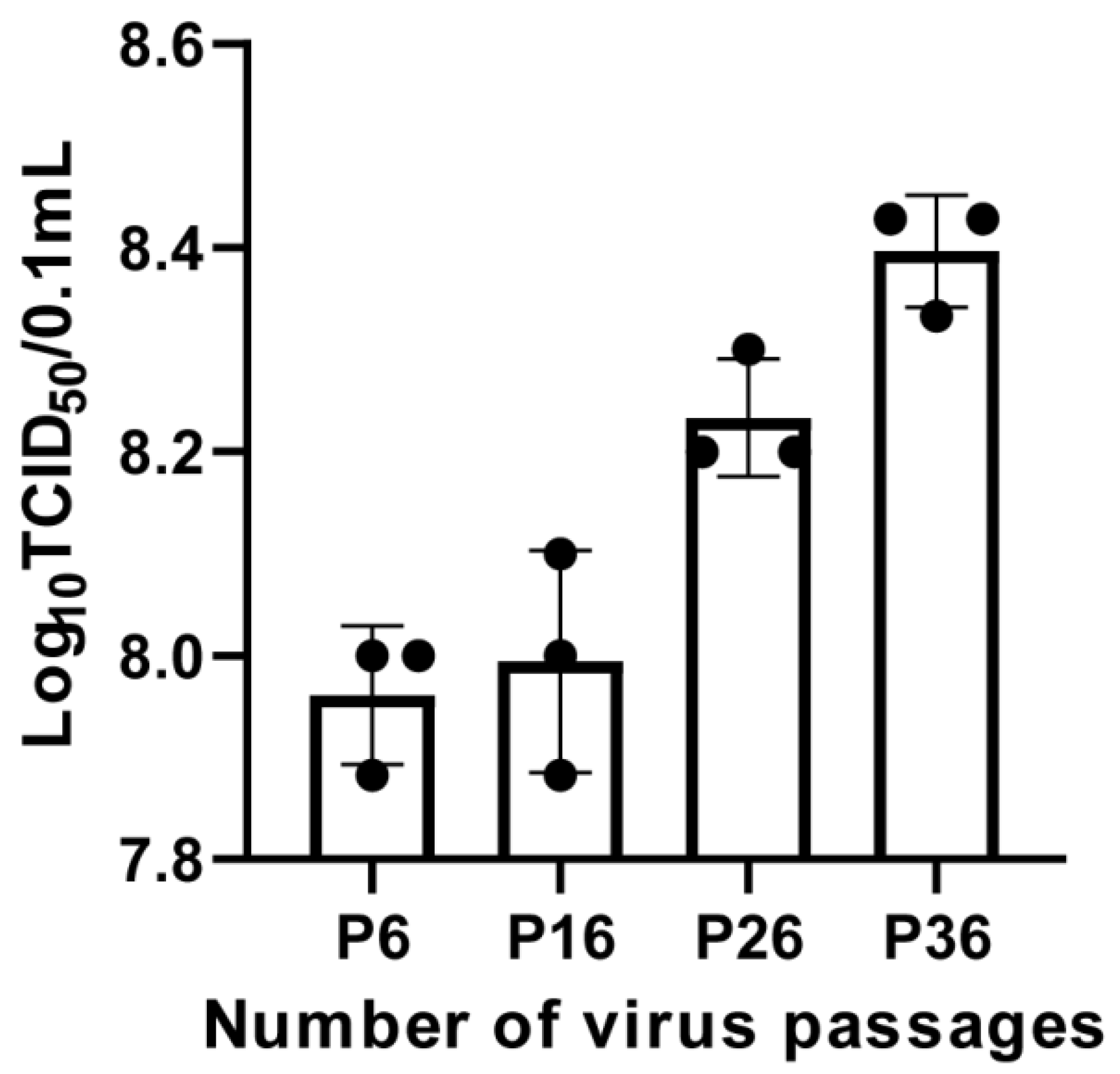
2.4. Viral Titer Determination for FCV Strain ZZ202306
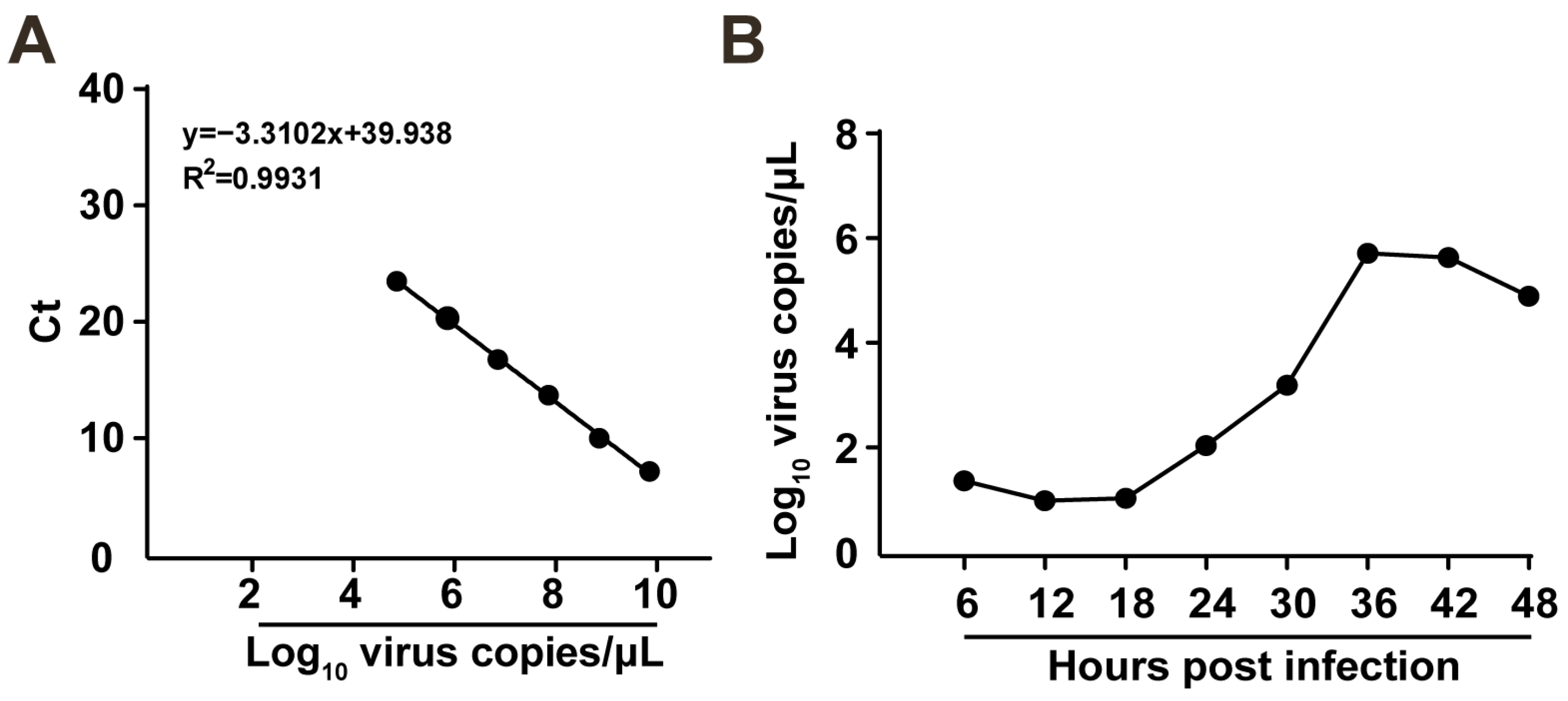
2.5. Proliferation Dynamics of FCV Strain ZZ202306 in CRFK Cells
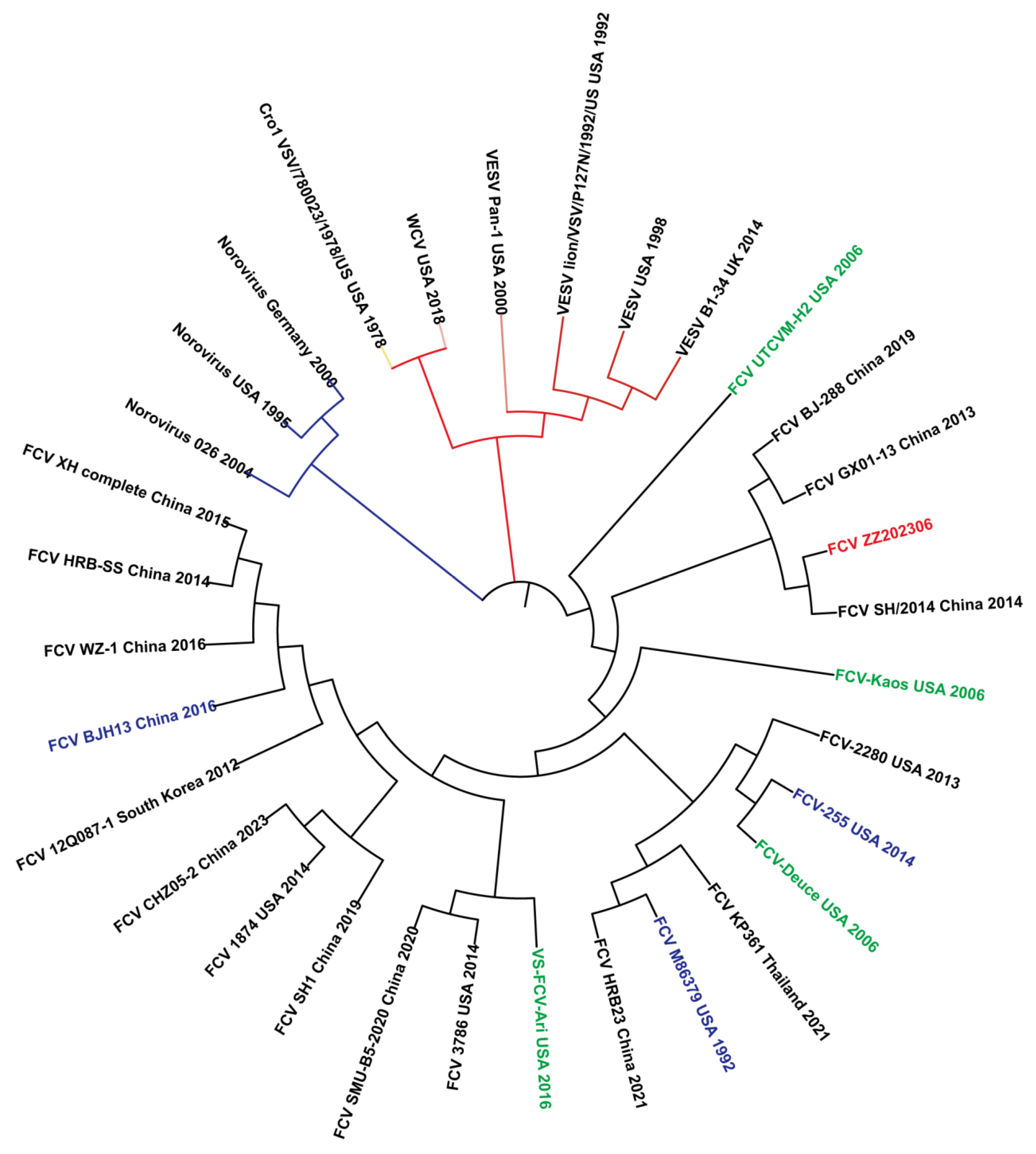
2.6. Amino Acid Homology and Phylogenetic Analysis of FCV Strain ZZ202306-VP1Gene
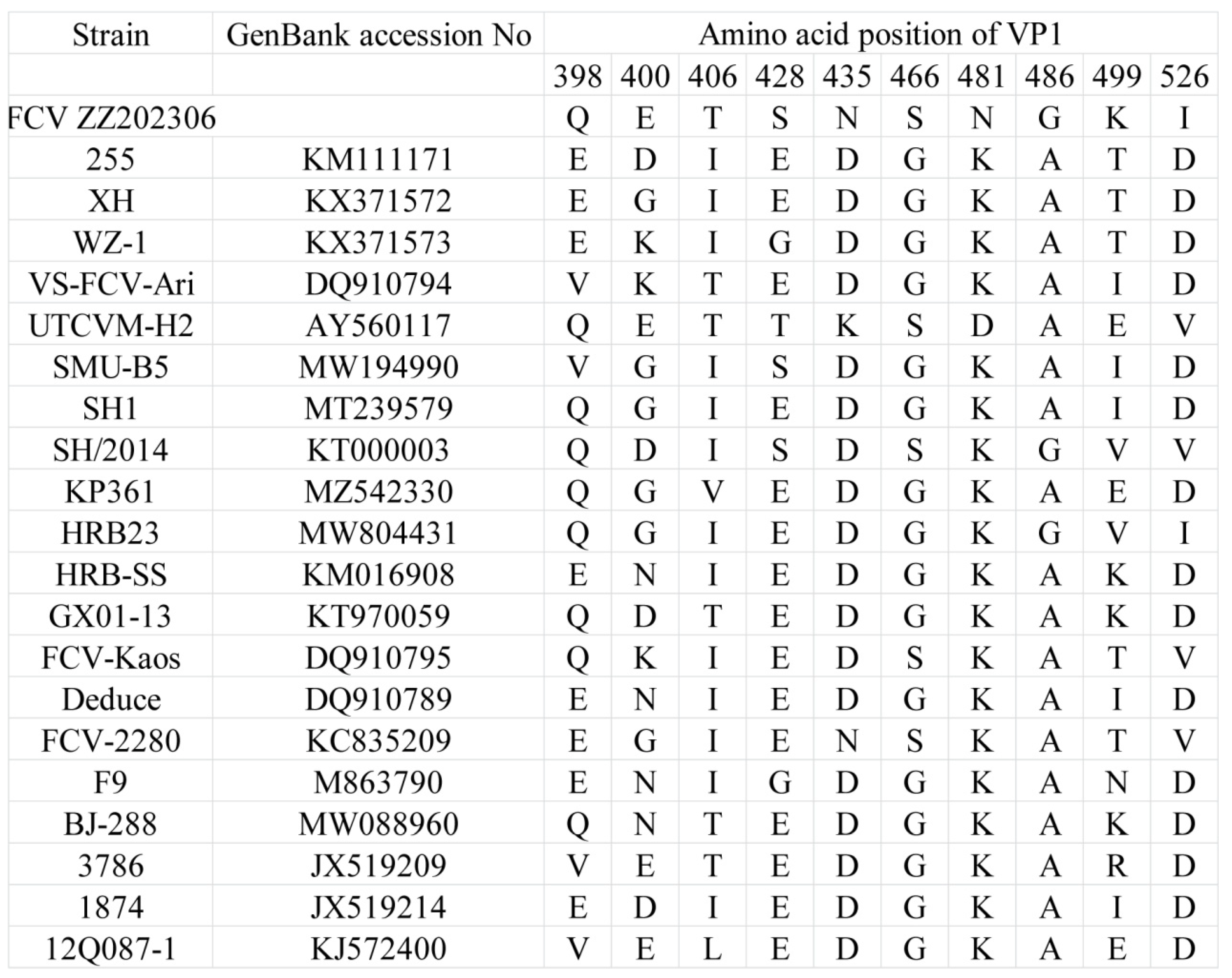
2.7. Analysis of Amino Acid Mutations in Highly Mutated Region of FCV Strain ZZ202306-VP1 Gene
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. Sample Processing
4.3. Specimen Testing
4.4. Virus Isolation and Cultivation
4.5. Western Blotting
4.6. Immunofluorescence Assay
4.7. TCID50 Assay
4.8. Viral Growth Kinetics Analysis
4.9. Electron Microscopy
4.10. Statistics
4.11. Statement of Informed Consent
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hosie, M.J.; Conley, M.J. Feline Calicivirus (Caliciviridae). In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: Oxford, UK, 2021; pp. 294–299. [Google Scholar]
- Hou, J.; Sánchez-Vizcaíno, F.; McGahie, D.; Lesbros, C.; Almeras, T.; Howarth, D.; O’Hara, V.; Dawson, S.; Radford, A.D. European molecular epidemiology and strain diversity of feline calicivirus. Vet. Rec. 2016, 178, 114–115. [Google Scholar] [CrossRef]
- Cao, L.; Liu, J.; Li, Y.; Xie, D.; Yan, Q.; Li, Q.; Cao, Y.; Du, W.; Li, J.; Ye, Z.; et al. Biological Characteristics of Feline Calicivirus Epidemic Strains in China and Screening of Broad-Spectrum Protective Vaccine Strains. Vaccines 2023, 11, 1858. [Google Scholar] [CrossRef] [PubMed]
- Hofmann-Lehmann, R.; Hosie, M.J.; Hartmann, K.; Egberink, H.; Truyen, U.; Tasker, S.; Belák, S.; Boucraut-Baralon, C.; Frymus, T.; Lloret, A.; et al. Calicivirus Infection in Cats. Viruses 2022, 14, 937. [Google Scholar] [CrossRef] [PubMed]
- Bhella, D.; Gatherer, D.; Chaudhry, Y.; Pink, R.; Goodfellow, I.G. Structural Insights into Calicivirus Attachment and Uncoating. J. Virol. 2008, 82, 8051–8058. [Google Scholar] [CrossRef] [PubMed]
- Sosnovtsev, S.V.; Sosnovtseva, S.A.; Green, K.Y. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 1998, 72, 3051–3059. [Google Scholar] [CrossRef]
- Fastier, L.B. A new feline virus isolated in tissue culture. Am. J. Vet. Res. 1957, 18, 382–389. [Google Scholar]
- Kadoi, K.; Kiryu, M.; Iwabuchi, M.; Kamata, H.; Yukawa, M.; Inaba, Y. A strain of calicivirus isolated from lions with vesicular lesions on tongue and snout. New Microbiol. 1997, 20, 141–148. [Google Scholar]
- Barbara, D.M.; Camillo, D.R.; Chiara, C.; Fulvio, M. Characterization of a strain of feline calicivirus isolated from a dog faecal sample. Vet. Microbiol. 2009, 139, 52–57. [Google Scholar]
- Hoover, E.A.; Kahn, D.E. Experimentally induced feline calicivirus infection: Clinical signs and lesions. J. Am. Vet. Med. Assoc. 1975, 166, 463–468. [Google Scholar]
- Neill, J.D. Nucleotide sequence of the capsid protein gene of two serotypes of San Miguel sea lion virus: Identification of conserved and non-conserved amino acid sequences among calicivirus capsid proteins. Virus Res. 1992, 24, 211–222. [Google Scholar] [CrossRef]
- Levy, J.K.; Marsh, A. Isolation of calicivirus from the joint of a kitten with arthritis. J. Am. Vet. Med. Assoc. 1992, 201, 753–755. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Qi, R.; Chen, M.; Feng, K.; Liu, Z.; Kang, H.; Jiang, Q.; Qu, L.; Liu, J. Screening and Immune Efficacy Evaluation of Antigens with Protection Against Feline Calicivirus. Vaccines 2024, 12, 1205. [Google Scholar] [CrossRef] [PubMed]
- Conley, M.J.; McElwee, M.; Azmi, L.; Gabrielsen, M.; Byron, O.; Goodfellow, I.G.; Bhella, D. Calicivirus VP2 forms a portal-like assembly following receptor engagement. Nature 2019, 565, 377–381. [Google Scholar] [CrossRef]
- Makino, A.; Shimojima, M.; Miyazawa, T.; Kato, K.; Tohya, Y.; Akashi, H. Junctional adhesion molecule 1 is a functional receptor for feline calicivirus. J. Virol. 2006, 80, 4482–4490. [Google Scholar] [CrossRef]
- Urban, C.; Luttermann, C. Major Capsid Protein Synthesis from the Genomic RNA of Feline Calicivirus. J. Virol. 2020, 94, e00280-20. [Google Scholar] [CrossRef]
- Ossiboff, R.J.; Zhou, Y.; Lightfoot, P.J.; Prasad, B.V.; Parker, J.S. Conformational changes in the capsid of a calicivirus upon interaction with its functional receptor. J. Virol. 2010, 84, 5550–5564. [Google Scholar] [CrossRef]
- Bhella, D.; Goodfellow, I.G. The cryo-electron microscopy structure of feline calicivirus bound to junctional adhesion molecule A at 9-angstrom resolution reveals receptor-induced flexibility and two distinct conformational changes in the capsid protein VP1. J. Virol. 2011, 85, 11381–11390. [Google Scholar] [CrossRef] [PubMed]
- Brunet, S.; Sigoillot-Claude, C.; Pialot, D.; Poulet, H. Multiple Correspondence Analysis on Amino Acid Properties within the Variable Region of the Capsid Protein Shows Differences between Classical and Virulent Systemic Feline Calicivirus Strains. Viruses 2019, 11, 1090. [Google Scholar] [CrossRef]
- Zhang, Z.-H.; Ji, Z.H.; Zhao, X.; Su, Y.Z.; Dong, H.; Si, Y.F.; Wang, J.M.; Shi, W.D. Isolation and Identification of Feline Calicivirus BJH13 Strain and Sequence Analysis of VP1 Gene. Prog. Progress. Vet. Med. 2024, 45, 29–35. [Google Scholar]
- Jian, L.; BAl Yilan, L.X.; Yaping, G.; Houbin, J.; Guohua., G.; Luming., X.; Weifeng., C.; Xiaoying., Z.; Xiaojing., C.; Zengqiang., L.; et al. VP1 Sequence and Pathogenicity Analysis of Feline Calicivirus lsolates. Acta Vet. Zootech. Sin. 2022, 53, 3530–3539. [Google Scholar]
- Guo, J.; Ding, Y.; Sun, F.; Zhou, H.; He, P.; Chen, J.; Guo, J.; Zeng, H.; Long, J.; Wei, Z.; et al. Co-circulation and evolution of genogroups I and II of respiratory and enteric feline calicivirus isolates in cats. Transbound. Emerg. Dis. 2022, 69, 2924–2937. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo-Lara, R.W.; Villabruna, N.; Hesselink, D.A.; Schapendonk, C.M.E.; Ribó Pons, S.; Nieuwenhuijse, D.; Meier, J.I.J.; Goodfellow, I.; Dalm, V.; Fraaij, P.L.A.; et al. Patterns of the within-host evolution of human norovirus in immunocompromised individuals and implications for treatment. EBioMedicine 2024, 109, 105391. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Cheng, Y.; Fang, Z.; Liu, X.; Zhongqi, Q.; Weidong, Y.; Yilmaz, A.; Yilmaz, H.; Umar, S. Molecular epidemiology and phylogenetic analysis of feline calicivirus in Kunshan, China. Virol. J. 2024, 21, 50. [Google Scholar] [CrossRef]
- Spiri, A.M. An Update on Feline Calicivirus. Schweiz. Arch. Tierheilkd. 2022, 164, 225–241. [Google Scholar]
- Li, L.; Liu, Z.; Shi, J.; Yang, M.; Yan, Y.; Fu, Y.; Shen, Z.; Peng, G. The CDE region of feline Calicivirus VP1 protein is a potential candidate subunit vaccine. BMC Vet. Res. 2024, 20, 80. [Google Scholar] [CrossRef]
- Cubillos-Zapata, C.; Angulo, I.; Almanza, H.; Borrego, B.; Zamora-Ceballos, M.; Castón, J.R.; Mena, I.; Blanco, E.; Bárcena, J. Precise location of linear epitopes on the capsid surface of feline calicivirus recognized by neutralizing and non-neutralizing monoclonal antibodies. Vet. Res. 2020, 51, 59. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Ledgerwood, E.D.; Hinchman, M.M.; Dick, R.; Parker, J.S.L. Conserved Surface Residues on the Feline Calicivirus Capsid Are Essential for Interaction with Its Receptor Feline Junctional Adhesion Molecule A (fJAM-A). J. Virol. 2018, 92, e00035-18. [Google Scholar] [CrossRef]
- Wang, J.; Ding, H.T.; Liao, J.L.; Qian, P.; Liu, Y.X.; Xi, X.F.; Zhang, X.K.; Tian, K.G. Isolation and Identification of Feline Calicivirus and Analysis of Its VP1 Gene Sequences. Prog. Progress. Vet. Med. 2021, 42, 50–56. [Google Scholar]

| Primer Name | Primer Sequence 5’-3’ | Fragment Size (bp) |
|---|---|---|
| 09-FCoV-P205 | GGCAACCCGATGTTTAAAACTGG | 214 |
| 09-FCoV-P211 | CACTAGATCCAGACGTTAGCTC | |
| 56-FCV-F | TGTACTTTGCGGGACTTGGT | 401 |
| 56-FCV-R | ACATTACCCACAGCTTGTGCT | |
| 57-FHV-F | GACGTGGTGAATTATCAGC | 288 |
| 57-FHV-R | CAACTAGATTTCCACCAGGA | |
| 58-FCV-F | CAARGGAGAAAATTCDGACGA | 321 |
| 58-FCV-R | GTATTTWAGCACGTTAGCGCAGGT | |
| 85-FCV-F | cagcaaatgggtcgcggatccTGCTCAACCTGCGCTAAC | 2010 |
| 85-FCV-R | ggtggtggtggtggtgctcgagTAATTTAGTCATTGAACTCC |
| Number | Strain | GenBank Accession No. | Isolation Location | Isolation Time |
|---|---|---|---|---|
| 1 | SMU-B5 | MW194990 | CHINA | 2020 |
| 2 | HRB23 | MW804431 | CHINA | 2020 |
| 3 | SH1 | MT239579 | CHINA | 2019 |
| 4 | BJ-288 | MW088960 | CHINA | 2019 |
| 5 | WZ-1 | KX371573 | CHINA | 2016 |
| 6 | XH | KX371572 | CHINA | 2015 |
| 7 | HRB-SS | KM016908 | CHINA | 2014 |
| 8 | SH/2014 | KT000003 | CHINA | 2014 |
| 9 | GX01-13 | KT970059 | CHINA | 2013 |
| 10 | CH-JL3 | KJ495730 | CHINA | 2013 |
| 11 | KP361 | MZ542330 | Thailand | 2021 |
| 12 | 12Q087-1 | KJ572400 | Korea | 2012 |
| 13 | 12Q087-5 | KJ572401 | Korea | 2012 |
| 14 | FCV-2280 | KC835209 | USA(VSD) | 2013 |
| 15 | FCV-Kaos | DQ910795 | USA(VSD) | 2006 |
| 16 | 3786 | JX519209 | USA | 1996 |
| 17 | 1874 | JX519214 | USA | 1996 |
| 18 | 20879 | JX519211 | USA | 1996 |
| 19 | 255 | KM111171 | USA | 2014 |
| 20 | F9 | M863790 | UK | 1992 |
| 21 | 5789 | JX519210 | USA | 2014 |
| 22 | UTCVM-H2 | AY560117 | USA | 2006 |
| 23 | VS-FCV-Ari | DQ910794 | USA | 2007 |
| 24 | Deduce | DQ910789 | USA | 2016 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.-J.; Su, D.; Zhao, S.-B.; Xing, J.-Y.; Zeng, L.; Wang, J.; Ming, S.-L.; Chu, B.-B. Isolation, Identification, and Genetic Evolution Analysis of VP1 Gene of Feline Calicivirus Strain ZZ202306. Int. J. Mol. Sci. 2025, 26, 2565. https://doi.org/10.3390/ijms26062565
Zhang S-J, Su D, Zhao S-B, Xing J-Y, Zeng L, Wang J, Ming S-L, Chu B-B. Isolation, Identification, and Genetic Evolution Analysis of VP1 Gene of Feline Calicivirus Strain ZZ202306. International Journal of Molecular Sciences. 2025; 26(6):2565. https://doi.org/10.3390/ijms26062565
Chicago/Turabian StyleZhang, Shi-Jun, Dan Su, Shi-Bo Zhao, Jia-You Xing, Lei Zeng, Jiang Wang, Sheng-Li Ming, and Bei-Bei Chu. 2025. "Isolation, Identification, and Genetic Evolution Analysis of VP1 Gene of Feline Calicivirus Strain ZZ202306" International Journal of Molecular Sciences 26, no. 6: 2565. https://doi.org/10.3390/ijms26062565
APA StyleZhang, S.-J., Su, D., Zhao, S.-B., Xing, J.-Y., Zeng, L., Wang, J., Ming, S.-L., & Chu, B.-B. (2025). Isolation, Identification, and Genetic Evolution Analysis of VP1 Gene of Feline Calicivirus Strain ZZ202306. International Journal of Molecular Sciences, 26(6), 2565. https://doi.org/10.3390/ijms26062565






