Elf1 Deficiency Impairs Macrophage Development in Zebrafish Model Organism
Abstract
1. Introduction
2. Results
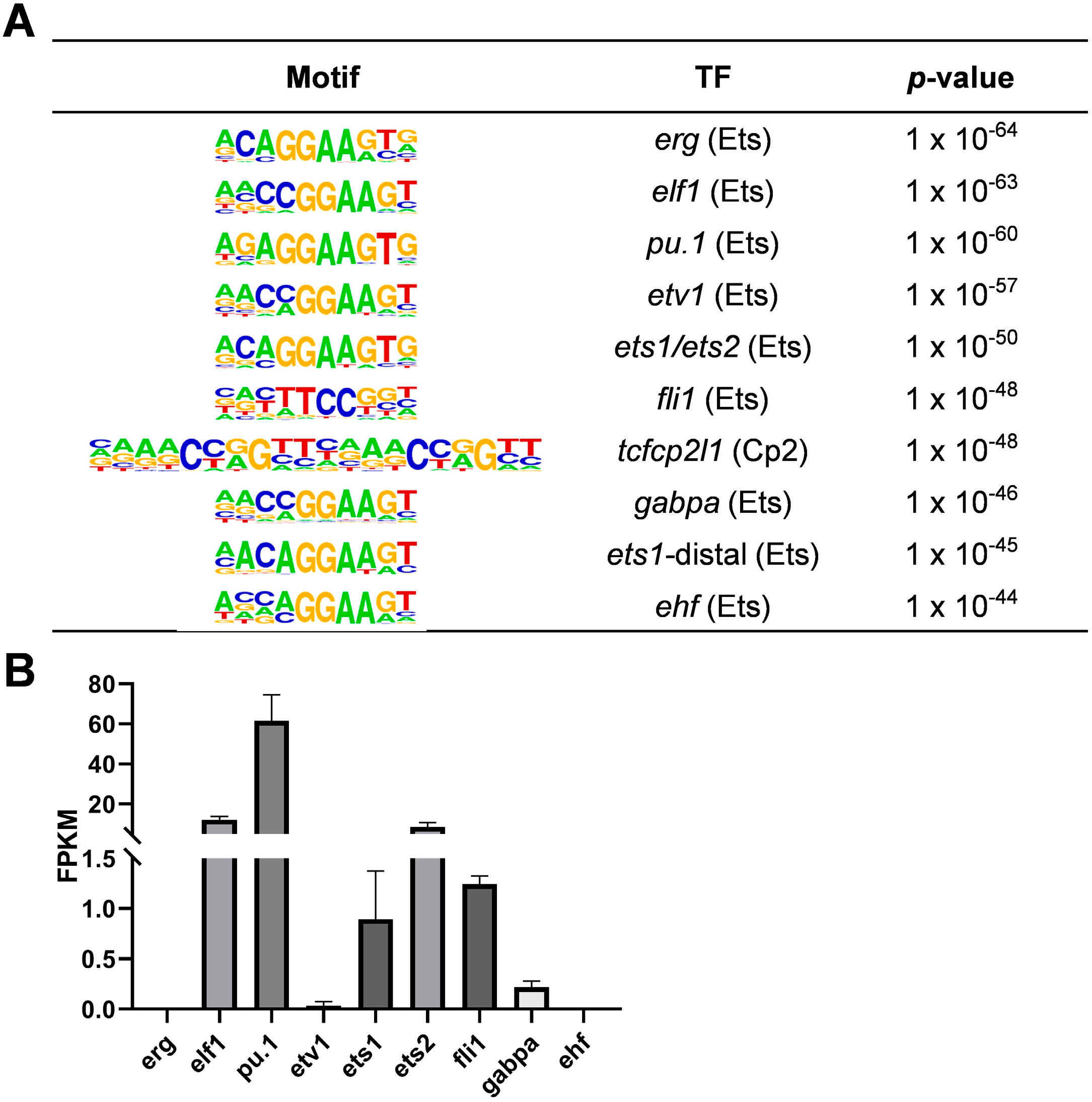
2.1. Enrichment of Ets Family Binding Motifs in Zebrafish Macrophage-Accessible Chromatin
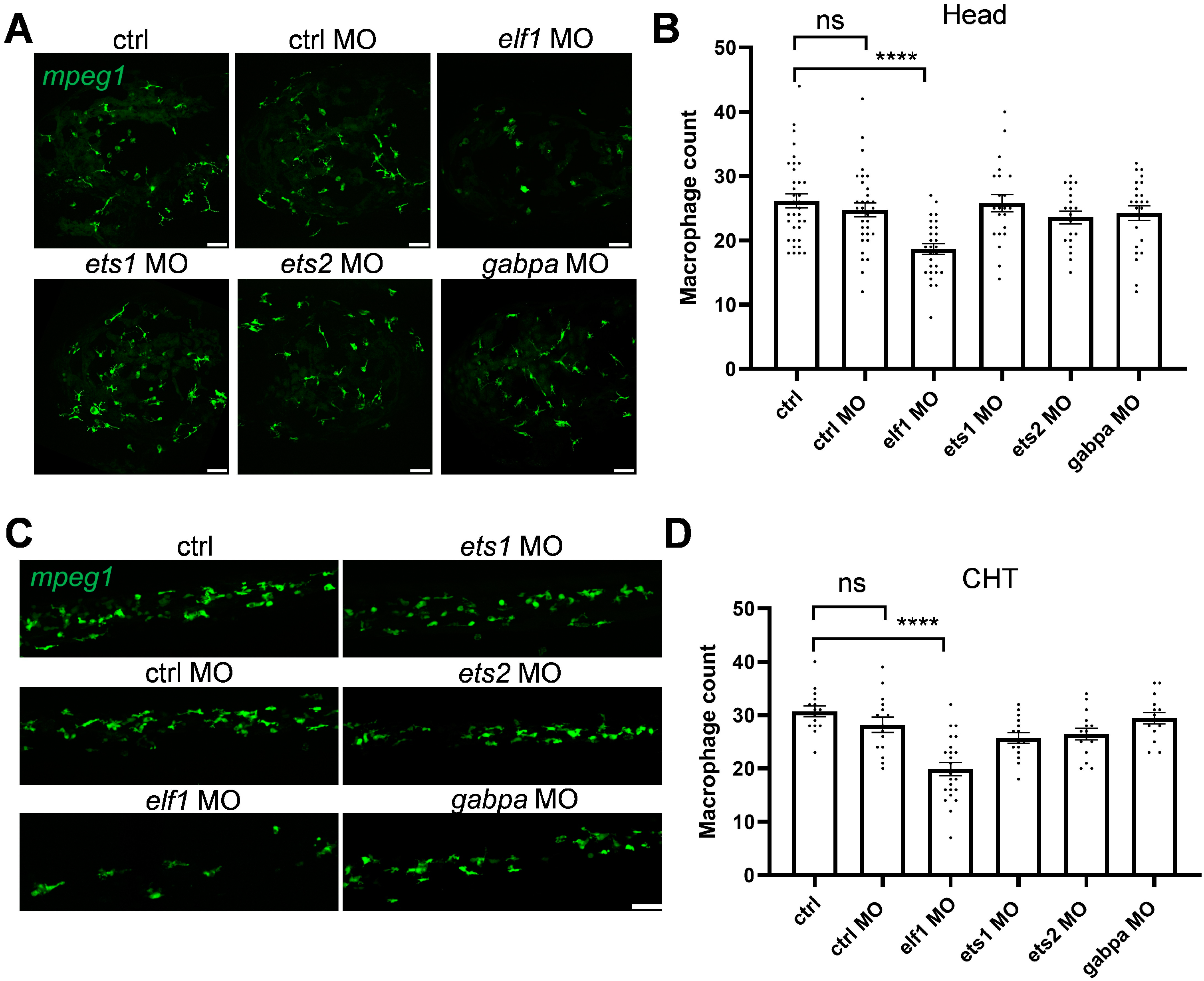
2.2. Selective Regulation of Macrophage Development by Elf1
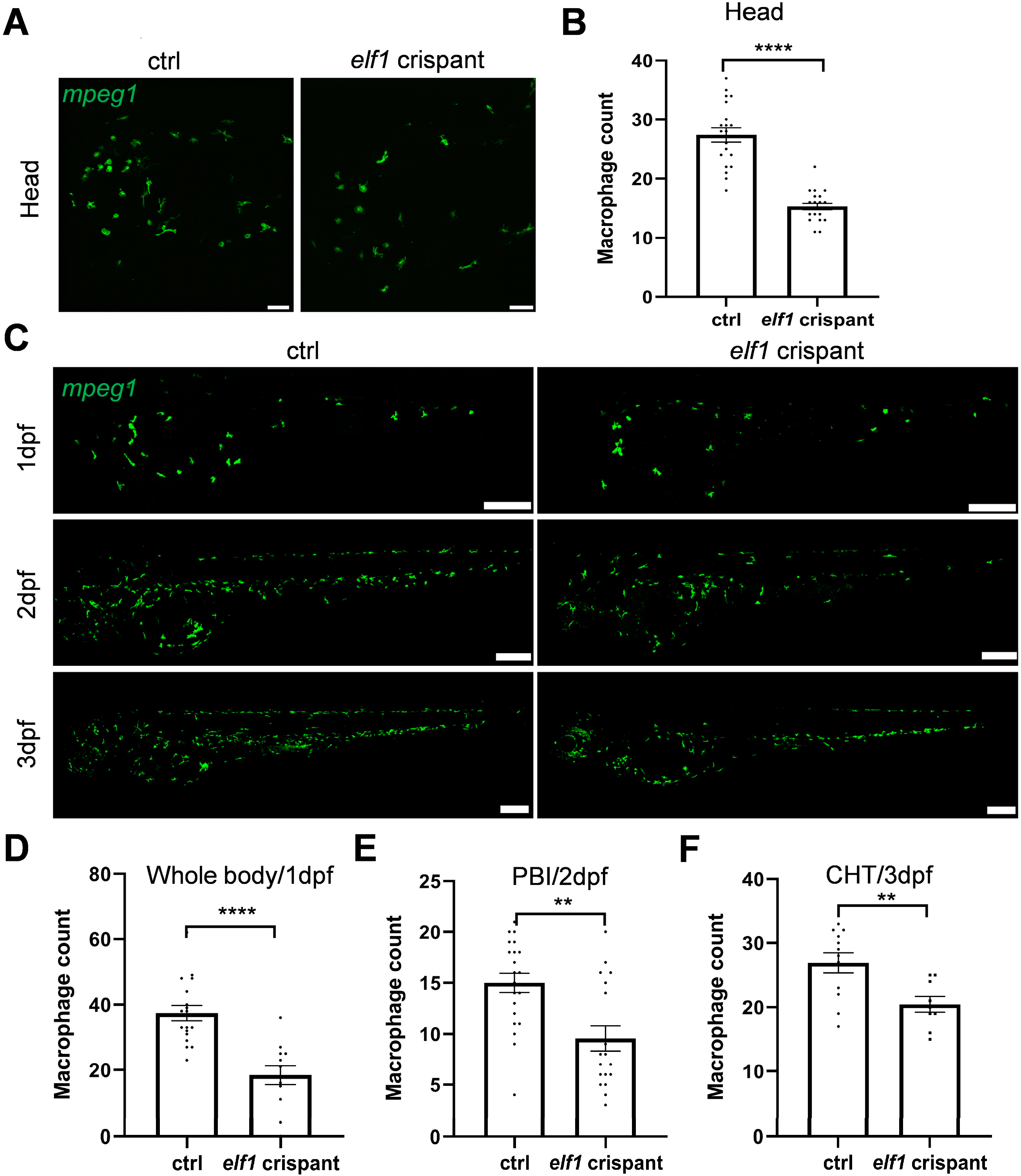
2.3. Macrophage Reduction in Elf1 Crispants
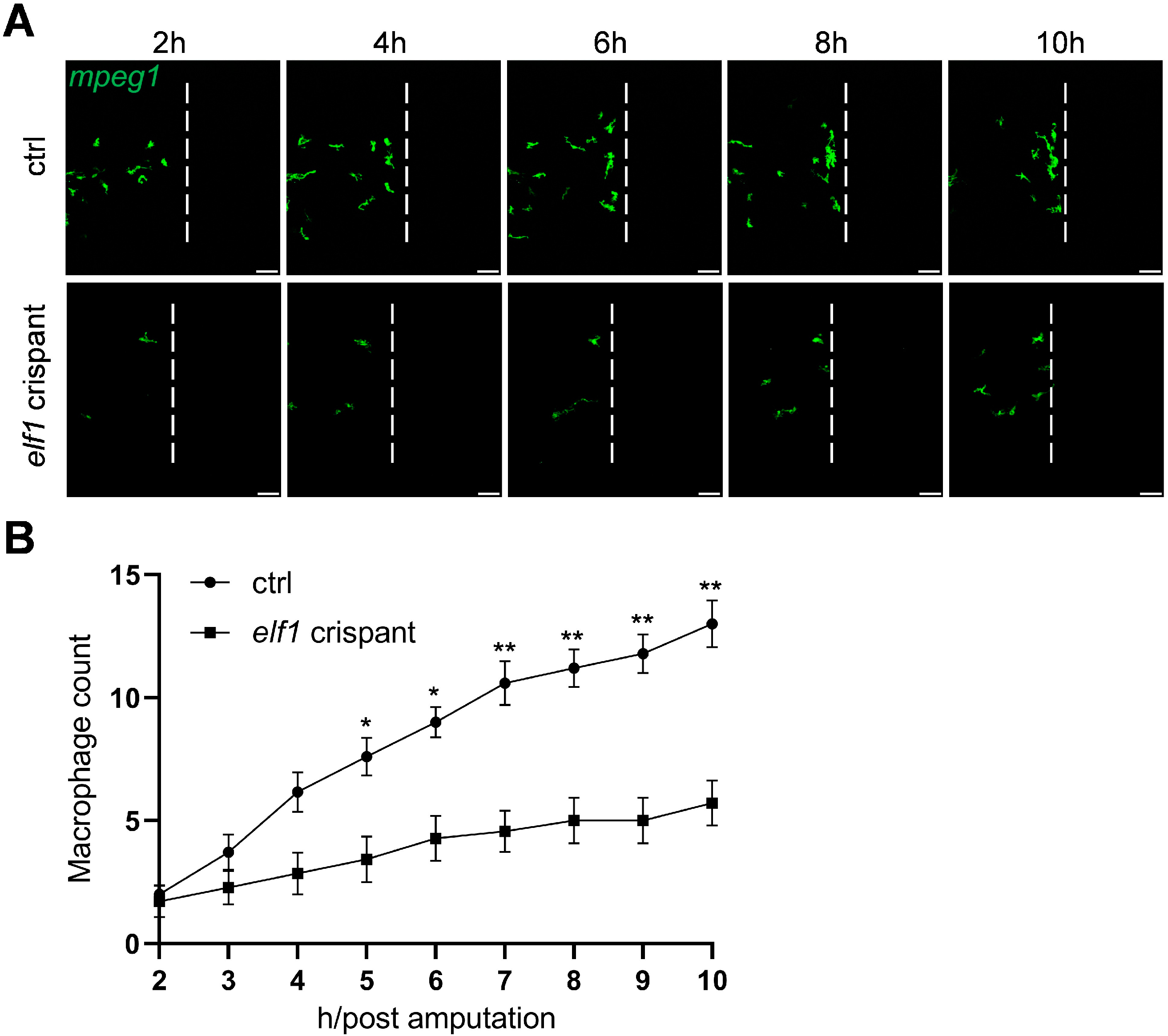
2.4. Impaired Macrophage Response to Injury in Elf1 Crispants
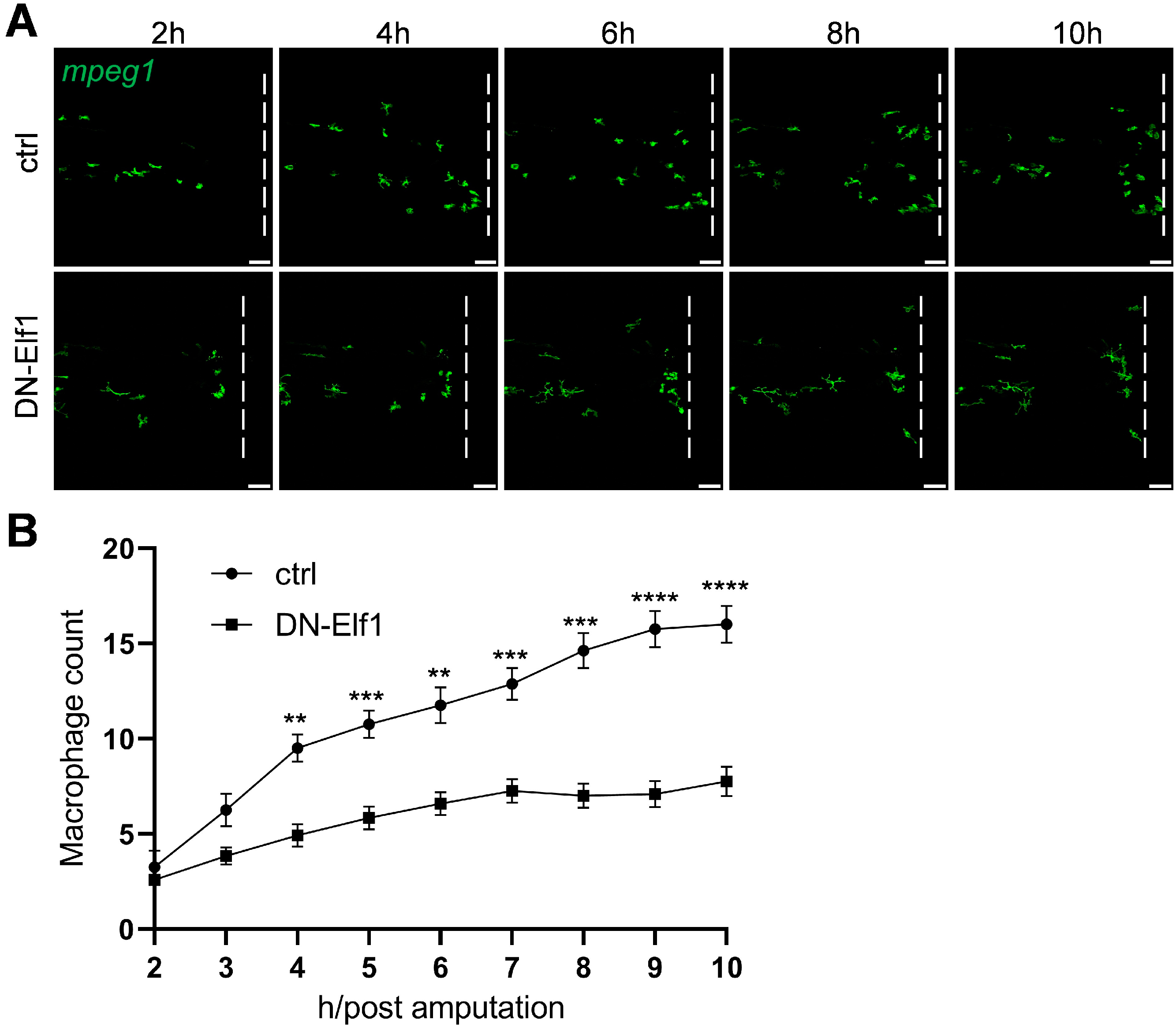
2.5. Dominant-Negative Elf1 Reduces Macrophage Infiltration in a Cell-Autonomous Manner
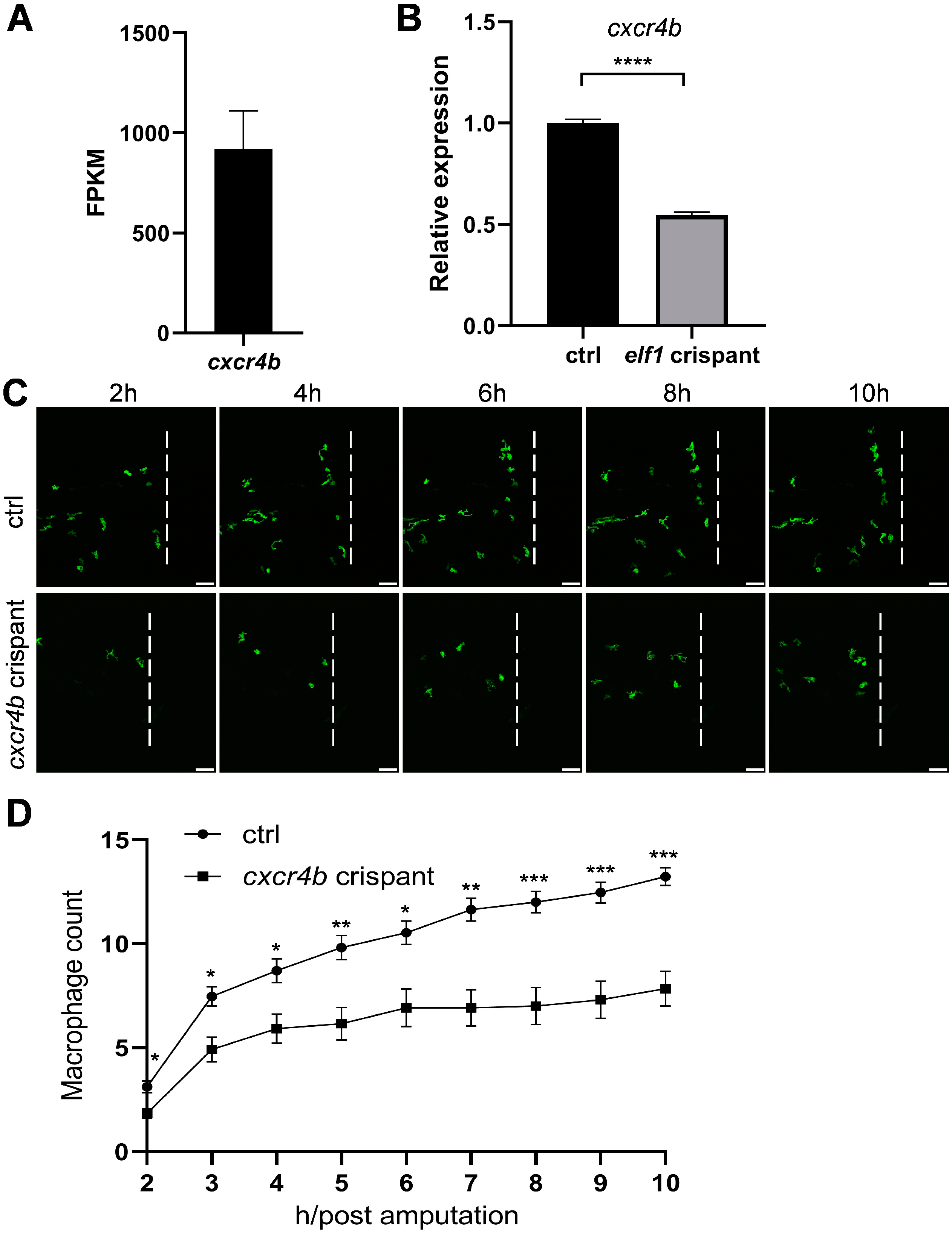
2.6. Cxcr4b Mediates Elf1-Regulated Macrophage Infiltration
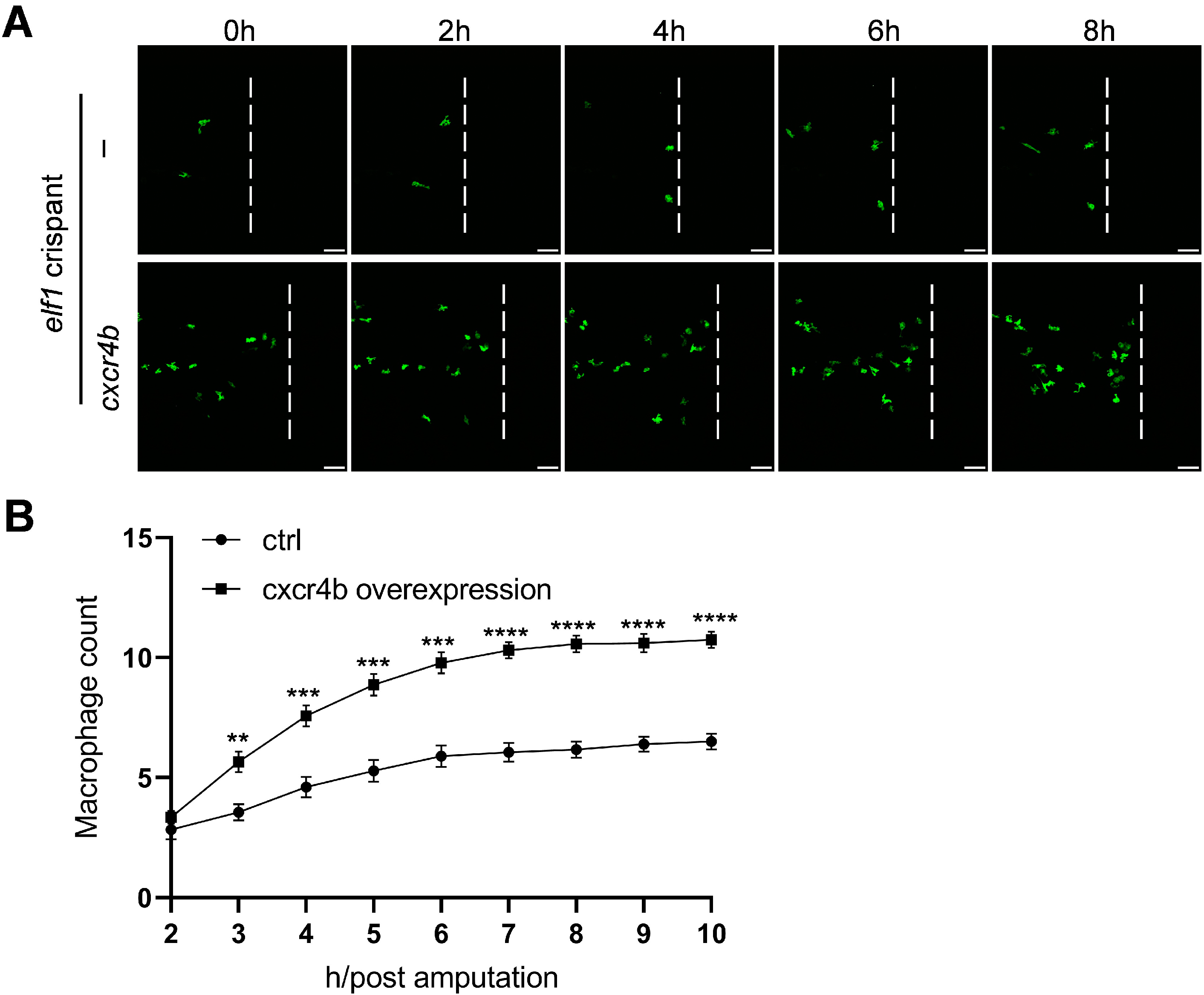
2.7. Rescue of Macrophage Infiltration via Cxcr4b Overexpression
3. Discussion
4. Materials and Methods
4.1. Zebrafish Husbandry and Transgenic Lines
4.2. Sorting of Zebrafish Macrophages
4.3. ATAC and RNA Sequencing
4.4. Morpholino Preparation and Microinjection
4.5. Crispant Generation
4.6. Plasmid Construction and Microinjection
4.7. Total RNA Extraction and qPCR
4.8. Imaging and Count
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Varol, C.; Mildner, A.; Jung, S. Macrophages: Development and tissue specialization. Annu. Rev. Immunol. 2015, 33, 643–675. [Google Scholar] [CrossRef]
- Vannella, K.M.; Wynn, T.A. Mechanisms of Organ Injury and Repair by Macrophages. Annu. Rev. Physiol. 2017, 79, 593–617. [Google Scholar] [CrossRef]
- Zhang, C.; Guo, Z.M. Multiple functions of Maf in the regulation of cellular development and differentiation. Diabetes Metab. Res. Rev. 2015, 31, 773–778. [Google Scholar] [CrossRef]
- Taniguchi, T.; Ogasawara, K.; Takaoka, A.; Tanaka, N. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 2001, 19, 623–655. [Google Scholar] [CrossRef]
- Toobian, D.; Ghosh, P.; Katkar, G.D. Parsing the Role of PPARs in Macrophage Processes. Front. Immunol. 2021, 12, 783780. [Google Scholar] [CrossRef]
- Yamanaka, R.; Lekstrom-Himes, J.; Barlow, C.; Wynshaw-Boris, A.; Xanthopoulos, K.G. CCAAT/enhancer binding proteins are critical components of the transcriptional regulation of hematopoiesis (Review). Int. J. Mol. Med. 1998, 1, 213–221. [Google Scholar] [CrossRef]
- Sharrocks, A.D. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2001, 2, 827–837. [Google Scholar] [CrossRef]
- Kurotaki, D.; Yamamoto, M.; Nishiyama, A.; Uno, K.; Ban, T.; Ichino, M.; Sasaki, H.; Matsunaga, S.; Yoshinari, M.; Ryo, A.; et al. IRF8 inhibits C/EBPα activity to restrain mononuclear phagocyte progenitors from differentiating into neutrophils. Nat. Commun. 2014, 5, 4978. [Google Scholar] [CrossRef]
- Hamada, M.; Tsunakawa, Y.; Jeon, H.; Yadav, M.K.; Takahashi, S. Role of MafB in macrophages. Exp. Anim. 2020, 69, 1–10. [Google Scholar] [CrossRef]
- Gažová, I.; Lefevre, L.; Bush, S.J.; Clohisey, S.; Arner, E.; De Hoon, M.; Severin, J.; van Duin, L.; Andersson, R.; Lengeling, A.; et al. The Transcriptional Network That Controls Growth Arrest and Macrophage Differentiation in the Human Myeloid Leukemia Cell Line THP-1. Front. Cell Dev. Biol. 2020, 8, 498. [Google Scholar] [CrossRef]
- Findlay, V.J.; LaRue, A.C.; Turner, D.P.; Watson, P.M.; Watson, D.K. Understanding the role of ETS-mediated gene regulation in complex biological processes. Adv. Cancer Res. 2013, 119, 1–61. [Google Scholar]
- Yang, Y.; Han, X.; Sun, L.; Shao, F.; Yin, Y.; Zhang, W. ETS Transcription Factors in Immune Cells and Immune-Related Diseases. Int. J. Mol. Sci. 2024, 25, 10004. [Google Scholar] [CrossRef]
- Laudet, V.; Hänni, C.; Stéhelin, D.; Duterque-Coquillaud, M. Molecular phylogeny of the ETS gene family. Oncogene 1999, 18, 1351–1359. [Google Scholar] [CrossRef]
- Scott, E.W.; Simon, M.C.; Anastasi, J.; Singh, H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 1994, 265, 1573–1577. [Google Scholar] [CrossRef]
- Song, G.; Zhang, Z.; Chen, Y.; Hou, W.; Zhong, W.; Zhou, Y.; Zhang, A.; Xu, Y. PU.1 induces tumor-associated macrophages promoting glioma progression through BTK-mediated Akt/mTOR pathway activation. Am. J. Cancer Res. 2024, 14, 1139–1156. [Google Scholar] [CrossRef]
- Guo, X.; Li, Y.; Wan, B.; Lv, Y.; Wang, X.; Liu, G.; Wang, P. ETV1 inhibition depressed M2 polarization of tumor-associated macrophage and cell process in gastrointestinal stromal tumor via down-regulating PDE3A. J. Clin. Biochem. Nutr. 2023, 72, 139–146. [Google Scholar] [CrossRef]
- Thompson, C.C.; Brown, T.A.; McKnight, S.L. Convergence of Ets- and notch-related structural motifs in a heteromeric DNA binding complex. Science 1991, 253, 762–768. [Google Scholar] [CrossRef]
- Wang, C.Y.; Bassuk, A.G.; Boise, L.H.; Thompson, C.B.; Bravo, R.; Leiden, J.M. Activation of the granulocyte-macrophage colony-stimulating factor promoter in T cells requires cooperative binding of Elf-1 and AP-1 transcription factors. Mol. Cell Biol. 1994, 14, 1153–1159. [Google Scholar]
- Wurster, A.L.; Siu, G.; Leiden, J.M.; Hedrick, S.M. Elf-1 binds to a critical element in a second CD4 enhancer. Mol. Cell Biol. 1994, v14, 6452–6463. [Google Scholar]
- Heydemann, A.; Juang, G.; Hennessy, K.; Parmacek, M.S.; Simon, M.C. The myeloid-cell-specific c-fes promoter is regulated by Sp1, PU.1, and a novel transcription factor. Mol. Cell Biol. 1996, 16, 1676–1686. [Google Scholar] [CrossRef] [PubMed]
- Bassuk, A.G.; Barton, K.P.; Anandappa, R.T.; Lu, M.M.; Leiden, J.M. Expression pattern of the Ets-related transcription factor Elf-1. Mol. Med. 1998, 4, 392–401.s. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Geng, Y.; Cho, H.; Li, S.; Giri, P.K.; Felio, K.; Wang, C.-R. Differential requirements for the Ets transcription factor Elf-1 in the development of NKT cells and NK cells. Blood 2011, 117, 1880–1887. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Brown, C.; Ni, W.; Maynard, E.; Rigby, A.C.; Oettgen, P. Critical role for the Ets transcription factor ELF-1 in the development of tumor angiogenesis. Blood 2006, 107, 3153–3160. [Google Scholar] [CrossRef][Green Version]
- Xu, J.; Du, L.; Wen, Z. Myelopoiesis during zebrafish early development. J. Genet. Genom. 2012, 39, 435–442. [Google Scholar] [CrossRef]
- Liu, F.; Patient, R. Genome-wide analysis of the zebrafish ETS family identifies three genes required for hemangioblast differentiation or angiogenesis. Circ. Res. 2008, 103, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Li, L.; Xu, J.; Zhen, F.; Zhu, L.; Liu, P.P.; Zhang, M.; Zhang, W.; Wen, Z. Runx1 regulates embryonic myeloid fate choice in zebrafish through a negative feedback loop inhibiting Pu.1 expression. Blood 2012, 119, 5239–5249. [Google Scholar] [CrossRef]
- Liu, F.; Walmsley, M.; Rodaway, A.; Patient, R. Fli1 acts at the top of the transcriptional network driving blood and endothelial development. Curr. Biol. 2008, 18, 1234–1240. [Google Scholar] [CrossRef]
- Zhang, A.; Lu, J.; Feng, S.; Yu, H.; Yu, T.; Zhao, S.; Chen, K.; Huang, Z.; Xu, J.; Qu, J.Y.; et al. Fli1 acts in parallel with Pu.1 to control macrophage and neutrophil fate in zebrafish. J. Genet. Genom. 2024, 51, 359–362. [Google Scholar] [CrossRef]
- Li, L.; Yan, B.; Shi, Y.Q.; Zhang, W.Q.; Wen, Z.L. Live imaging reveals differing roles of macrophages and neutrophils during zebrafish tail fin regeneration. J. Biol. Chem. 2012, 287, 25353–25360. [Google Scholar] [CrossRef]
- Ellett, F.; Pase, L.; Hayman, J.W.; Andrianopoulos, A.; Lieschke, G.J. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 2011, 117, e49–e56. [Google Scholar] [CrossRef]
- Werner, Y.; Mass, E.; Ashok Kumar, P.; Ulas, T.; Händler, K.; Horne, A.; Klee, K.; Lupp, A.; Schütz, D.; Saaber, F.; et al. Cxcr4 distinguishes HSC-derived monocytes from microglia and reveals monocyte immune responses to experimental stroke. Nat. Neurosci. 2020, 23, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Moulton, J.D. Making a Morpholino Experiment Work: Controls, Favoring Specificity, Improving Efficacy, Storage, and Dose. Methods Mol. Biol. 2017, 1565, 17–29. [Google Scholar]
- Bolukbasi, M.F.; Gupta, A.; Wolfe, S.A. Creating and evaluating accurate CRISPR-Cas9 scalpels for genomic surgery. Nat. Methods 2016, 13, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J.; Hagen, A.; Hsu, K.; Deng, M.; Liu, T.X.; Look, A.; Kanki, J.P. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev. Cell 2005, 8, 97–108. [Google Scholar] [CrossRef] [PubMed]
- DeKoter, R.P.; Singh, H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 2000, 288, 1439–1441. [Google Scholar] [CrossRef]
- Calero-Nieto, F.J.; Wood, A.D.; Wilson, N.K.; Kinston, S.; Landry, J.R.; Göttgens, B. Transcriptional regulation of Elf-1: Locus-wide analysis reveals four distinct promoters, a tissue-specific enhancer, control by PU.1 and the importance of Elf-1 downregulation for erythroid maturation. Nucleic Acids Res. 2010, 38, 6363–6374. [Google Scholar] [CrossRef]
- Ross, I.L.; Yue, X.; Ostrowski, M.C.; Hume, D.A. Interaction between PU.1 and another Ets family transcription factor promotes macrophage-specific Basal transcription initiation. J. Biol. Chem. 1998, 273, 6662–6669. [Google Scholar] [CrossRef]
- Ushach, I.; Zlotnik, A. Biological role of granulocyte macrophage colony-stimulating factor (GM-CSF) and macrophage colony-stimulating factor (M-CSF) on cells of the myeloid lineage. J. Leukoc. Biol. 2016, 100, 481–489. [Google Scholar] [CrossRef]
- Hackenmiller, R.; Kim, J.; Feldman, R.A.; Simon, M.C. Abnormal Stat activation, hematopoietic homeostasis, and innate immunity in c-fes-/- mice. Immunity 2000, 13, 397–407. [Google Scholar] [CrossRef]
- Hume, D.A.; Caruso, M.; Ferrari-Cestari, M.; Summers, K.M.; Pridans, C.; Irvine, K.M. Phenotypic impacts of CSF1R deficiencies in humans and model organisms. J. Leukoc. Biol. 2020, 107, 205–219. [Google Scholar] [CrossRef]
- Ellett, F.; Lieschke, G.J. Zebrafish as a model for vertebrate hematopoiesis. Curr. Opin. Pharmacol. 2010, 10, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Zon, L.I. Zebrafish as a model for normal and malignant hematopoiesis. Dis. Model. Mech. 2011, 4, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Herbomel, P.; Thisse, B.; Thisse, C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev. Biol. 2001, 238, 274–288. [Google Scholar] [CrossRef]
- Bertrand, J.Y.; Kim, A.D.; Violette, E.P.; Stachura, D.L.; Cisson, J.L.; Traver, D. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development 2007, 134, 4147–4156. [Google Scholar] [CrossRef]
- Murayama, E.; Kissa, K.; Zapata, A.; Mordelet, E.; Briolat, V.; Lin, H.-F.; Handin, R.I.; Herbomel, P. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity 2006, 25, 963–975. [Google Scholar] [CrossRef]
- Larsen, S.; Kawamoto, S.; Tanuma, S.; Uchiumi, F. The hematopoietic regulator, ELF-1, enhances the transcriptional response to Interferon-β of the OAS1 anti-viral gene. Sci. Rep. 2015, 5, 17497. [Google Scholar] [CrossRef]
- Chia, K.; Mazzolini, J.; Mione, M.; Sieger, D. Tumor initiating cells induce Cxcr4-mediated infiltration of pro-tumoral macrophages into the brain. eLife 2018, 7, e31918. [Google Scholar] [CrossRef] [PubMed]
- Torraca, V.; Tulotta, C.; Snaar-Jagalska, B.E.; Meijer, A.H. The chemokine receptor CXCR4 promotes granuloma formation by sustaining a mycobacteria-induced angiogenesis programme. Sci. Rep. 2017, 7, 45061. [Google Scholar] [CrossRef]
- Nguyen-Chi, M.; Laplace-Builhe, B.; Travnickova, J.; Luz-Crawford, P.; Tejedor, G.; Phan, Q.T.; Duroux-Richard, I.; Levraud, J.-P.; Kissa, K.; Lutfalla, G.; et al. Identification of polarized macrophage subsets in zebrafish. eLife 2015, 4, e07288. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Jin, H.; Huang, Z.; Chi, Y.; Wu, M.; Zhou, R.; Zhao, L.; Xu, J.; Zhen, F.; Lan, Y.; Li, L.; et al. c-Myb acts in parallel and cooperatively with Cebp1 to regulate neutrophil maturation in zebrafish. Blood 2016, 128, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, K.; Kawakami, K. The Tol2-mediated Gal4-UAS method for gene and enhancer trapping in zebrafish. Methods 2009, 49, 275–281. [Google Scholar] [CrossRef]
- Davison, J.M.; Akitake, C.M.; Goll, M.G.; Rhee, J.M.; Gosse, N.; Baier, H.; Halpern, M.E.; Leach, S.D.; Parsons, M.J. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev. Biol. 2007, 304, 811–824. [Google Scholar] [CrossRef]
- Pham, V.N.; Lawson, N.D.; Mugford, J.W.; Dye, L.; Castranova, D.; Lo, B.; Weinstein, B.M. Combinatorial function of ETS transcription factors in the developing vasculature. Dev. Biol. 2007, 303, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Pham, V.N.; Stratman, A.N.; Castranova, D.; Kamei, M.; Kidd, K.R.; Lo, B.D.; Shaw, K.M.; Torres-Vazquez, J.; Mikelis, C.M.; et al. CDP-diacylglycerol synthetase-controlled phosphoinositide availability limits VEGFA signaling and vascular morphogenesis. Blood 2012, 120, 489–498. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Labuhn, M.; Adams, F.F.; Ng, M.; Knoess, S.; Schambach, A.; Charpentier, E.M.; Schwarzer, A.; Mateo, J.L.; Klusmann, J.H.; Heckl, D. Refined sgRNA efficacy prediction improves large- and small-scale CRISPR-Cas9 applications. Nucleic Acids Res. 2018, 46, 1375–1385. [Google Scholar] [CrossRef] [PubMed]
- Fei, J.; Zhao, D.; Pang, C.; Li, J.; Li, S.; Qiao, W.; Tan, J.; Bi, C.; Zhang, X. Mismatch prime editing gRNA increased efficiency and reduced indels. Nat. Commun. 2025, 16, 139. [Google Scholar] [CrossRef]
- Wu, R.S.; Lam, I.I.; Clay, H.; Duong, D.N.; Deo, R.C.; Coughlin, S.R. A Rapid Method for Directed Gene Knockout for Screening in G0 Zebrafish. Dev. Cell 2018, 46, 112–125.e4. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, Q.; Wang, J.; Hao, Y.; Yang, S.; Cao, B.; Pan, W.; Cao, M. Elf1 Deficiency Impairs Macrophage Development in Zebrafish Model Organism. Int. J. Mol. Sci. 2025, 26, 2537. https://doi.org/10.3390/ijms26062537
Tan Q, Wang J, Hao Y, Yang S, Cao B, Pan W, Cao M. Elf1 Deficiency Impairs Macrophage Development in Zebrafish Model Organism. International Journal of Molecular Sciences. 2025; 26(6):2537. https://doi.org/10.3390/ijms26062537
Chicago/Turabian StyleTan, Qianli, Jing Wang, Yimei Hao, Shizeng Yang, Biao Cao, Weijun Pan, and Mengye Cao. 2025. "Elf1 Deficiency Impairs Macrophage Development in Zebrafish Model Organism" International Journal of Molecular Sciences 26, no. 6: 2537. https://doi.org/10.3390/ijms26062537
APA StyleTan, Q., Wang, J., Hao, Y., Yang, S., Cao, B., Pan, W., & Cao, M. (2025). Elf1 Deficiency Impairs Macrophage Development in Zebrafish Model Organism. International Journal of Molecular Sciences, 26(6), 2537. https://doi.org/10.3390/ijms26062537





