Reactions of Plasmodium falciparum Type II NADH: Ubiquinone Oxidoreductase with Nonphysiological Quinoidal and Nitroaromatic Oxidants
Abstract
1. Introduction
2. Results
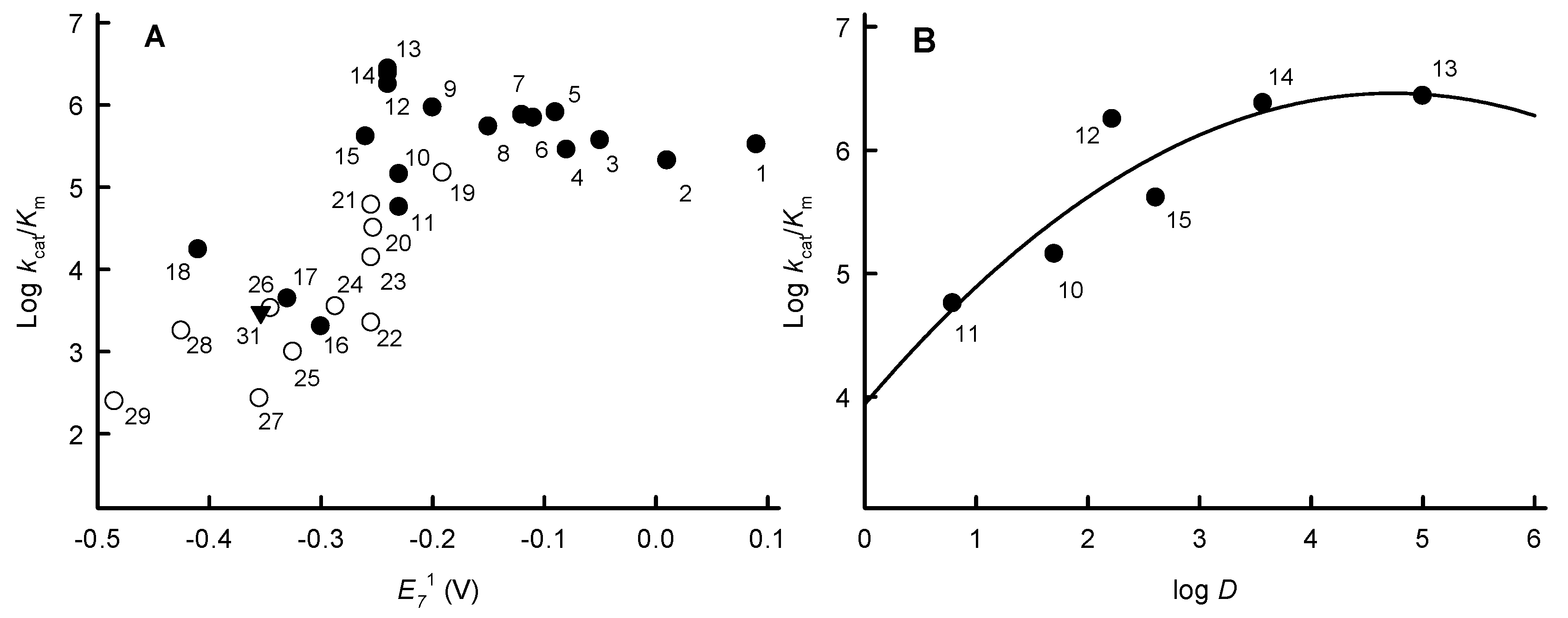
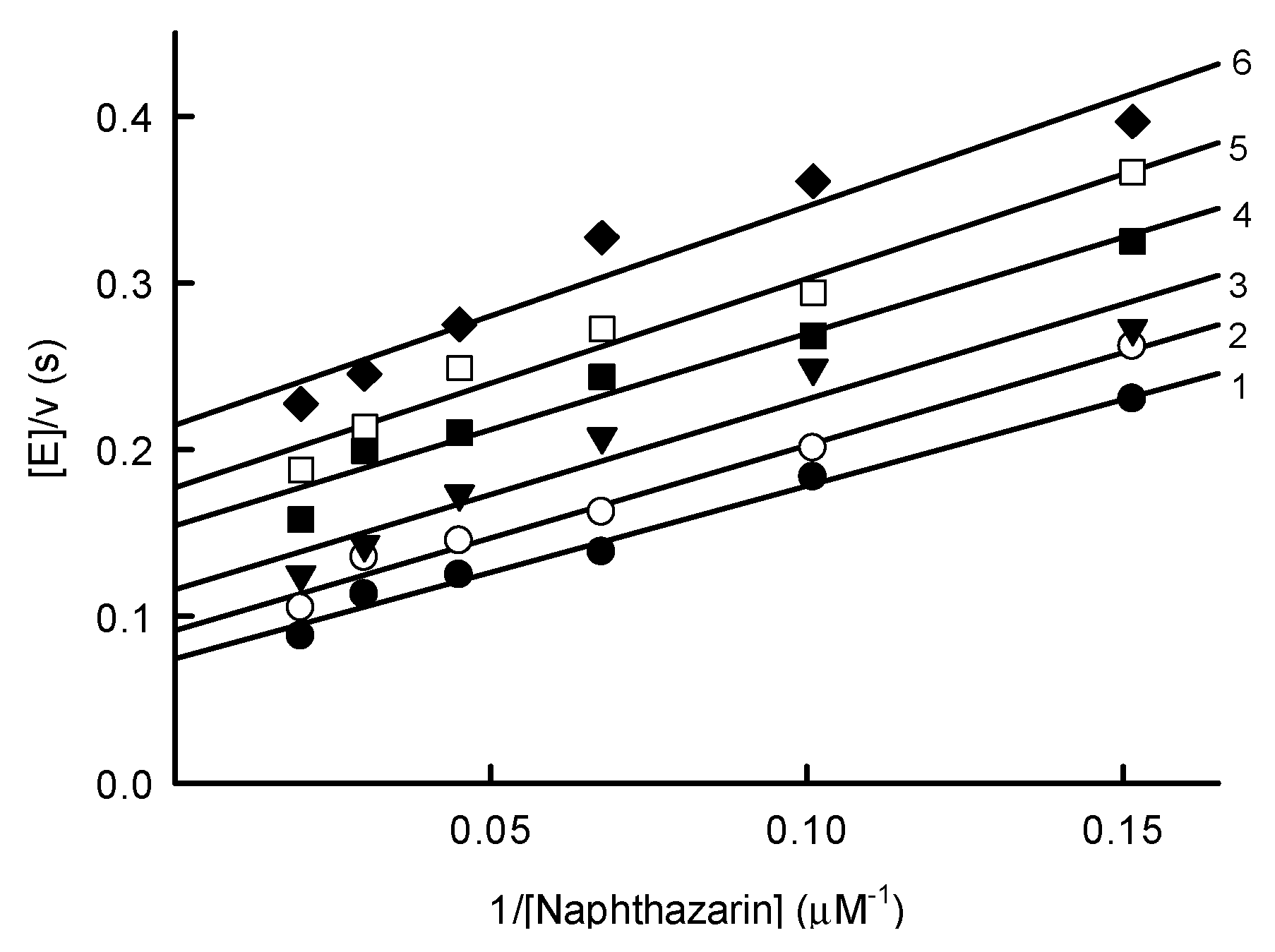
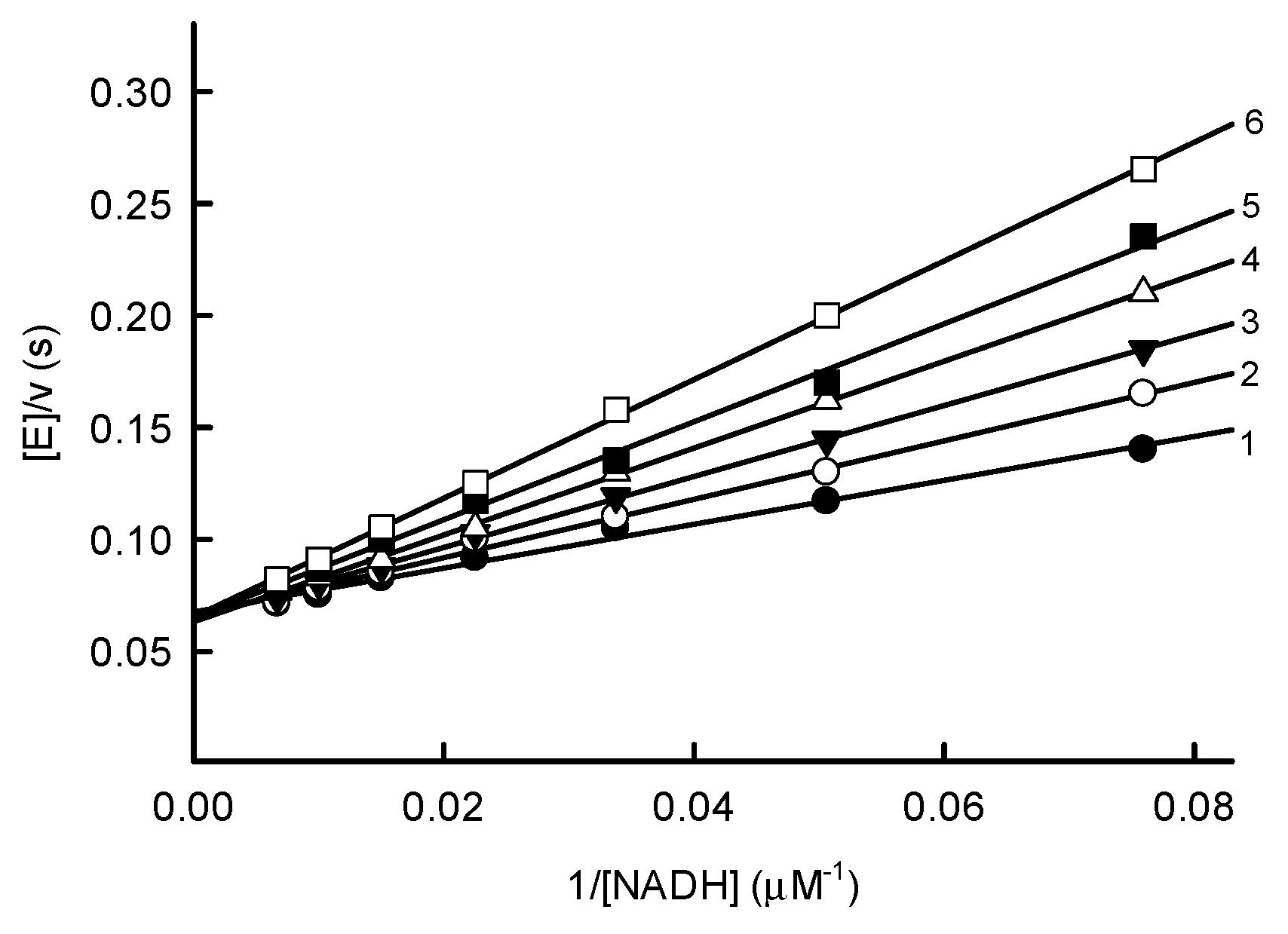
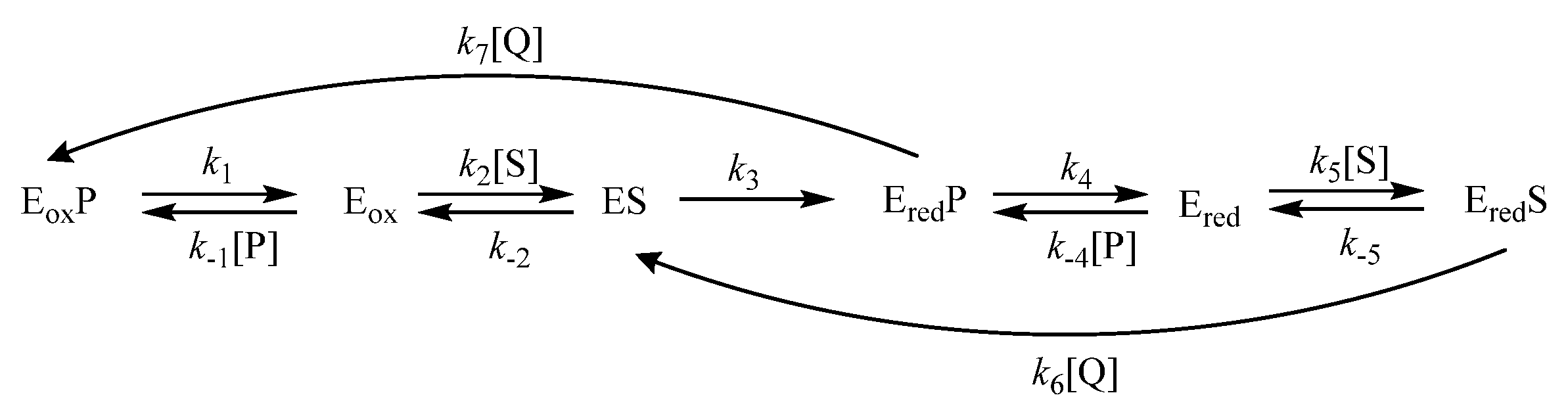
2.1. Steady-State Reactions of PfNDH2
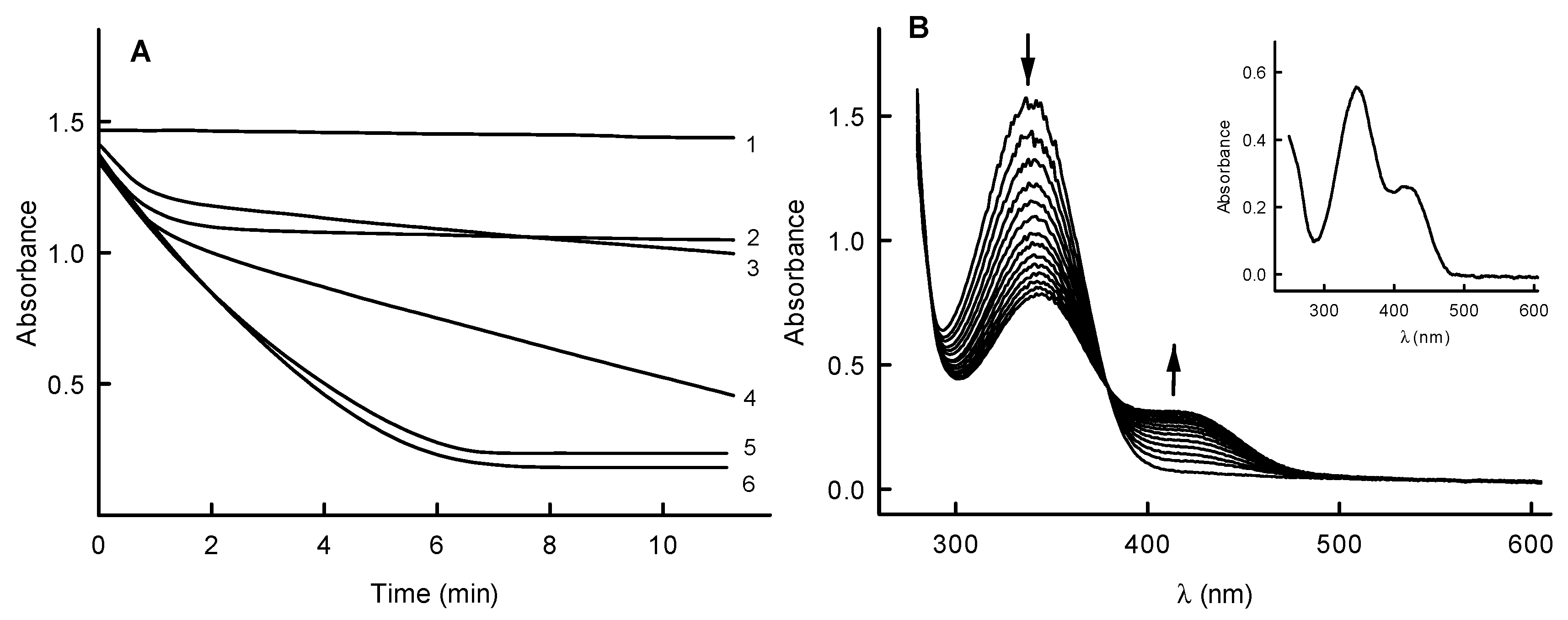
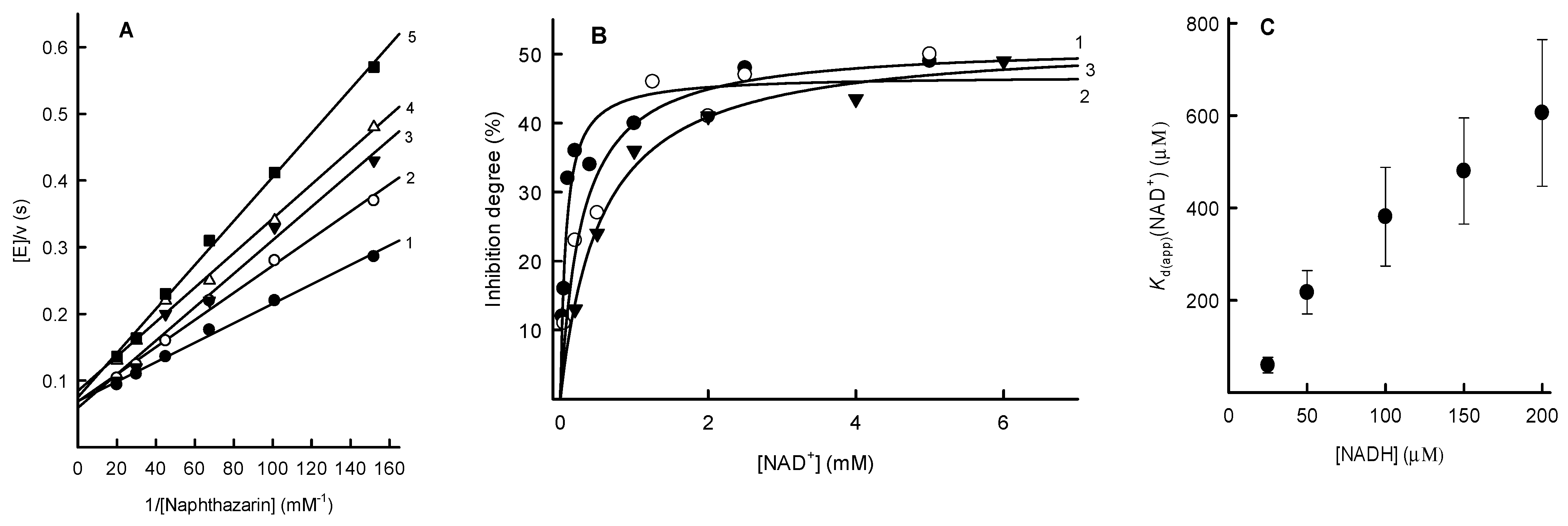
2.2. Inhibition Studies of PfNDH2
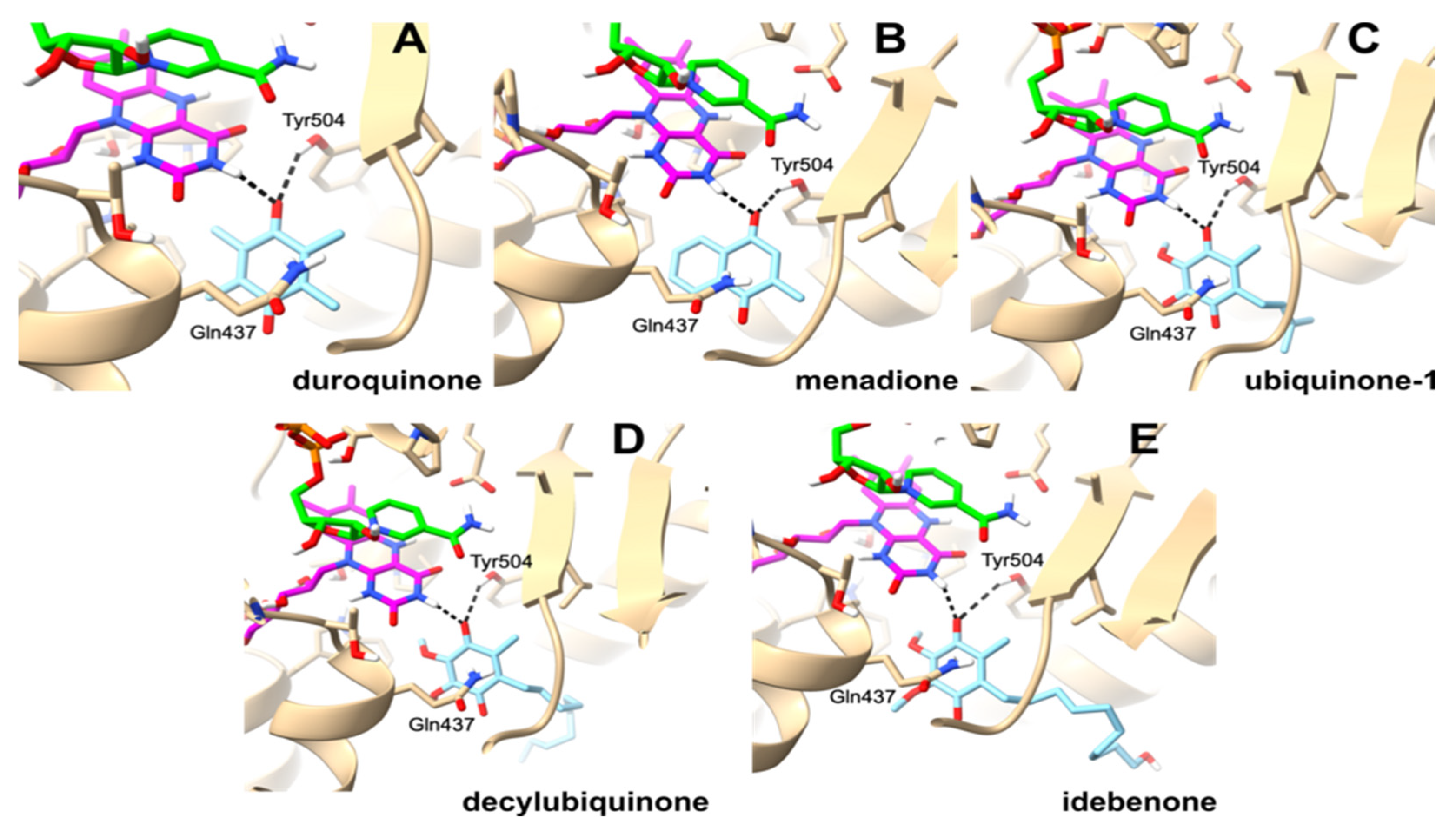
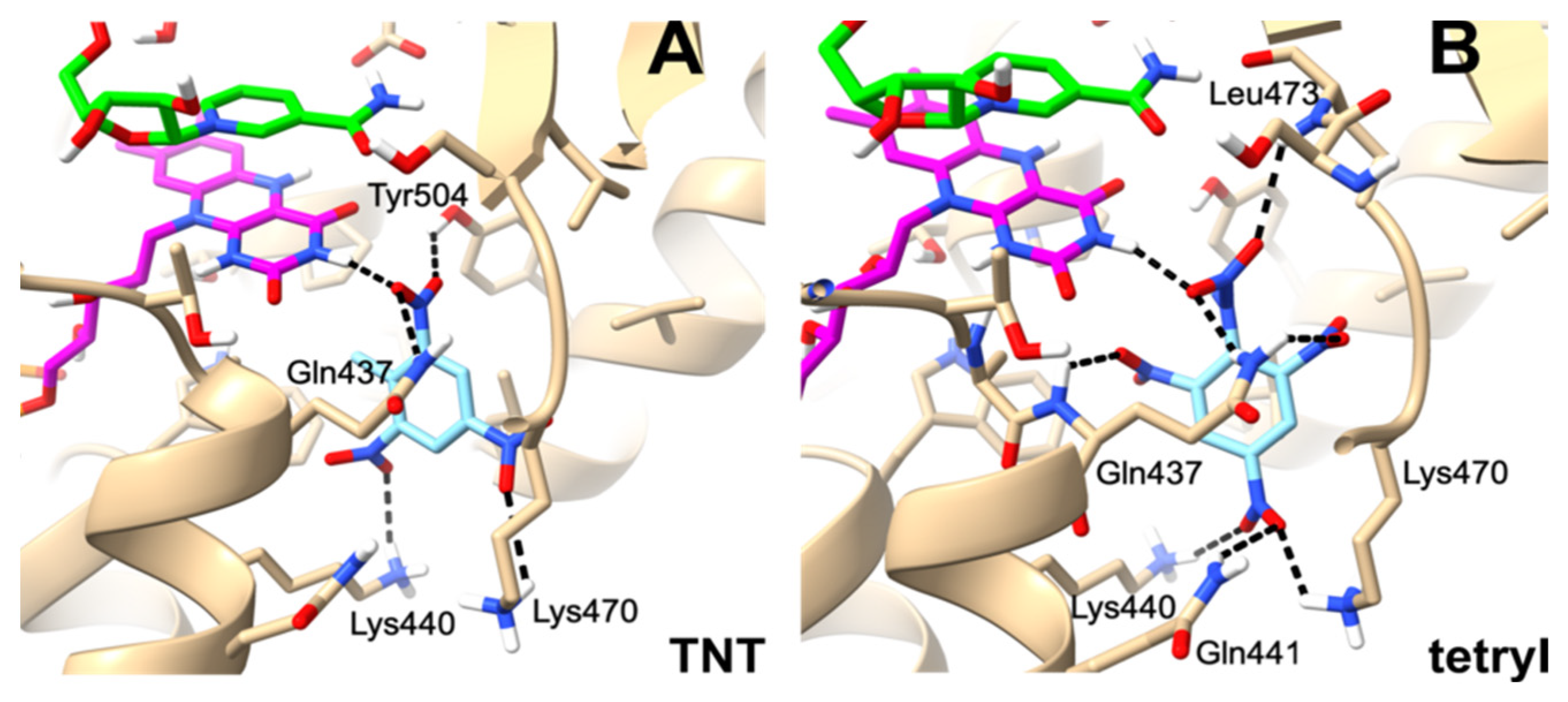
2.3. Molecular Modeling of Oxidant Binding in the Active Center of PfNDH2
3. Discussion
4. Materials and Methods
4.1. Expression and Purification of PfNDH2
4.2. Reagents and Enzymes
4.3. Steady-State Kinetics Studies
4.4. Molecular Docking Studies
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhatt, S.; Weiss, D.J.; Cameron, E.; Bisancia, D.; Mappin, B.; Dalrymple, U.; Battle, K.; Moyes, C.L.; Henry, A.; Eckhoff, P.A.; et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 2015, 526, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Haldar, K.; Bhattacharjee, S.; Safeukui, I. Drug resistance in Plasmodium. Nat. Rev. Microbiol. 2018, 16, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Pal, C. Redox modulating small molecules having antimalarial activity. Biochem. Pharmacol. 2023, 218, 115927. [Google Scholar] [CrossRef]
- Lin, T.S.; Zhu, L.Y.; Xu, S.P.; Divo, A.A.; Sartorelli, A.C. Synthesis and antimalarial activity of 2-aziridinyl- and 2,3-bis(aziridinyl)-1,4-naphthoquinone sulfonate and acylate derivatives. J. Med. Chem. 1991, 34, 1634–1639. [Google Scholar] [CrossRef]
- Grellier, P.; Marozienė, A.; Nivinskas, H.; Šarlauskas, J.; Aliverti, A.; Čėnas, N. Antiplasmodial activity of quinones: Roles of aziridinyl substituents and inhibition of Plasmodium falciparum glutathione reductase. Arch. Biochem. Biophys. 2010, 494, 32–39. [Google Scholar] [CrossRef]
- Marozienė, A.; Lesanavičius, M.; Davioud-Charvet, E.; Aliverti, A.; Grellier, P.; Šarlauskas, J.; Čėnas, N. Antiplasmodial activity of nitroaromatic compounds: Correlation with their reduction potential and inhibitory activity on Plasmodium falciparum glutathione reductase. Molecules 2019, 24, 4509. [Google Scholar] [CrossRef]
- Navidpour, L.; Chibale, K.; Esmaeili, S.; Ghiaee, A.; Hadj-Esfandiari, N.; Irami, M.; Ahmadi-Koulaei, S.; Yassa, N. Antimicrobial activities of (Z)-2-(nitroheteroarylmethylene)-3-(2H)-benzofuranone derivatives: In vitro and in vivo assessment and β-hematin formation ihibition activity. Antimicrob. Agents Chemother. 2021, 65, e0268320. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Müller, J.; Kunz, S.; Siderius, M.; Maes, L.; Caljon, G.; Müller, N.; Hemphill, A.; Sterk, G.J.; Leurs, R. 3-Nitroimidazo{1,2-b]pyridazine as a novel scaffold for antiparasitics with sub-nanomolar anti-Giardia liamblia activity. Int. J. Parasitol. Drugs Drug Resist. 2022, 19, 47–55. [Google Scholar] [CrossRef]
- Shafi, S.; Gupta, S.; Jain, R.; Shoaib, R.; Munjal, A.; Maurya, P.; Kumar, P.; Najmi, K.; Singh, S. Tackling the emerging artemsiin-resistant malaria parasite by modulation of defensive oxido-reductase mechanism via nitrofurantoin repurposing. Biochem. Pharmacol. 2023, 215, 115756. [Google Scholar] [CrossRef]
- Lopez-Mercado, S.; Enriquez, C.; Valderrama, J.A.; Pino-Rios, R.; Ruis-Vasquea, L.; Ruiz Mesia, L.; Vargas-Arana, G.; Buc Calderon, P.; Benitez, J. Exploring the antibacterial and antiparasitic activity of phenylaminonaphthoquinones–green synthesis, biologivcal evaluations and computational studies. Int. J. Mol. Sci. 2024, 25, 10670. [Google Scholar] [CrossRef]
- Painter, H.J.; Morrissey, J.M.; Mather, M.W.; Vaidya, A.B. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature 2007, 446, 88–91. [Google Scholar] [CrossRef]
- Biagini, G.A.; Fisher, N.; Shone, A.E.; Mubaraki, M.A.; Srivastava, A.; Hill, A.; Antoine, T.; Warman, A.J.; Davies, J.; Pidathala, C.; et al. Generation of quinolone antimalarials targeting the Plasmodium falciparum mitochondrial respiratory chain for the treatment and prophylaxis of malaria. Proc. Natl. Acad. Sci. USA 2012, 109, 8298–8303. [Google Scholar] [CrossRef] [PubMed]
- Pidathala, C.; Amewu, R.; Pacorel, B.; Nixon, G.L.; Gibbons, P.; Hong, W.D.; Leung, S.C.; Berry, N.G.; Sharma, R.; Stocks, P.A.; et al. Identification, design and biological evaluation of bisaryl quinolones targeting Plasmodium falciparum type II NADH:quinone oxidoreductase (PfNDH2). J. Med. Chem. 2012, 55, 1831–1843. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yu, Y.; Li, X.; Li, J.; Wu, Y.; Yu, Y.; Ge, J.; Huang, Z.; Jiang, L.; Rao, Y.; et al. Target elucidation by cocrystal structures of NADH-ubiquinone oxidoreductase of Plasmodium falciparum with small molecule to eliminate drug-resistant malaria. J. Med. Chem. 2017, 60, 1994–2005. [Google Scholar] [CrossRef]
- Velazquez, I.; Pardo, J.P. Kinetic characterization of the rotenone-insensitive internal NADH:ubiquinone oxidoreductase of mitochondria of Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2001, 389, 7–14. [Google Scholar] [CrossRef]
- Yamashita, T.; Nakamaru-Ogiso, E.; Miyoshi, H.; Matsuno-Yagi, A.; Yagi, T. Roles of bound quinone in the single subunit NADH-quinone oxidoreductase (Ndi1) from Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 6012–6020. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yamashita, T.; Nakamaru-Ogiso, E.; Hashimoto, T.; Murai, M.; Igarashi, J.; Miyoshi, H.; Mori, N.; Matsuno-Yagi, A.; Yagi, T.; et al. Reaction mechanism of single subunit NADH-ubiquinone oxidoreductase (Ndi1) from Saccharomyces cerevisiae. Evidence for a ternary complex mechanism. J. Biol. Chem. 2011, 286, 9287–9297. [Google Scholar] [CrossRef]
- Feng, Y.; Li, W.; Li, J.; Wang, J.; Ge, J.; Xu, D.; Liu, Y.; Wu, K.; Zeng, Q.; Wu, J.-W.; et al. Structural insights into the type-II mitochondrial NADH dehydrogenases. Nature 2012, 491, 478–482. [Google Scholar] [CrossRef]
- Yano, T.; Rahimian, M.; Aneja, K.K.; Schechter, N.M.; Rubin, H.; Scott, C.P. Mycobacterium tuberculosis type II NADH-menaquinone oxidoreductase catalyzes electron transfer through a two-site ping-pong mechanism and has two quinone-binding sites. Biochemistry 2014, 53, 1179–1190. [Google Scholar] [CrossRef]
- Heikal, A.; Nakatani, Y.; Dunn, E.; Weimar, M.R.; Day, C.L.; Baker, E.N.; Lott, J.S.; Sazanov, L.A.; Cook, G.M. Structure of the bacterial type II NADH dehydrogenase: A monotopic membrane protein with an essential role in energy generation. Mol. Microbiol. 2014, 91, 950–964. [Google Scholar] [CrossRef]
- Sena, F.V.; Batista, A.P.; Catarino, T.; Brito, J.A.; Archer, M.; Viertler, M.; Madl, T.; Cabrita, E.J.; Pereira, M.M. Type-II NADH:quinone oxidoreductase from Staphylococcus aureus has two distinct binding sites and is rate limited by quinone reduction. Mol. Microbiol. 2015, 98, 272–288. [Google Scholar] [CrossRef]
- Blaza, J.N.; Bridges, H.R.; Aragao, D.; Dunn, E.A.; Heikal, A.; Cook, G.M.; Nakatani, Y.; Hirst, J. The mechanism of catalysis by type-II NADH:quinone oxidoreductases. Sci. Rep. 2017, 7, 40165. [Google Scholar] [CrossRef]
- Yamashita, T.; Inaoka, D.K.; Shiba, T.; Oohashi, T.; Iwata, S.; Yagi, T.; Kosaka, T.; Miyoshi, H.; Harada, S.; Kita, K.; et al. Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors. Sci. Rep. 2018, 8, 2427. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Åkelbladh, L.; Ahmad, S.; Konda, V.; Cao, S.; Vocat, A.; Maes, L.; Cole, S.T.; Hughes, D.; Larhed, M.; et al. Synthesis and in vitro biological evaluation of quinolinyl pyrimidines targeting type II NADH-dehydrogenases (NDH-2). ACS Infect. Dis. 2022, 8, 482–498. [Google Scholar] [CrossRef] [PubMed]
- Saha, P.; Sau, S.; Kalia, N.P.; Sharma, D.K. 2-Aryl-benzoimidazoles as type II NADH dehydrogenase inhibitors of Mycobacterium tuberculosis. ACS Infect. Dis. 2024, 10, 3699–3711. [Google Scholar] [CrossRef]
- Ke, H.; Ganesan, S.M.; Dass, S.; Morrisey, J.M.; Pou, S.; Nilsen, A.; Riscoe, M.K.; Mather, M.W.; Vaidya, A.B. Mitochondrial type II NADH dehydrogenase of Plasmodium falciparum (PfNDH2) is dispensable in the asexual blood stages. PLoS ONE 2019, 14, e0214023. [Google Scholar] [CrossRef] [PubMed]
- Lesanavičius, M.; Aliverti, A.; Šarlauskas, J.; Čėnas, N. Reactions of Plasmodium falciparum ferredoxin:NADP+ oxidoreductase with redox cycling xenobiotics: A mechanistic study. Int. J. Mol. Sci. 2020, 21, 3234. [Google Scholar] [CrossRef]
- Wardman, P.; Dennis, M.F.; Everett, S.A.; Patel, K.B.; Stratford, M.R.L.; Tracy, M. Radicals from one-electron reduction of nitro compounds, aromatic N-oxides and quinones: The kinetic basis for hypoxia-selective, bioreductive drugs. Biochem. Soc. Symp. 1995, 61, 171–194. [Google Scholar]
- Vidakovic, M.; Crossnoe, C.R.; Neidre, C.; Kim, K.; Krause, K.L.; Germanas, J.P. Reactivity of reduced [2Fe-2S] ferredoxin parallels host susceptibility to nitroimidazoles. Antimicrob. Agents Chemother. 2003, 47, 302–308. [Google Scholar] [CrossRef]
- Williams, E.M.; Little, R.F.; Mowday, A.M.; Rich, M.H.; Chan-Hyams, J.V.; Copp, J.N.; Smaill, J.B.; Patterson, A.V.; Ackerley, D.F. Nitroreductase gene-directed enzyme prodrug therapy: Insights and advances towards clinical utility. Biochem. J. 2015, 471, 131–153. [Google Scholar] [CrossRef]
- Nepali, K.; Lee, H.-J.; Liou, J.-P. Nitro-group-containing drugs. J. Med. Chem. 2019, 62, 2851–2893. [Google Scholar] [CrossRef] [PubMed]
- Di Francesco, A.; Ward, T.H.; Butler, J. Diaziridinylbenzoquinones. Methods Enzymol. 2004, 382B, 174–193. [Google Scholar]
- Morin, C.; Besset, T.; Moutet, J.C.; Fayolle, M.; Brückner, M.; Limosin, D.; Becker, K.; Davioud-Charvet, E. The aza-analogues of 1,4-naphthoquinones are potent substrates and inhibitors of plasmodial thioredoxin and glutathione reductases and of human erythrocyte glutathione reductase. Org. Biomol. Chem. 2008, 6, 2731–2742. [Google Scholar] [CrossRef]
- Dong, C.K.; Patel, V.; Yang, J.C.; Dvorin, J.D.; Duraisingh, M.T.; Clardy, J.; Wirth, D.F. Type II NADH dehydrogenase of the respiratory chain of Plasmodium falciparum and its inhibitors. Bioorg. Med. Chem. Lett. 2009, 19, 972–975. [Google Scholar] [CrossRef] [PubMed]
- Wardman, P. Reduction potentials of one-electron couples involving free radicals in aqueous solution. J. Phys. Chem. Ref. Data 1989, 18, 1637–1755. [Google Scholar] [CrossRef]
- Čėnas, N.; Anusevičius, Ž.; Nivinskas, H.; Misevičienė, L.; Šarlauskas, J. Structure-activity relationships in two-electron reduction of quinones. Methods Enzymol. 2004, 382B, 258–277. [Google Scholar]
- Buffinton, G.D.; Öllinger, K.; Brunmark, A.; Cadenas, E. DT-diaphorase-catalyzed reduction of 1,4-naphthoquinone derivatives and glutathionyl-quinone conjugates. Effects of substituents on autoxidation rates. Biochem. J. 1989, 257, 561–571. [Google Scholar] [CrossRef]
- O’Brien, P.J. Molecular mechanisms of quinone cytotoxicity. Chem. Biol. Interact. 1991, 80, 1–41. [Google Scholar] [CrossRef]
- Nivinskas, H.; Staškevičienė, S.; Šarlauskas, J.; Koder, R.L.; Miller, A.F.; Čėnas, N. Two-electron reduction of quinones by Entero Enterobacter cloacae NAD(P)H:nitroreductase: Quantitative structure-activity relationships. Arch. Biochem. Biophys. 2002, 403, 249–258. [Google Scholar] [CrossRef]
- Iyanagi, T. On the mechanism of one-electron reduction of quinones by microsomal flavin enzymes. The kinetic analysis between cytochrome b5 and menadione. Free Radic. Res. Commun. 1990, 8, 259–268. [Google Scholar] [CrossRef]
- Shah, M.M.; Spain, J.C. Elimination of nitrite from the explosive 2,4,6-trinitrophenylmethylnitramine (tetryl) catalyzed by ferredoxin NADP+ oxidoreductase from spinach. Biochem. Biophys. Res. Commun. 1996, 220, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDoc Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- McNutt, A.T.; Francoeur, P.; Aggarwal, R.; Masuda, T.; Meli, R.; Ragoza, M.; Sunseri, J.; Koes, D.R. GNINA 1:0: Molecular docking with deap learning. J. Cheminform. 2021, 13, 43. [Google Scholar] [CrossRef] [PubMed]
- Day, M.A.; Jarrom, D.; Christofferson, A.J.; Graziano, A.E.; Anderson, J.L.R.; Searle, P.F.; Hyde, E.I.; White, S.A. The structures of E. coli NfsA bound to the antibiotic nitrofurantoin, to 1,4-benzoquinone and to FMN. Biochem. J. 2021, 478, 2601–2617. [Google Scholar] [CrossRef]
- Valiauga, B.; Bagdžiūnas, G.; Sharrock, A.V.; Ackerley, D.F.; Čėnas, N. The catalysis mechanism of E. coli nitroreductase A, a candidate for gene-directed prodrug therapy: Potentiometric and substrate specificity studies. Int. J. Mol. Sci. 2024, 25, 4413. [Google Scholar] [CrossRef]
- Bulger, J.E.; Brandt, K.G. Yeast glutathione reductase II. Interaction of oxidized and 2-electron reduced enzyme with reduced and oxidized nicotinamide adenine dinucleotide phosphate. J. Biol. Chem. 1971, 246, 5578–5587. [Google Scholar] [CrossRef]
- Bironaitė, D.; Anusevičius, Ž.; Jacquot, J.-P.; Čėnas, N. Interaction of quinones with Arabidopsis thaliana thioredoxin reductase. Biochim. Biophys. Acta 1998, 1383, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Salewski, J.; Batista, A.P.; Sena, F.V.; Millo, D.; Zebger, I.; Pereira, M.M.; Hildebrandt, P. Substrate-protein interactions of type II NADH:quinone oxidoreductase from Escherichia coli. Biochemistry 2016, 55, 2722–2734. [Google Scholar] [CrossRef]
- Huang, C.Y. Derivation and initial velocity and isotope exchange rate equations. Methods Enzymol. 1979, 63, 54–84. [Google Scholar]
- Tejero, J.; Peregrina, J.R.; Martínez-Júlvez, M.; Gutiérrez, A.; Gómez-Moreno, C.; Scrutton, N.S.; Medina, M. Catalytic mechanism of hydride transfer between NADP+/H and ferredoxin-NADP+ reductase from Anabaena PCC 7119. Arch. Biochem. Biophys. 2007, 459, 79–90. [Google Scholar] [CrossRef]
- Čėnas, N.; Nemeikaitė-Čėnienė, A.; Kosychova, L. Single- and two-electron reduction of nitroaromatic compounds by flavoezymes: Mechanisms and implications for cytotoxicity. Int. J. Mol. Sci. 2021, 22, 8534. [Google Scholar] [CrossRef] [PubMed]
- Marcus, R.A.; Sutin, N. Electron transfers in chemistry and biology. Biochim. Biophys. Acta-Rev. Bioenerg. 1985, 811, 265–322. [Google Scholar] [CrossRef]
- Bianchet, M.A.; Faig, M.; Amzel, L.M. Structure and mechanism of NAD(P)H:quinone acceptor oxidoreductases (NQO). Methods Enzymol. 2004, 382B, 144–174. [Google Scholar]
- Xie, T.; Wu, Z.; Gu, J.; Guo, R.; Yan, X.; Duan, H.; Liu, X.; Liu, W.; Liang, L.; Wan, H.; et al. The global motion affecting electron transfer in Plasmodium falciparum type II NADH dehydrogenase: A novel non-competitive mechanism for quinolone ketone derivative inhibitors. Phys. Chem. Chem. Phys. 2019, 21, 18105–18118. [Google Scholar] [CrossRef]
- Monti, D.; Basilico, N.; Parapini, S.; Pasini, E.; Olliaro, P.; Taramelli, D. Does chloroquine really act through oxidative stress? FEBS Lett. 2002, 522, 3–5. [Google Scholar] [CrossRef]
- Becker, K.; Tilley, L.; Vennerstrom, J.L.; Roberts, D.; Rogerson, S.; Ginsburg, H. Oxidative stress in malaria parasite-infected erythrocytes: Host-parasite interactions. Int. J. Parasitol. 2004, 34, 163–189. [Google Scholar] [CrossRef] [PubMed]
- Čėnas, N.; Nemeikaitė-Čėnienė, A.; Sergedienė, E.; Nivinskas, H.; Anusevičius, Ž.; Šarlauskas, J. Quantitative structure-activity relationships in enzymatic single-electron reduction of nitroaromatic explosives: Implications for their cytotoxicity. Biochim. Biophys. Acta 2001, 1528, 31–38. [Google Scholar] [CrossRef]
- Montgomery, H.A.C.; Dymock, J. Determination of nitrite in water. Analyst 1961, 86, 414–416. [Google Scholar]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. CSF ChimeraX: Tools for structural building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. Pubchem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Halgren, T.A. MMFFVI, MMFF94s options for energy minimization studies. J. Comput. Chem. 1999, 20, 720–729. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, E.D.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advance semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Moriss, C.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.S. AutoDock4 and AutoDockTools4: Automated doc king with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Oper Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Bell, E.W.; Zhang, Y. DockRMSD: An open-source tool for atom mapping and RMSD calculation of symmetric molecules. J. Cheminform. 2019, 11, 40. [Google Scholar] [CrossRef]
- Olechnovič, K.; Venclovas, Č. Voronota: A fast and reliable tool for computing the vertices of the Voronoi diagram of atomic balls. J. Comput. Chem. 2014, 35, 672–681. [Google Scholar] [CrossRef]
- Koes, D.R.; Baumgartner, M.P.; Camacho, C.J. Lesons learned in empirical scoring with smina from the CSAR 2011. J. Chem. Inf. Model. 2013, 53, 1893–1904. [Google Scholar] [CrossRef]
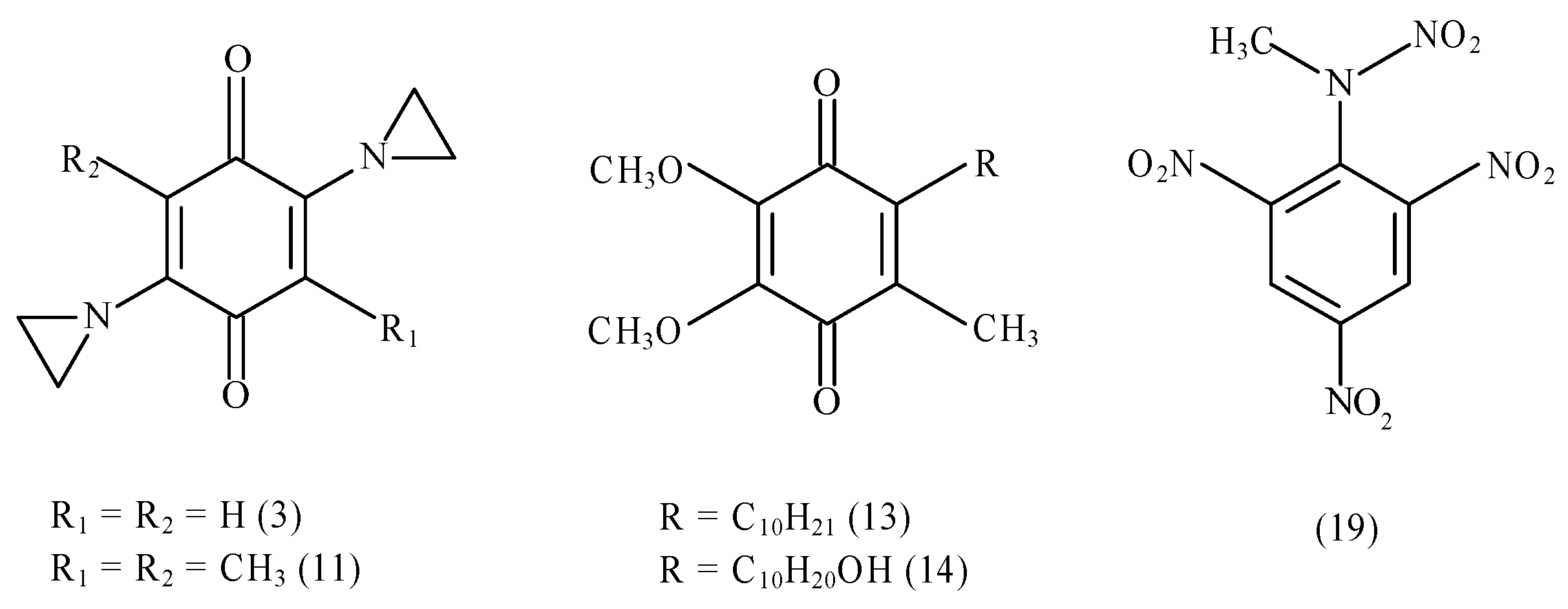
| No. | Compound | (V) | log D | (s−1) | kcat/Km (M−1s−1) |
|---|---|---|---|---|---|
| Quinones | |||||
| 1 | 1,4-Benzoquinone | 0.09 | 1.02 | 14.00 ± 0.92 | 3.30 ± 0.21 × 105 |
| 2 | 2-Methyl-1,4-benzoquinone | 0.01 | 1.42 | 8.50 ± 0.87 | 2.10 ± 0.22 × 105 |
| 3 | 2,5-Diaziridinyl-1,4-benzoquinone (DZQ) | −0.05 | 0.00 | 9.00 ± 0.81 | 3.72 ± 0.32 × 105 |
| 4 | 2,6-Dimethyl-1,4-benzoquinone | −0.08 | 1.82 | 10.80 ± 0.71 | 2.85 ± 0.19 × 105 |
| 5 | 5-Hydroxy-1,4-naphthoquinone (juglone) | −0.09 | 1.82 | 5.85 ± 0.32 | 8.11 ± 0.58 × 105 |
| 6 | 5,8-Dihydroxy-1,4-naphthoquinone (naphthazarin) | −0.11 | 2.17 | 14.30 ± 1.01 | 6.95 ± 0.39 × 105 |
| 7 | 9,10-Phenanthrene quinone | −0.12 | 2.92 | 15.55 ± 0.98 | 7.55 ± 0.43 × 105 |
| 8 | 1,4-Naphthoquinone | −0.15 | 1.50 | 17.50 ± 1.05 | 5.45 ± 0.24 × 105 |
| 9 | 2-Methyl-1,4-naphthoquinone (menadione) | −0.20 | 1.89 | 7.05 ± 0.25 | 9.25 ± 0.45 × 105 |
| 10 | Trimethyl-aziridinyl-1,4-benzoquinone | −0.23 | 1.70 | 20.90 ± 1.29 | 1.44 ± 0.11 × 105 |
| 11 | 2,5-Dimethyl-3,6-diaziridinyl-1,4-benzoquinone (MeDZQ) | −0.23 | 0.79 | 1.60 ± 0.09 | 5.72 ± 0.31 × 104 |
| 12 | Ubiquinone (Q1) | −0.24 | 2.22 | 36.70 ± 2.23 | 1.78 ± 0.09 × 106 |
| 13 | Decylubiquinone | −0.24 | 5.00 | 32.10 ± 2.02 | 2.75 ± 0.19 × 106 |
| 14 | Idebenone | −0.24 | 3.57 | 41.00 ± 2.54 | 2.40 ± 0.16 × 106 |
| 15 | Tetramethyl-1,4-benzoquinone (duroquinone) | −0.26 | 2.61 | 13.00 ± 0.58 | 4.12 ± 0.26 × 105 |
| 16 | 1,4-Dihydroxy-9,10-anthraquinone | −0.30 | 3.60 | - | 2.02 ± 0.14 × 103 |
| 17 | 1,8-Dihydroxy-9,10-anthraquinone | −0.33 | 3.60 | - | 4.41 ± 0.27 × 103 |
| 18 | 2-Hydroxy-1,4-naphthoquinone | −0.41 | −0.70 | 0.42 ± 0.06 | 1.75 ± 0.19 × 104 |
| Nitroaromatic compounds | |||||
| 19 | 2,4,6-Trinitrophenyl-N-methylnitramine (tetryl) | −0.191 | 1.38 | 7.40 ± 0.55 | 1.50 ± 0.11 × 105 |
| 20 | 2,4,6-Trinitrotoluene | −0.253 | 2.31 | 2.40 ± 0.55 | 3.20 ± 0.19 × 104 |
| 21 | p-Dinitrobenzene | −0.255 | 1.85 | 7.35 ± 0.49 | 6.09 ± 0.29 × 104 |
| 22 | Nitrofurantoin | −0.255 | −0.25 | 0.60 ± 0.05 | 2.25 ± 0.18 × 103 |
| 23 | Nifuroxime | −0.255 | −0.35 | 1.30 ± 0.08 | 1.40 ± 0.11 × 104 |
| 24 | o-Dinitrobenzene | −0.287 | 1.85 | 1.10 ± 0.06 | 3.54 ± 0.15 × 103 |
| 25 | p-Nitrobenzaldehyde | −0.325 | 1.63 | 1.50 ± 0.07 | 9.95 ± 0.08 × 102 |
| 26 | m-Dinitrobenzene | −0.345 | 1.85 | 0.60 ± 0.04 | 3.37 ± 0.12 × 103 |
| 27 | p-Nitroacetophenone | −0.355 | 1.47 | - | 2.70 ± 0.13 × 102 |
| 28 | p-Nitrobenzoic acid | −0.425 | −1.66 | 0.20 ± 0.02 | 1.79 ± 0.11 × 103 |
| 29 | Nitrobenzene | −0.485 | 1.91 | 0.80 ± 0.10 | 2.49 ± 0.20 × 102 |
| Single-electron acceptors | |||||
| 30 | Ferricyanide | 0.410 | - | 6.80 ± 0.32 | 5.10 ± 0.24 × 104 |
| 31 | Benzylviologen | −0.354 | - | 0.50 ± 0.03 | 3.00 ± 0.16 × 103 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Misevičienė, L.; Golinelli-Cohen, M.-P.; Kairys, V.; Marozienė, A.; Lesanavičius, M.; Čėnas, N. Reactions of Plasmodium falciparum Type II NADH: Ubiquinone Oxidoreductase with Nonphysiological Quinoidal and Nitroaromatic Oxidants. Int. J. Mol. Sci. 2025, 26, 2509. https://doi.org/10.3390/ijms26062509
Misevičienė L, Golinelli-Cohen M-P, Kairys V, Marozienė A, Lesanavičius M, Čėnas N. Reactions of Plasmodium falciparum Type II NADH: Ubiquinone Oxidoreductase with Nonphysiological Quinoidal and Nitroaromatic Oxidants. International Journal of Molecular Sciences. 2025; 26(6):2509. https://doi.org/10.3390/ijms26062509
Chicago/Turabian StyleMisevičienė, Lina, Marie-Pierre Golinelli-Cohen, Visvaldas Kairys, Audronė Marozienė, Mindaugas Lesanavičius, and Narimantas Čėnas. 2025. "Reactions of Plasmodium falciparum Type II NADH: Ubiquinone Oxidoreductase with Nonphysiological Quinoidal and Nitroaromatic Oxidants" International Journal of Molecular Sciences 26, no. 6: 2509. https://doi.org/10.3390/ijms26062509
APA StyleMisevičienė, L., Golinelli-Cohen, M.-P., Kairys, V., Marozienė, A., Lesanavičius, M., & Čėnas, N. (2025). Reactions of Plasmodium falciparum Type II NADH: Ubiquinone Oxidoreductase with Nonphysiological Quinoidal and Nitroaromatic Oxidants. International Journal of Molecular Sciences, 26(6), 2509. https://doi.org/10.3390/ijms26062509







