Posttranslational Regulation of Mammalian Sulfur Amino Acid Metabolism
Abstract
1. Introduction
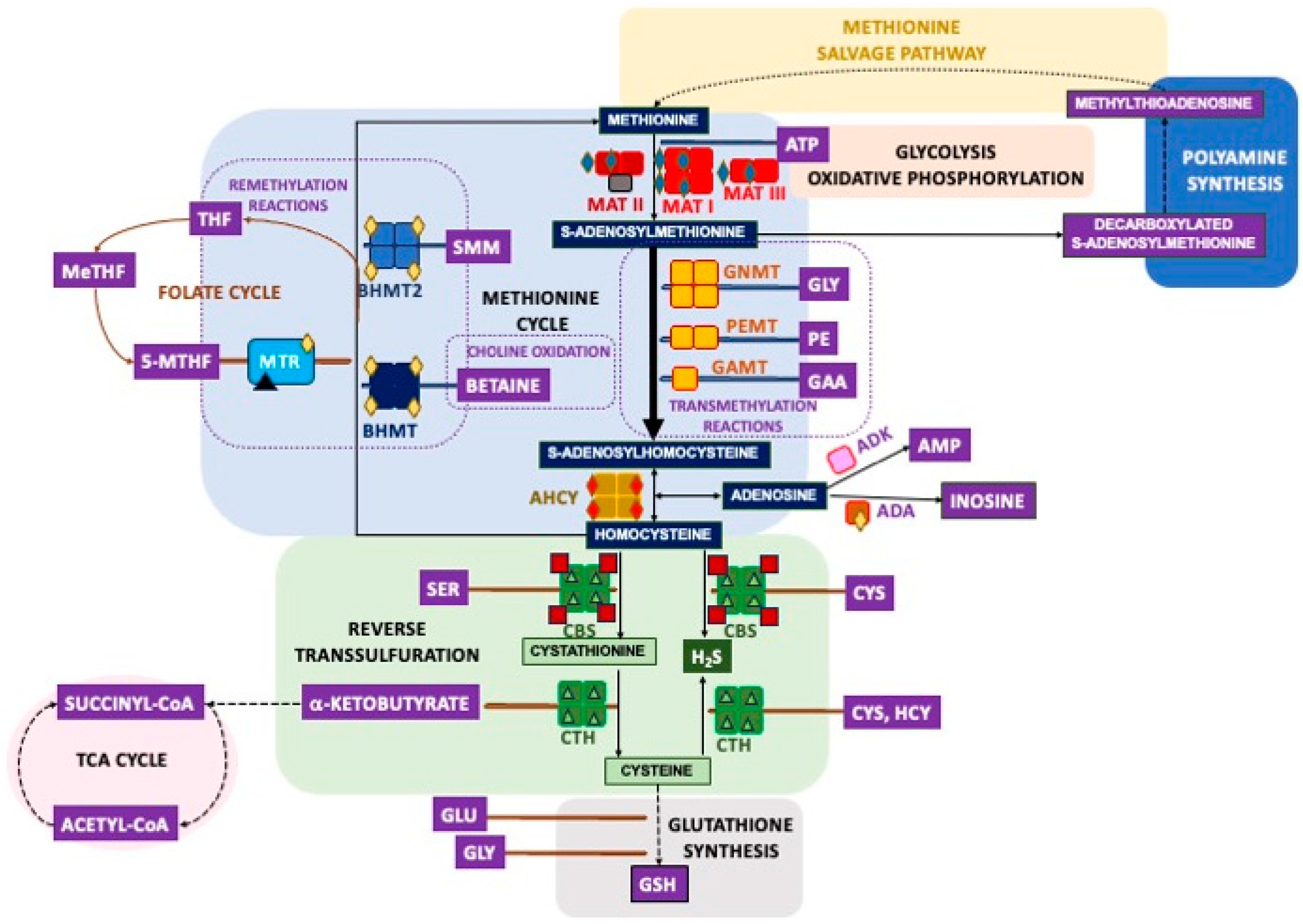
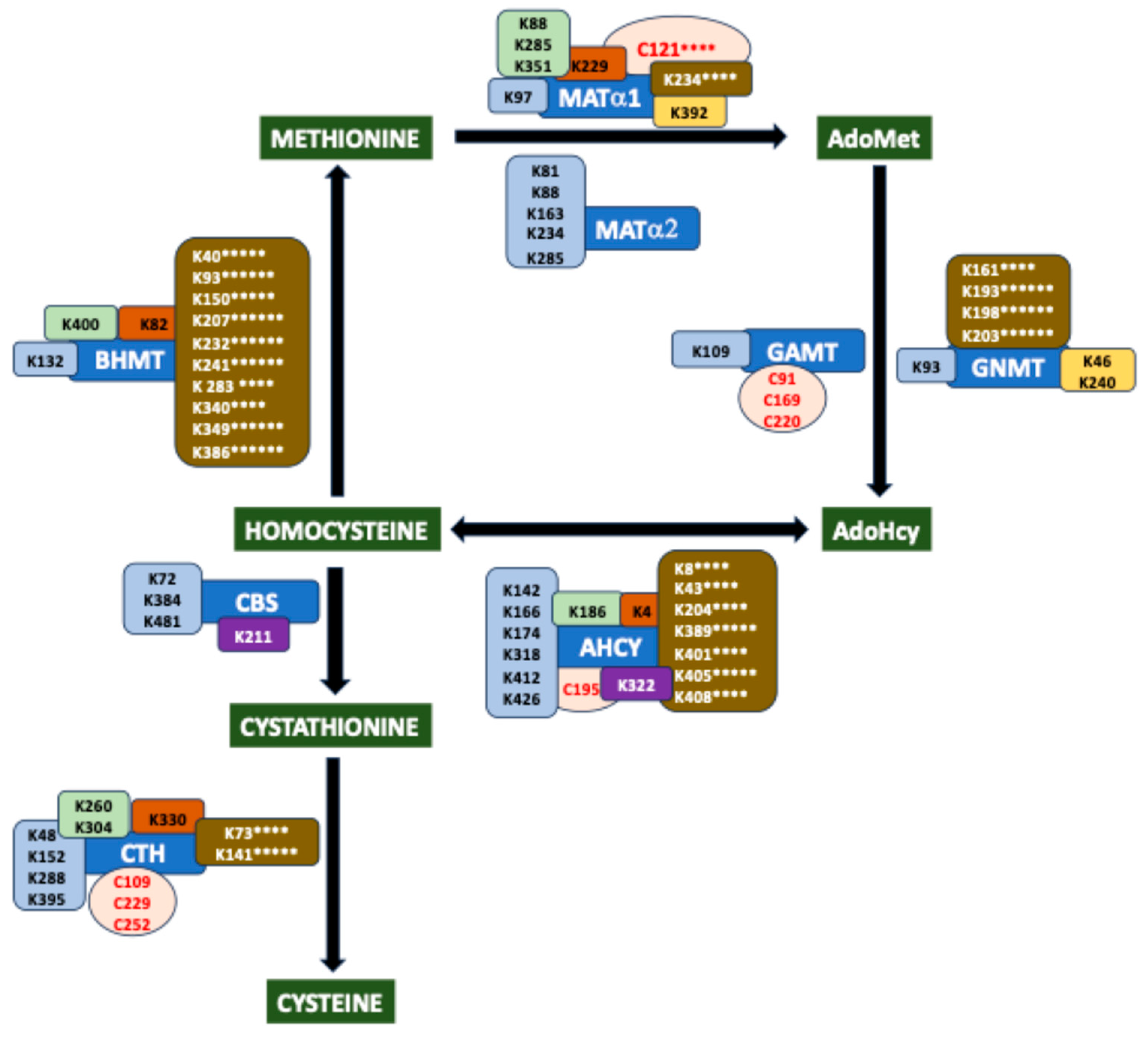
2. Overview of the Mammalian Sulfur Amino Acid Metabolism
3. Methods and Sample Types Used for the Identification of Posttranslational Modifications in the Main Enzymes of Sulfur Amino Acid Metabolism
4. Regulation by Phosphorylation
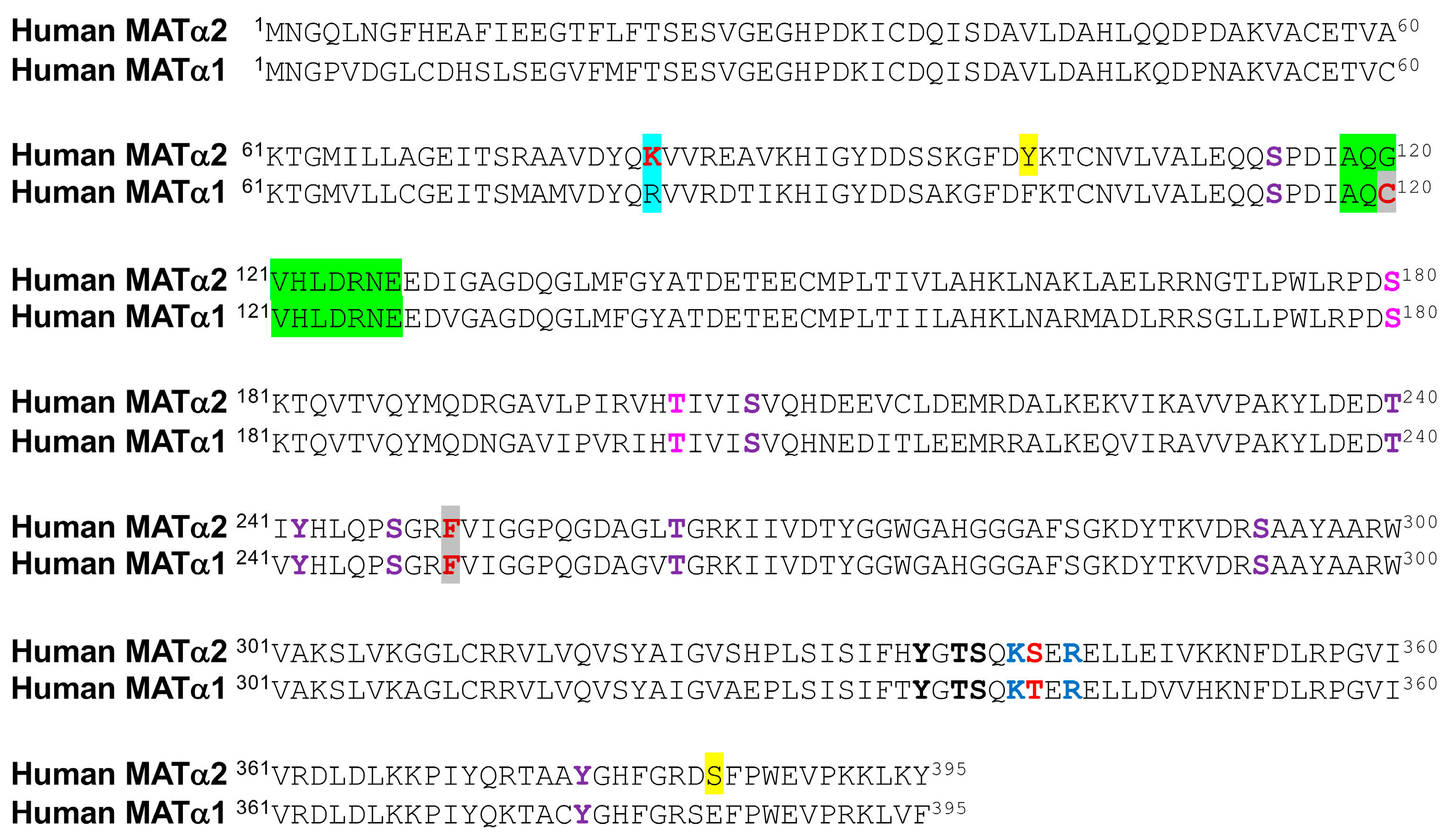
4.1. Phosphorylation of Methionine Adenosyltransferases
4.2. Phosphorylation of Main Hepatic Methyltransferases
4.3. Phosphorylation of S-Adenosylhomocysteine Hydrolase and Remethylation Enzymes
4.4. Phosphorylation of Reverse Transsulfuration Enzymes
5. Regulation by Ubiquitylation and Sumoylation
5.1. Modification by Ubiquitylation
5.2. Regulation by Sumoylation
| PTM [Ref] 4 | Gene Name | Modification Site [Ref] 5 |
|---|---|---|
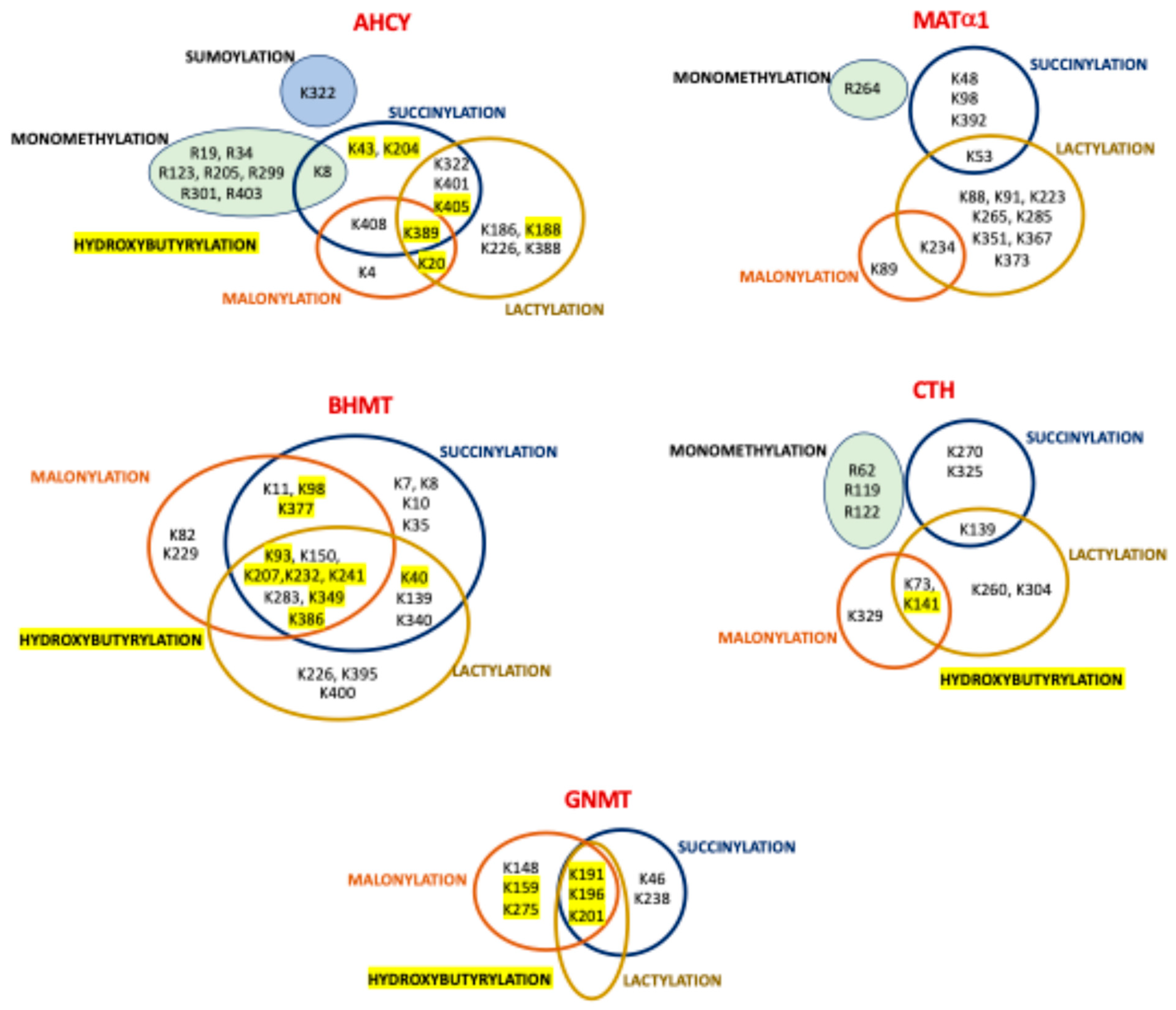
| Sumoylation 1 (cell lines) [57] | AHCY | K322 |
| N-glycosylation (mouseL) [101] | MAT1A | N106 |
| AHCY | N126, N181 | |
| BHMT | N69 | |
| Monomethylation (cell lines) | MAT1A | R264 [64,65] |
| MAT2A | R192 [65] | |
| MAT2B | R29 [65], R30 [65] | |
| AHCY | K8 [66], R19 [65], R34 [65], R123 2 [65], R205 [65], R299 2 [65], R301 2 [65], R403 2 [65] | |
| MTR | K817 [66], R1132 [65] | |
| CBS | R18 [65], R164 [65], R190 [65], R336 [65], R389 [65] | |
| CTH | R62 [65], R119 [65], R122 [65] | |
| Succinylation 3 (mouseL) | MAT1A | K48 [164], K54 [164], K98 [59], K392 [59] |
| AHCY | K8 [63], K43 [59], K204 (cells, mouseL) [59], K322 [59], K389 [59], K401 [164], K405 (cells, mouseL) [59,164], K408 [164] | |
| GNMT | K46 [63], K191 [63], K196 [59,63], K201 [59,63], K238 [59,63] | |
| MTR | K327 [59] | |
| BHMT | K7 [164], K8 [164], K10 [164], K11 [164], K35 [164], K40 [63], K93 [59,63], K98 [63,164], K139 [164], K150 [59], K207 [63,164], K232 [59,63], K241 [63], K283 [59,63], K340 [63], K349 [59,63], K377 [63], K386 [59,63] | |
| BHMT2 | K274 [59,63] | |
| CBS | K485 (cells) [59], K174 [59] | |
| CTH | K138 [63], K270 [59], K325 [63] | |
| Malonylation (mouseL) [107] | MAT1A | K89, K235 |
| AHCY | K4, K20, K389, K408 | |
| GNMT | K148, K159, K191, K196, K201, K275 | |
| BHMT | K11, K82, K93, K98, K150, K207, K229, K232, K241, K283, K349, K377, K386 | |
| CTH | K72, K140, K329 | |
| Lactylation (humanL) [106] | MAT1A | K53, K88, K91, K223, K234, K265, K285, K351, K367, K373 |
| AHCY | K20, K186, K188, K226, K322, K388, K389, K401, K405 | |
| GAMT | K109 | |
| GNMT | K193, K198, K203 | |
| BHMT | K40, K93, K139, K150, K207, K226, K232, K241, K283, K340, K349, K369, K386, K395, K400 | |
| BHMT2 | K104, K120, K198 | |
| CBS | K25, K30, K177, K281 | |
| CTH | K73, K139, K141, K260, K304 | |
| Hydroxybutyrylation (mouseL) [105] | AHCY | K20, K43, K188, K204, K389, K405 |
| GNMT | K159, K191, K196, K201, K275 | |
| BHMT | K40, K93, K98, K207, K232, K241, K349, K377, K386 | |
| CTH | K140 | |
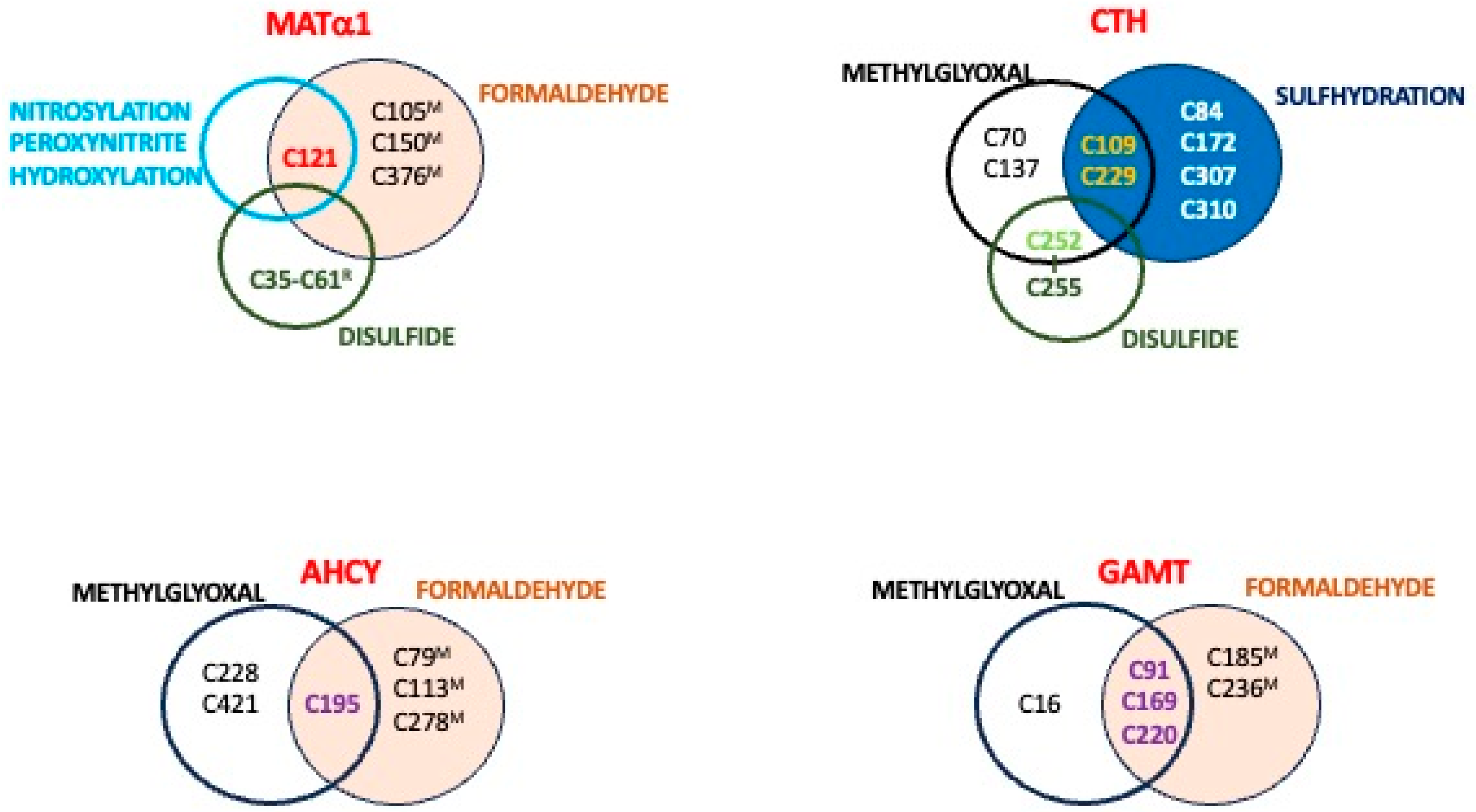
| Methylglyoxal (Cell lines) [67] | MAT2A | C56, C104 6, C214 |
| MAT2B | C58 6, C297 6 | |
| AHCY | C195 6, C228, C421 6 | |
| GAMT | C16, C91 6, C169 6, C220 6 | |
| PEMT | C70 6 | |
| CBS | C15 6, C52, C103, C109, C165, C370, C431 | |
| CTH | C70, C109, C137, C229 | |
| Formaldehyde (mouseL) [108] | MAT1A | C105, C121, C150, C376 7 |
| AHCY | C79, C113, C195, C278 | |
| GAMT | C92, C169, C185, C220, C236 | |
| GNMT | C147, C186, C247 | |
| BHMT | C104, C131, C217, C256, C299, C300 | |
| BHMT2 | C290, C391 | |
| CBS | C427 |
6. Posttranslational Modification by Products of Several Metabolic Pathways
6.1. Regulation by Acetylation
6.2. Regulation by Glycosylation
6.3. Regulation by Amidation and Crosslinking
6.4. Regulation by Malonylation
6.5. Regulation by Lactylation
6.6. Regulation by Succinylation
6.7. Regulation by Hydroxybutyrylation
6.8. Regulation by ADP-Ribosylation
6.9. Regulation by Methylglyoxal
6.10. Regulation by Formylation
7. Redox Regulation and Associated Posttranslational Modifications
7.1. Disulfide Bonds
7.2. Glutathionylation
7.3. Nitration and Sulfhydration
8. Hotspots for Posttranslational Modification
9. Influence of Posttranslational Modification in Pathological States
10. Concluding Remarks and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Bradley, D. The evolution of post-translational modifications. Curr. Opin. Genet. Dev. 2022, 76, 101956. [Google Scholar] [CrossRef] [PubMed]
- Keenan, E.K.; Zachman, D.K.; Hirschey, M.D. Discovering the landscape of protein modifications. Mol. Cell 2021, 81, 1868–1878. [Google Scholar] [CrossRef] [PubMed]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef] [PubMed]
- Ramazi, S.; Zahiri, J. Post-translational modifications in proteins: Resources, tools and prediction methods. Database 2021, 2021, baab012. [Google Scholar] [CrossRef]
- Zhong, Q.; Xiao, X.; Qiu, Y.; Xu, Z.; Chen, C.; Chong, B.; Zhao, X.; Hai, S.; Li, S.; An, Z.; et al. Protein posttranslational modifications in health and diseases: Functions, regulatory mechanisms, and therapeutic implications. MedComm 2023, 4, e261. [Google Scholar] [CrossRef]
- Conigrave, A.D.; Van Der Weyden, L.; Holt, L.; Jiang, L.; Wilson, P.; Christopherson, R.I.; Morris, M.B. Extracellular ATP-dependent suppression of proliferation and induction of differentiation of human HL-60 leukemia cells by distinct mechanisms. Biochem. Pharmacol. 2000, 60, 1585–1591. [Google Scholar] [CrossRef]
- Pajares, M.A.; Perez-Sala, D. Mammalian Sulfur Amino Acid Metabolism: A Nexus Between Redox Regulation, Nutrition, Epigenetics, and Detoxification. Antioxid. Redox Signal. 2018, 29, 408–452. [Google Scholar] [CrossRef]
- Finkelstein, J.D. Methionine metabolism in mammals. J. Nutr. Biochem. 1990, 1, 228–237. [Google Scholar] [CrossRef]
- Mato, J.M.; Alvarez, L.; Ortiz, P.; Pajares, M.A. S-adenosylmethionine synthesis: Molecular mechanisms and clinical implications. Pharmacol. Ther. 1997, 73, 265–280. [Google Scholar] [CrossRef]
- Jakubowski, H. Homocysteine Modification in Protein Structure/Function and Human Disease. Physiol. Rev. 2019, 99, 555–604. [Google Scholar] [CrossRef]
- Womack, M.; Kemmerer, K.S.; Rose, W.C. The relation of dietary methionine and cysteine to growth. J. Biol. Chem. 1937, 121, 403–410. [Google Scholar] [CrossRef]
- Sowers, M.L.; Herring, J.; Zhang, W.; Tang, H.; Ou, Y.; Gu, W.; Zhang, K. Analysis of glucose-derived amino acids involved in one-carbon and cancer metabolism by stable-isotope tracing gas chromatography mass spectrometry. Anal. Biochem. 2019, 566, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Maddocks, O.D.; Labuschagne, C.F.; Adams, P.D.; Vousden, K.H. Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Mol. Cell 2016, 61, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, T.S.; Benevenga, N.J.; Harper, A.E. Effect of dietary methionine level on its metabolism in rats. J. Nutr. 1974, 104, 761–771. [Google Scholar] [CrossRef]
- Zeisel, S.H.; Wurtman, R.J. Dietary intake of methionine: Influence of brain S-adenosylmethionine. In Transmethylation; Usdin, E., Borchardt, R.T., Creveling, C.R., Eds.; Elsevier: New York, NY, USA, 1979; pp. 59–68. [Google Scholar]
- Sugimura, T.; Birnbaum, S.M.; Winitz, M.; Greenstein, J.P. Quantitative nutritional studies with water-soluble, chemically defined diets. VIII. The forced feeding of diets each lacking in one essential amino acid. Arch. Biochem. Biophys. 1959, 81, 448–455. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Erbe, R.W. High in vivo rates of methionine biosynthesis in transformed human and malignant rat cells auxotrophic for methionine. Proc. Natl. Acad. Sci. USA 1976, 73, 1523–1527. [Google Scholar] [CrossRef]
- Reytor, E.; Perez-Miguelsanz, J.; Alvarez, L.; Perez-Sala, D.; Pajares, M.A. Conformational signals in the C-terminal domain of methionine adenosyltransferase I/III determine its nucleocytoplasmic distribution. FASEB J. 2009, 23, 3347–3360. [Google Scholar] [CrossRef]
- Perez-Miguelsanz, J.; Vallecillo, N.; Garrido, F.; Reytor, E.; Perez-Sala, D.; Pajares, M.A. Betaine homocysteine S-methyltransferase emerges as a new player of the nuclear methionine cycle. Biochim. Biophys. Acta 2017, 1864, 1165–1182. [Google Scholar] [CrossRef]
- Katoh, Y.; Ikura, T.; Hoshikawa, Y.; Tashiro, S.; Ito, T.; Ohta, M.; Kera, Y.; Noda, T.; Igarashi, K. Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol. Cell 2011, 41, 554–566. [Google Scholar] [CrossRef]
- Yeo, E.J.; Wagner, C. Tissue distribution of glycine N-methyltransferase, a major folate-binding protein of liver. Proc. Natl. Acad. Sci. USA 1994, 91, 210–214. [Google Scholar] [CrossRef]
- Radomski, N.; Kaufmann, C.; Dreyer, C. Nuclear accumulation of S-adenosylhomocysteine hydrolase in transcriptionally active cells during development of Xenopus laevis. Mol. Biol. Cell 1999, 10, 4283–4298. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kabil, O.; Zhou, Y.; Banerjee, R. Human Cystathionine β-Synthase Is a Target for Sumoylation. Biochemistry 2006, 45, 13528–13536. [Google Scholar] [CrossRef] [PubMed]
- Krupenko, N.I.; Wagner, C. Transport of rat liver glycine N-methyltransferase into rat liver nuclei. J. Biol. Chem. 1997, 272, 27140–27146. [Google Scholar] [CrossRef]
- Murray, B.; Peng, H.; Barbier-Torres, L.; Robinson, A.; Li, T.W.H.; Fan, W.; Tomasi, M.L.; Gottlieb, R.A.; Van Eyk, J.; Lu, Z.; et al. Methionine Adenosyltransferase alpha1 is targeted to the mitochondrial matrix and interacts with cytochrome P450 2E1 to lower its expression. Hepatology 2019, 70, 2018–2034. [Google Scholar] [CrossRef]
- Glass, J.I.; Assad-Garcia, N.; Alperovich, N.; Yooseph, S.; Lewis, M.R.; Maruf, M.; Hutchison, C.A.; Smith, H.O.; Venter, J.C. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA 2006, 103, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, C.A.; Chuang, R.Y.; Noskov, V.N.; Assad-Garcia, N.; Deerinck, T.J.; Ellisman, M.H.; Gill, J.; Kannan, K.; Karas, B.J.; Ma, L.; et al. Design and synthesis of a minimal bacterial genome. Science 2016, 351, aad6253. [Google Scholar] [CrossRef]
- Gonzalez, B.; Pajares, M.A.; Hermoso, J.A.; Alvarez, L.; Garrido, F.; Sufrin, J.R.; Sanz-Aparicio, J. The crystal structure of tetrameric methionine adenosyltransferase from rat liver reveals the methionine-binding site. J. Mol. Biol. 2000, 300, 363–375. [Google Scholar] [CrossRef]
- Gonzalez, B.; Pajares, M.A.; Hermoso, J.A.; Guillerm, D.; Guillerm, G.; Sanz-Aparicio, J. Crystal structures of methionine adenosyltransferase complexed with substrates and products reveal the methionine-ATP recognition and give insights into the catalytic mechanism. J. Mol. Biol. 2003, 331, 407–416. [Google Scholar] [CrossRef]
- Murray, B.; Antonyuk, S.V.; Marina, A.; Van Liempd, S.M.; Lu, S.C.; Mato, J.M.; Hasnain, S.S.; Rojas, A.L. Structure and function study of the complex that synthesizes S-adenosylmethionine. IUCrJ 2014, 1, 240–249. [Google Scholar] [CrossRef]
- Gonzalez, B.; Garrido, F.; Ortega, R.; Martinez-Julvez, M.; Revilla-Guarinos, A.; Perez-Pertejo, Y.; Velazquez-Campoy, A.; Sanz-Aparicio, J.; Pajares, M.A. NADP+ binding to the regulatory subunit of methionine adenosyltransferase II increases intersubunit binding affinity in the hetero-trimer. PLoS ONE 2012, 7, e50329. [Google Scholar] [CrossRef]
- Pajares, M.A.; Markham, G.D. Methionine adenosyltransferase (s-adenosylmethionine synthetase). Adv. Enzymol. Relat. Areas Mol. Biol. 2011, 78, 449–521. [Google Scholar] [PubMed]
- Markham, G.D.; Pajares, M.A. Structure-function relationships in methionine adenosyltransferases. Cell. Mol. Life Sci. 2009, 66, 636–648. [Google Scholar] [CrossRef] [PubMed]
- Delgado, M.; Garrido, F.; Perez-Miguelsanz, J.; Pacheco, M.; Partearroyo, T.; Perez-Sala, D.; Pajares, M.A. Acute liver injury induces nucleocytoplasmic redistribution of hepatic methionine metabolism enzymes. Antioxid. Redox Signal. 2014, 20, 2541–2554. [Google Scholar] [CrossRef] [PubMed]
- Perez, C.; Perez-Zuniga, F.J.; Garrido, F.; Reytor, E.; Portillo, F.; Pajares, M.A. The Oncogene PDRG1 Is an Interaction Target of Methionine Adenosyltransferases. PLoS ONE 2016, 11, e0161672. [Google Scholar] [CrossRef]
- Pajares, M.Á. PDRG1 at the interface between intermediary metabolism and oncogenesis. World J. Biol. Chem. 2017, 8, 175–186. [Google Scholar] [CrossRef]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169, 824–835. [Google Scholar] [CrossRef]
- Yang, H.; Ara, A.I.; Magilnick, N.; Xia, M.; Ramani, K.; Chen, H.; Lee, T.D.; Mato, J.M.; Lu, S.C. Expression pattern, regulation, and functions of methionine adenosyltransferase 2beta splicing variants in hepatoma cells. Gastroenterology 2008, 134, 281–291. [Google Scholar] [CrossRef]
- Cantoni, G.L. Biological methylation: Selected aspects. Annu. Rev. Biochem. 1975, 44, 435–451. [Google Scholar] [CrossRef]
- Pegg, A.E. Functions of Polyamines in Mammals. J. Biol. Chem. 2016, 291, 14904–14912. [Google Scholar] [CrossRef]
- Broderick, J.B.; Duffus, B.R.; Duschene, K.S.; Shepard, E.M. Radical S -Adenosylmethionine Enzymes. Chem. Rev. 2014, 114, 4229–4317. [Google Scholar] [CrossRef]
- Broderick, J.B.; Broderick, W.E.; Hoffman, B.M. Radical SAM enzymes: Nature’s choice for radical reactions. FEBS Lett. 2023, 597, 92–101. [Google Scholar] [CrossRef]
- Agrimi, G.; Di Noia, M.A.; Marobbio, C.M.; Fiermonte, G.; Lasorsa, F.M.; Palmieri, F. Identification of the human mitochondrial S-adenosylmethionine transporter: Bacterial expression, reconstitution, functional characterization and tissue distribution. Biochem. J. 2004, 379, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Shields, D.J.; Agellon, L.B.; Vance, D.E. Structure, expression profile and alternative processing of the human phosphatidylethanolamine N-methyltransferase (PEMT) gene1Sequence data from this article have been deposited with the GenBank Data Library under accession numbers AF294460–AF294468 inclusive.1. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2001, 1532, 105–114. [Google Scholar] [CrossRef]
- Muratore, C.R.; Hodgson, N.W.; Trivedi, M.S.; Abdolmaleky, H.M.; Persico, A.M.; Lintas, C.; De La Monte, S.; Deth, R.C. Age-Dependent Decrease and Alternative Splicing of Methionine Synthase mRNA in Human Cerebral Cortex and an Accelerated Decrease in Autism. PLoS ONE 2013, 8, e56927. [Google Scholar] [CrossRef] [PubMed]
- Kraus, J.P.; Oliveriusová, J.; Sokolová, J.; Kraus, E.; Vlček, Č.; De Franchis, R.; Maclean, K.N.; Bao, L.; Bukovská, G.; Patterson, D.; et al. The Human Cystathionine β-Synthase (CBS) Gene: Complete Sequence, Alternative Splicing, and Polymorphisms. Genomics 1998, 52, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Mudd, S.H.; Skovby, F.; Levy, H.L.; Pettigrew, K.D.; Wilcken, B.; Pyeritz, R.E.; Andria, G.; Boers, G.H.; Bromberg, I.L.; Cerone, R. The natural history of homocystinuria due to cystathionine beta-synthase deficiency. Am. J. Hum. Genet. 1985, 37, 1–31. [Google Scholar]
- Filipovic, M.R.; Zivanovic, J.; Alvarez, B.; Banerjee, R. Chemical Biology of H2 S Signaling through Persulfidation. Chem. Rev. 2018, 118, 1253–1337. [Google Scholar] [CrossRef]
- Kabil, O.; Banerjee, R. Enzymology of H2 S Biogenesis, Decay and Signaling. Antioxid. Redox Signal. 2014, 20, 770–782. [Google Scholar] [CrossRef]
- Zuhra, K.; Augsburger, F.; Majtan, T.; Szabo, C. Cystathionine-β-synthase: Molecular Regulation and Pharmacological Inhibition. Biomolecules 2020, 10, 697. [Google Scholar] [CrossRef]
- Akimov, V.; Barrio-Hernandez, I.; Hansen, S.V.F.; Hallenborg, P.; Pedersen, A.-K.; Bekker-Jensen, D.B.; Puglia, M.; Christensen, S.D.K.; Vanselow, J.T.; Nielsen, M.M.; et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 2018, 25, 631–640. [Google Scholar] [CrossRef]
- Boeing, S.; Williamson, L.; Encheva, V.; Gori, I.; Saunders, R.E.; Instrell, R.; Aygün, O.; Rodriguez-Martinez, M.; Weems, J.C.; Kelly, G.P.; et al. Multiomic Analysis of the UV-Induced DNA Damage Response. Cell Rep. 2016, 15, 1597–1610. [Google Scholar] [CrossRef]
- Wagner, S.A.; Beli, P.; Weinert, B.T.; Nielsen, M.L.; Cox, J.; Mann, M.; Choudhary, C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteom. 2011, 10, M111-013284. [Google Scholar] [CrossRef] [PubMed]
- Mertins, P.; Qiao, J.W.; Patel, J.; Udeshi, N.D.; Clauser, K.R.; Mani, D.R.; Burgess, M.W.; Gillette, M.A.; Jaffe, J.D.; Carr, S.A. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods 2013, 10, 634–637. [Google Scholar] [CrossRef]
- Udeshi, N.D.; Svinkina, T.; Mertins, P.; Kuhn, E.; Mani, D.R.; Qiao, J.W.; Carr, S.A. Refined Preparation and Use of Anti-diglycine Remnant (K-ε-GG) Antibody Enables Routine Quantification of 10,000 s of Ubiquitination Sites in Single Proteomics Experiments. Mol. Cell. Proteom. 2013, 12, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J.; et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 2011, 44, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Lumpkin, R.J.; Gu, H.; Zhu, Y.; Leonard, M.; Ahmad, A.S.; Clauser, K.R.; Meyer, J.G.; Bennett, E.J.; Komives, E.A. Site-specific identification and quantitation of endogenous SUMO modifications under native conditions. Nat. Commun. 2017, 8, 1171. [Google Scholar] [CrossRef]
- Povlsen, L.K.; Beli, P.; Wagner, S.A.; Poulsen, S.L.; Sylvestersen, K.B.; Poulsen, J.W.; Nielsen, M.L.; Bekker-Jensen, S.; Mailand, N.; Choudhary, C. Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat. Cell Biol. 2012, 14, 1089–1098. [Google Scholar] [CrossRef]
- Weinert, B.T.; Scholz, C.; Wagner, S.A.; Iesmantavicius, V.; Su, D.; Daniel, J.A.; Choudhary, C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013, 4, 842–851. [Google Scholar] [CrossRef]
- Beli, P.; Lukashchuk, N.; Wagner, S.A.; Weinert, B.T.; Olsen, J.V.; Baskcomb, L.; Mann, M.; Jackson, S.P.; Choudhary, C. Proteomic Investigations Reveal a Role for RNA Processing Factor THRAP3 in the DNA Damage Response. Mol. Cell 2012, 46, 212–225. [Google Scholar] [CrossRef]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef]
- Bienvenut, W.V.; Sumpton, D.; Martinez, A.; Lilla, S.; Espagne, C.; Meinnel, T.; Giglione, C. Comparative Large Scale Characterization of Plant versus Mammal Proteins Reveals Similar and Idiosyncratic N-α-Acetylation Features. Mol. Cell. Proteom. 2012, 11, M111.015131. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Chen, Y.; Tishkoff, D.X.; Peng, C.; Tan, M.; Dai, L.; Xie, Z.; Zhang, Y.; Zwaans, B.M.M.; Skinner, M.E.; et al. SIRT5-Mediated Lysine Desuccinylation Impacts Diverse Metabolic Pathways. Mol. Cell 2013, 50, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Sylvestersen, K.B.; Horn, H.; Jungmichel, S.; Jensen, L.J.; Nielsen, M.L. Proteomic Analysis of Arginine Methylation Sites in Human Cells Reveals Dynamic Regulation During Transcriptional Arrest. Mol. Cell. Proteom. 2014, 13, 2072–2088. [Google Scholar] [CrossRef] [PubMed]
- Larsen, S.C.; Sylvestersen, K.B.; Mund, A.; Lyon, D.; Mullari, M.; Madsen, M.V.; Daniel, J.A.; Jensen, L.J.; Nielsen, M.L. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal. 2016, 9, rs9. [Google Scholar] [CrossRef]
- Olsen, J.B.; Cao, X.-J.; Han, B.; Chen, L.H.; Horvath, A.; Richardson, T.I.; Campbell, R.M.; Garcia, B.A.; Nguyen, H. Quantitative Profiling of the Activity of Protein Lysine Methyltransferase SMYD2 Using SILAC-Based Proteomics. Mol. Cell. Proteom. 2016, 15, 892–905. [Google Scholar] [CrossRef]
- Coukos, J.S.; Lee, C.W.; Pillai, K.S.; Liu, K.J.; Moellering, R.E. Widespread, Reversible Cysteine Modification by Methylglyoxal Regulates Metabolic Enzyme Function. ACS Chem. Biol. 2023, 18, 91–101. [Google Scholar] [CrossRef]
- Franchin, C.; Cesaro, L.; Salvi, M.; Millioni, R.; Iori, E.; Cifani, P.; James, P.; Arrigoni, G.; Pinna, L. Quantitative analysis of a phosphoproteome readily altered by the protein kinase CK2 inhibitor quinalizarin in HEK-293T cells. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2015, 1854, 609–623. [Google Scholar] [CrossRef]
- Kettenbach, A.N.; Schweppe, D.K.; Faherty, B.K.; Pechenick, D.; Pletnev, A.A.; Gerber, S.A. Quantitative Phosphoproteomics Identifies Substrates and Functional Modules of Aurora and Polo-Like Kinase Activities in Mitotic Cells. Sci. Signal. 2011, 4, rs5. [Google Scholar] [CrossRef]
- Santamaria, A.; Wang, B.; Elowe, S.; Malik, R.; Zhang, F.; Bauer, M.; Schmidt, A.; Silljé, H.H.W.; Körner, R.; Nigg, E.A. The Plk1-dependent Phosphoproteome of the Early Mitotic Spindle. Mol. Cell. Proteom. 2011, 10, M110.004457. [Google Scholar] [CrossRef]
- Rolland, D.; Basrur, V.; Conlon, K.; Wolfe, T.; Fermin, D.; Nesvizhskii, A.I.; Lim, M.S.; Elenitoba-Johnson, K.S.J. Global Phosphoproteomic Profiling Reveals Distinct Signatures in B-Cell Non-Hodgkin Lymphomas. Am. J. Pathol. 2014, 184, 1331–1342. [Google Scholar] [CrossRef]
- Stuart, S.A.; Houel, S.; Lee, T.; Wang, N.; Old, W.M.; Ahn, N.G. A Phosphoproteomic Comparison of B-RAFV600E and MKK1/2 Inhibitors in Melanoma Cells*. Mol. Cell. Proteom. 2015, 14, 1599–1615. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Xiao, C.-L.; Lu, C.-H.; Jia, H.-T.; Ge, F.; Wang, W.; Yin, X.-F.; Jia, H.-L.; He, J.-X.; He, Q.-Y. Phosphoproteome profile of human lung cancer cell line A549. Mol. BioSyst. 2011, 7, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Dephoure, N.; Zhou, C.; Villén, J.; Beausoleil, S.A.; Bakalarski, C.E.; Elledge, S.J.; Gygi, S.P. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 2008, 105, 10762–10767. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; D’Souza, R.C.; Tyanova, S.; Schaab, C.; Wisniewski, J.R.; Cox, J.; Mann, M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014, 8, 1583–1594. [Google Scholar] [CrossRef]
- Wu, F.; Wang, P.; Zhang, J.; Young, L.C.; Lai, R.; Li, L. Studies of phosphoproteomic changes induced by nucleophosmin-anaplastic lymphoma kinase (ALK) highlight deregulation of tumor necrosis factor (TNF)/Fas/TNF-related apoptosis-induced ligand signaling pathway in ALK-positive anaplastic large cell lymphoma. Mol. Cell. Proteom. 2010, 9, 1616–1632. [Google Scholar] [CrossRef]
- Shiromizu, T.; Adachi, J.; Watanabe, S.; Murakami, T.; Kuga, T.; Muraoka, S.; Tomonaga, T. Identification of Missing Proteins in the neXtProt Database and Unregistered Phosphopeptides in the PhosphoSitePlus Database As Part of the Chromosome-Centric Human Proteome Project. J. Proteome Res. 2013, 12, 2414–2421. [Google Scholar] [CrossRef]
- Brill, L.M.; Xiong, W.; Lee, K.-B.; Ficarro, S.B.; Crain, A.; Xu, Y.; Terskikh, A.; Snyder, E.Y.; Ding, S. Phosphoproteomic Analysis of Human Embryonic Stem Cells. Cell Stem Cell 2009, 5, 204–213. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, F.; Fu, Y.; Huang, X.; Wang, W.; Jiang, X.; Gritsenko, M.A.; Zhao, R.; Monore, M.E.; Pertz, O.C.; et al. Spatial Phosphoprotein Profiling Reveals a Compartmentalized Extracellular Signal-regulated Kinase Switch Governing Neurite Growth and Retraction. J. Biol. Chem. 2011, 286, 18190–18201. [Google Scholar] [CrossRef]
- Mayya, V.; Lundgren, D.H.; Hwang, S.-I.; Rezaul, K.; Wu, L.; Eng, J.K.; Rodionov, V.; Han, D.K. Quantitative Phosphoproteomic Analysis of T Cell Receptor Signaling Reveals System-Wide Modulation of Protein-Protein Interactions. Sci. Signal. 2009, 2, ra46. [Google Scholar] [CrossRef]
- Tsai, C.-F.; Wang, Y.-T.; Yen, H.-Y.; Tsou, C.-C.; Ku, W.-C.; Lin, P.-Y.; Chen, H.-Y.; Nesvizhskii, A.I.; Ishihama, Y.; Chen, Y.-J. Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics. Nat. Commun. 2015, 6, 6622. [Google Scholar] [CrossRef]
- Wu, X.; Tian, L.; Li, J.; Zhang, Y.; Han, V.; Li, Y.; Xu, X.; Li, H.; Chen, X.; Chen, J.; et al. Investigation of receptor interacting protein (RIP3)-dependent protein phosphorylation by quantitative phosphoproteomics. Mol. Cell. Proteom. 2012, 11, 1640–1651. [Google Scholar] [CrossRef] [PubMed]
- Helou, Y.A.; Nguyen, V.; Beik, S.P.; Salomon, A.R. ERK Positive Feedback Regulates a Widespread Network of Tyrosine Phosphorylation Sites across Canonical T Cell Signaling and Actin Cytoskeletal Proteins in Jurkat T Cells. PLoS ONE 2013, 8, e69641. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.; Nardone, J.; Wang, Y.; Loriaux, M.; Villén, J.; Beausoleil, S.; Tucker, M.; Kornhauser, J.; Ren, J.; MacNeill, J.; et al. Survey of Activated FLT3 Signaling in Leukemia. PLoS ONE 2011, 6, e19169. [Google Scholar] [CrossRef] [PubMed]
- Ding, V.M.Y.; Boersema, P.J.; Foong, L.Y.; Preisinger, C.; Koh, G.; Natarajan, S.; Lee, D.-Y.; Boekhorst, J.; Snel, B.; Lemeer, S.; et al. Tyrosine Phosphorylation Profiling in FGF-2 Stimulated Human Embryonic Stem Cells. PLoS ONE 2011, 6, e17538. [Google Scholar] [CrossRef]
- Pinto, S.M.; Nirujogi, R.S.; Rojas, P.L.; Patil, A.H.; Manda, S.S.; Subbannayya, Y.; Roa, J.C.; Chatterjee, A.; Prasad, T.S.K.; Pandey, A. Quantitative phosphoproteomic analysis of IL-33-mediated signaling. Proteomics 2015, 15, 532–544. [Google Scholar] [CrossRef]
- Luo, W.; Slebos, R.J.; Hill, S.; Li, M.; Brabek, J.; Amanchy, R.; Chaerkady, R.; Pandey, A.; Ham, A.J.; Hanks, S.K. Global impact of oncogenic Src on a phosphotyrosine proteome. J. Proteome Res. 2008, 7, 3447–3460. [Google Scholar] [CrossRef]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R.; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR Substrate Analysis Reveals Extensive Protein Networks Responsive to DNA Damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef]
- Gnad, F.; Young, A.; Zhou, W.; Lyle, K.; Ong, C.C.; Stokes, M.P.; Silva, J.C.; Belvin, M.; Friedman, L.S.; Koeppen, H.; et al. Systems-wide Analysis of K-Ras, Cdc42, and PAK4 Signaling by Quantitative Phosphoproteomics. Mol. Cell. Proteom. 2013, 12, 2070–2080. [Google Scholar] [CrossRef]
- Reinartz, M.; Raupach, A.; Kaisers, W.; Gödecke, A. AKT1 and AKT2 Induce Distinct Phosphorylation Patterns in HL-1 Cardiac Myocytes. J. Proteome Res. 2014, 13, 4232–4245. [Google Scholar] [CrossRef]
- Gauci, S.; Helbig, A.O.; Slijper, M.; Krijgsveld, J.; Heck, A.J.R.; Mohammed, S. Lys-N and Trypsin Cover Complementary Parts of the Phosphoproteome in a Refined SCX-Based Approach. Anal. Chem. 2009, 81, 4493–4501. [Google Scholar] [CrossRef]
- Grosstessner-Hain, K.; Hegemann, B.; Novatchkova, M.; Rameseder, J.; Joughin, B.A.; Hudecz, O.; Roitinger, E.; Pichler, P.; Kraut, N.; Yaffe, M.B.; et al. Quantitative Phospho-proteomics to Investigate the Polo-like Kinase 1-Dependent Phospho-proteome. Mol. Cell. Proteom. 2011, 10, M111.008540. [Google Scholar] [CrossRef]
- Klammer, M.; Kaminski, M.; Zedler, A.; Oppermann, F.; Blencke, S.; Marx, S.; Müller, S.; Tebbe, A.; Godl, K.; Schaab, C. Phosphosignature Predicts Dasatinib Response in Non-small Cell Lung Cancer. Mol. Cell. Proteom. 2012, 11, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Di Palma, S.; Preisinger, C.; Peng, M.; Polat, A.N.; Heck, A.J.; Mohammed, S. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 2013, 12, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.V.; Vermeulen, M.; Santamaria, A.; Kumar, C.; Miller, M.L.; Jensen, L.J.; Gnad, F.; Cox, J.; Jensen, T.S.; Nigg, E.A.; et al. Quantitative Phosphoproteomics Reveals Widespread Full Phosphorylation Site Occupancy During Mitosis. Sci. Signal. 2010, 3, ra3. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, S.J.; Yang, G.; Yang, P.; Fazakerley, D.J.; Stöckli, J.; Yang, J.Y.; James, D.E. Dynamic Adipocyte Phosphoproteome Reveals that Akt Directly Regulates mTORC2. Cell Metab. 2013, 17, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.L.; Yang, G.; Humphrey, S.J.; Chaudhuri, R.; Ma, X.; Peterman, S.; James, D.E. Targeted phosphoproteomics of insulin signaling using data-independent acquisition mass spectrometry. Sci. Signal. 2015, 8, rs6. [Google Scholar] [CrossRef]
- Rigbolt, K.T.G.; Prokhorova, T.A.; Akimov, V.; Henningsen, J.; Johansen, P.T.; Kratchmarova, I.; Kassem, M.; Mann, M.; Olsen, J.V.; Blagoev, B. System-Wide Temporal Characterization of the Proteome and Phosphoproteome of Human Embryonic Stem Cell Differentiation. Sci. Signal. 2011, 4, rs3. [Google Scholar] [CrossRef]
- Moritz, A.; Li, Y.; Guo, A.; Villen, J.; Wang, Y.; MacNeill, J.; Kornhauser, J.; Sprott, K.; Zhou, J.; Possemato, A.; et al. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci. Signal. 2010, 3, ra64. [Google Scholar] [CrossRef]
- Wagner, S.A.; Beli, P.; Weinert, B.T.; Scholz, C.; Kelstrup, C.D.; Young, C.; Nielsen, M.L.; Olsen, J.V.; Brakebusch, C.; Choudhary, C. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell. Proteom. 2012, 11, 1578–1585. [Google Scholar] [CrossRef]
- Zielinska, D.F.; Gnad, F.; Wiśniewski, J.R.; Mann, M. Precision Mapping of an In Vivo N-Glycoproteome Reveals Rigid Topological and Sequence Constraints. Cell 2010, 141, 897–907. [Google Scholar] [CrossRef]
- Lundby, A.; Lage, K.; Weinert, B.T.; Bekker-Jensen, D.B.; Secher, A.; Skovgaard, T.; Kelstrup, C.D.; Dmytriyev, A.; Choudhary, C.; Lundby, C.; et al. Proteomic Analysis of Lysine Acetylation Sites in Rat Tissues Reveals Organ Specificity and Subcellular Patterns. Cell Rep. 2012, 2, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Xu, W.; Jiang, W.; Yu, W.; Lin, Y.; Zhang, T.; Yao, J.; Zhou, L.; Zeng, Y.; Li, H.; et al. Regulation of Cellular Metabolism by Protein Lysine Acetylation. Science 2010, 327, 1000–1004. [Google Scholar] [CrossRef] [PubMed]
- Simon, G.M.; Cheng, J.; Gordon, J.I. Quantitative assessment of the impact of the gut microbiota on lysine ε-acetylation of host proteins using gnotobiotic mice. Proc. Natl. Acad. Sci. USA 2012, 109, 11133–11138. [Google Scholar] [CrossRef] [PubMed]
- Koronowski, K.B.; Greco, C.M.; Huang, H.; Kim, J.-K.; Fribourgh, J.L.; Crosby, P.; Mathur, L.; Ren, X.; Partch, C.L.; Jang, C.; et al. Ketogenesis impact on liver metabolism revealed by proteomics of lysine β-hydroxybutyrylation. Cell Rep. 2021, 36, 109487. [Google Scholar] [CrossRef]
- Yang, Z.; Yan, C.; Ma, J.; Peng, P.; Ren, X.; Cai, S.; Shen, X.; Wu, Y.; Zhang, S.; Wang, X.; et al. Lactylome analysis suggests lactylation-dependent mechanisms of metabolic adaptation in hepatocellular carcinoma. Nat. Metab. 2023, 5, 61–79. [Google Scholar] [CrossRef]
- Nishida, Y.; Rardin, M.J.; Carrico, C.; He, W.; Sahu, A.K.; Gut, P.; Najjar, R.; Fitch, M.; Hellerstein, M.; Gibson, B.W.; et al. SIRT5 Regulates both Cytosolic and Mitochondrial Protein Malonylation with Glycolysis as a Major Target. Mol. Cell 2015, 59, 321–332. [Google Scholar] [CrossRef]
- Pham, V.N.; Bruemmer, K.J.; Toh, J.D.W.; Ge, E.J.; Tenney, L.; Ward, C.C.; Dingler, F.A.; Millington, C.L.; Garcia-Prieto, C.A.; Pulos-Holmes, M.C.; et al. Formaldehyde regulates S -adenosylmethionine biosynthesis and one-carbon metabolism. Science 2023, 382, eabp9201. [Google Scholar] [CrossRef]
- Bian, Y.; Song, C.; Cheng, K.; Dong, M.; Wang, F.; Huang, J.; Sun, D.; Wang, L.; Ye, M.; Zou, H. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J Proteom. 2014, 96, 253–262. [Google Scholar] [CrossRef]
- Gu, T.-L.; Deng, X.; Huang, F.; Tucker, M.; Crosby, K.; Rimkunas, V.; Wang, Y.; Deng, G.; Zhu, L.; Tan, Z.; et al. Survey of Tyrosine Kinase Signaling Reveals ROS Kinase Fusions in Human Cholangiocarcinoma. PLoS ONE 2011, 6, e15640. [Google Scholar] [CrossRef]
- Mertins, P.; Yang, F.; Liu, T.; Mani, D.R.; Petyuk, V.A.; Gillette, M.A.; Clauser, K.R.; Qiao, J.W.; Gritsenko, M.A.; Moore, R.J.; et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol. Cell. Proteom. 2014, 13, 1690–1704. [Google Scholar] [CrossRef]
- Dai, J.; Jin, W.-H.; Sheng, Q.-H.; Shieh, C.-H.; Wu, J.-R.; Zeng, R. Protein Phosphorylation and Expression Profiling by Yin-Yang Multidimensional Liquid Chromatography (Yin-Yang MDLC) Mass Spectrometry. J. Proteome Res. 2007, 6, 250–262. [Google Scholar] [CrossRef] [PubMed]
- Villen, J.; Beausoleil, S.A.; Gerber, S.A.; Gygi, S.P. Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. USA 2007, 104, 1488–1493. [Google Scholar] [CrossRef] [PubMed]
- Wilson-Grady, J.T.; Haas, W.; Gygi, S.P. Quantitative comparison of the fasted and re-fed mouse liver phosphoproteomes using lower pH reductive dimethylation. Methods 2013, 61, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Goswami, T.; Li, X.; Smith, A.M.; Luderowski, E.M.; Vincent, J.J.; Rush, J.; Ballif, B.A. Comparative phosphoproteomic analysis of neonatal and adult murine brain. Proteomics 2012, 12, 2185–2189. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Jedrychowski, M.P.; Elias, J.E.; Goswami, T.; Rad, R.; Beausoleil, S.A.; Villen, J.; Haas, W.; Sowa, M.E.; Gygi, S.P. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 2010, 143, 1174–1189. [Google Scholar] [CrossRef]
- Manes, N.P.; Dong, L.; Zhou, W.; Du, X.; Reghu, N.; Kool, A.C.; Choi, D.; Bailey, C.L.; Petricoin, E.F.; Liotta, L.A.; et al. Discovery of Mouse Spleen Signaling Responses to Anthrax using Label-Free Quantitative Phosphoproteomics via Mass Spectrometry. Mol. Cell. Proteom. 2011, 10, M110.000927. [Google Scholar] [CrossRef]
- Demirkan, G.; Yu, K.; Boylan, J.M.; Salomon, A.R.; Gruppuso, P.A. Phosphoproteomic Profiling of In Vivo Signaling in Liver by the Mammalian Target of Rapamycin Complex 1 (mTORC1). PLoS ONE 2011, 6, e21729. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Nagaraj, N.; Zougman, A.; Gnad, F.; Mann, M. Brain phosphoproteome obtained by a FASP-based method reveals plasma membrane protein topology. J. Proteome Res. 2010, 9, 3280–3289. [Google Scholar] [CrossRef]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016, 534, 55–62. [Google Scholar] [CrossRef]
- Robles, M.S.; Humphrey, S.J.; Mann, M. Phosphorylation Is a Central Mechanism for Circadian Control of Metabolism and Physiology. Cell Metab. 2017, 25, 118–127. [Google Scholar] [CrossRef]
- Lundby, A.; Secher, A.; Lage, K.; Nordsborg, N.B.; Dmytriyev, A.; Lundby, C.; Olsen, J.V. Quantitative maps of protein phosphorylation sites across 14 different rat organs and tissues. Nat. Commun. 2012, 3, 876. [Google Scholar] [CrossRef] [PubMed]
- Grimsrud, P.A.; Carson, J.J.; Hebert, A.S.; Hubler, S.L.; Niemi, N.M.; Bailey, D.J.; Jochem, A.; Stapleton, D.S.; Keller, M.P.; Westphall, M.S.; et al. A quantitative map of the liver mitochondrial phosphoproteome reveals posttranslational control of ketogenesis. Cell Metab. 2012, 16, 672–683. [Google Scholar] [CrossRef] [PubMed]
- Burnett, G.; Kennedy, E.P. The enzymatic phosphorylation of proteins. J. Biol. Chem. 1954, 211, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Pawson, T.; Scott, J.D. Protein phosphorylation in signaling—50 years and counting. Trends Biochem. Sci. 2005, 30, 286–290. [Google Scholar] [CrossRef]
- Pajares, M.A.; Duran, C.; Corrales, F.; Mato, J.M. Protein kinase C phosphorylation of rat liver S-adenosylmethionine synthetase: Dissociation and production of an active monomer. Biochem. J. 1994, 303 Pt 3, 949–955. [Google Scholar] [CrossRef][Green Version]
- Barbier-Torres, L.; Murray, B.; Yang, J.W.; Wang, J.; Matsuda, M.; Robinson, A.; Binek, A.; Fan, W.; Fernández-Ramos, D.; Lopitz-Otsoa, F.; et al. Depletion of mitochondrial methionine adenosyltransferase α1 triggers mitochondrial dysfunction in alcohol-associated liver disease. Nat. Commun. 2022, 13, 557. [Google Scholar] [CrossRef]
- Lu, L.; Zhang, J.; Fan, W.; Li, Y.; Wang, J.; Li, T.W.H.; Barbier-Torres, L.; Mato, J.M.; Liu, T.; Seki, E.; et al. Deregulated 14-3-3ζ and methionine adenosyltransferase α1 interplay promotes liver cancer tumorigenesis in mice and humans. Oncogene 2021, 40, 5866–5879. [Google Scholar] [CrossRef]
- Kotb, M.; Kredich, N.M. S-Adenosylmethionine synthetase from human lymphocytes. Purification and characterization. J. Biol. Chem. 1985, 260, 3923–3930. [Google Scholar] [CrossRef]
- Panayiotidis, M.I.; Stabler, S.P.; Ahmad, A.; Pappa, A.; Legros, L.H.; Hernandez-Saavedra, D.; Schneider, B.K.; Allen, R.H.; Vasiliou, V.; McCord, J.M.; et al. Activation of a novel isoform of methionine adenosyl transferase 2A and increased S-adenosylmethionine turnover in lung epithelial cells exposed to hyperoxia. Free. Radic. Biol. Med. 2006, 40, 348–358. [Google Scholar] [CrossRef]
- Ramani, K.; Donoyan, S.; Tomasi, M.L.; Park, S. Role of Methionine Adenosyltransferase alpha2 and beta Phosphorylation and Stabilization in Human Hepatic Stellate Cell Trans-Differentiation. J. Cell. Physiol. 2014, 230, 1075–1085. [Google Scholar] [CrossRef]
- Shafqat, N.; Muniz, J.R.; Pilka, E.S.; Papagrigoriou, E.; von Delft, F.; Oppermann, U.; Yue, W.W. Insight into S-adenosylmethionine biosynthesis from the crystal structures of the human methionine adenosyltransferase catalytic and regulatory subunits. Biochem. J. 2013, 452, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Herr, P.; Boström, J.; Rullman, E.; Rudd, S.G.; Vesterlund, M.; Lehtiö, J.; Helleday, T.; Maddalo, G.; Altun, M. Cell Cycle Profiling Reveals Protein Oscillation, Phosphorylation, and Localization Dynamics. Mol. Cell. Proteom. 2020, 19, 608–623. [Google Scholar] [CrossRef] [PubMed]
- Ndzeidze, R.; Leestemaker-Palmer, A.; Danelishvili, L.; Bermudez, L.E. Virulent Mycobacterium avium subspecies hominissuis subverts macrophages during early stages of infection. Microbiology 2022, 168, 001133. [Google Scholar] [CrossRef] [PubMed]
- Castaño, J.G.; Alemany, S.; Nieto, A.; Mato, J.M. Activation of phospholipid methyltransferase by glucagon in rat hepatocytes. J. Biol. Chem. 1980, 255, 9041–9043. [Google Scholar] [CrossRef]
- Villalba, M.; Varela, I.; Mérida, I.; Pajares, M.A.; Del Pozo, A.M.; Mato José, M. Modulation by the ratio S-adenosylmethionineS-adenosylhomocysteine of cyclic AMP-dependent phosphorylation of the 50 kDa protein of rat liver phospholipid methyltransferase. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 1985, 847, 273–279. [Google Scholar] [CrossRef]
- Villalba, M.; Pajares, M.A.; Renart, M.F.; Mato, J.M. Protein kinase C catalyses the phosphorylation and activation of rat liver phospholipid methyltransferase. Biochem. J. 1987, 241, 911–916. [Google Scholar] [CrossRef]
- Ridgway, N.D.; Vance, D.E. In vitro phosphorylation of phosphatidylethanolamine N-methyltransferase by cAMP-dependent protein kinase: Lack of in vivo phosphorylation in response to N6-2′-O-dibutryladenosine 3′,5′-cyclic monophosphate. Biochim. Biophys. Acta (BBA) Lipids Lipid Metab. 1989, 1004, 261–270. [Google Scholar] [CrossRef]
- Luka, Z.; Ham, A.L.; Norris, J.L.; Yeo, E.; Yermalitsky, V.; Glenn, B.; Caprioli, R.M.; Liebler, D.C.; Wagner, C. Identification of phosphorylation sites in glycine N-methyltransferase from rat liver. Protein Sci. 2006, 15, 785–794. [Google Scholar] [CrossRef]
- Wagner, C.; Decha-Umphai, W.; Corbin, J. Phosphorylation modulates the activity of glycine N-methyltransferase, a folate binding protein. In vitro phosphorylation is inhibited by the natural folate ligand. J. Biol. Chem. 1989, 264, 9638–9642. [Google Scholar] [CrossRef]
- Yang, M.-H.; Liao, C.-C.; Hung, J.-H.; Lai, X.-T.; Yen, C.-H.; Chen, Y.-M.A. Utilizing proteomic approach to identify nuclear translocation related serine kinase phosphorylation site of GNMT as downstream effector for benzo[a]pyrene. J. Food Drug Anal. 2019, 27, 603–609. [Google Scholar] [CrossRef]
- Alegre, S.; Pascual, J.; Trotta, A.; Angeleri, M.; Rahikainen, M.; Brosche, M.; Moffatt, B.; Kangasjärvi, S. Evolutionary conservation and post-translational control of S-adenosyl-L-homocysteine hydrolase in land plants. PLoS ONE 2020, 15, e0227466. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Liu, W.; Xiang, R.; Li, X.; Hou, S.; Xu, L.; Wang, L.; Zhao, D.; Liu, X.; Wang, G.; et al. Ribosomal modification protein rimK-like family member A activates betaine-homocysteine S-methyltransferase 1 to ameliorate hepatic steatosis. Signal Transduct. Target. Ther. 2024, 9, 214. [Google Scholar] [CrossRef] [PubMed]
- d’Emmanuele Di Villa Bianca, R.; Mitidieri, E.; Fusco, F.; Russo, A.; Pagliara, V.; Tramontano, T.; Donnarumma, E.; Mirone, V.; Cirino, G.; Russo, G.; et al. Urothelium muscarinic activation phosphorylates CBSSer227 via cGMP/PKG pathway causing human bladder relaxation through H2S production. Sci. Rep. 2016, 6, 31491. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.; Pardue, S.; Shen, X.; Glawe, J.D.; Yagi, T.; Bhuiyan, M.A.N.; Patel, R.P.; Dominic, P.S.; Virk, C.S.; Bhuiyan, M.S.; et al. Hypoxia increases persulfide and polysulfide formation by AMP kinase dependent cystathionine gamma lyase phosphorylation. Redox Biol. 2023, 68, 102949. [Google Scholar] [CrossRef]
- Renga, B.; Bucci, M.; Cipriani, S.; Carino, A.; Monti, M.C.; Zampella, A.; Gargiulo, A.; d’Emmanuele Di Villa Bianca, R.; Distrutti, E.; Fiorucci, S. Cystathionine γ-lyase, a H2 S-generating enzyme, is a GPBAR1-regulated gene and contributes to vasodilation caused by secondary bile acids. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H114–H126. [Google Scholar] [CrossRef]
- Renga, B.; Cipriani, S.; Carino, A.; Simonetti, M.; Zampella, A.; Fiorucci, S. Reversal of Endothelial Dysfunction by GPBAR1 Agonism in Portal Hypertension Involves a AKT/FOXOA1 Dependent Regulation of H2S Generation and Endothelin-1. PLoS ONE 2015, 10, e0141082. [Google Scholar] [CrossRef]
- Xu, X.; Yan, Q.; Liu, X.; Li, P.; Li, X.; Chen, Y.; Simoncini, T.; Liu, J.; Zhu, D.; Fu, X. 17β-Estradiol nongenomically induces vascular endothelial H2S release by promoting phosphorylation of cystathionine γ-lyase. J. Biol. Chem. 2019, 294, 15577–15592. [Google Scholar] [CrossRef]
- Bibli, S.-I.; Hu, J.; Sigala, F.; Wittig, I.; Heidler, J.; Zukunft, S.; Tsilimigras, D.I.; Randriamboavonjy, V.; Wittig, J.; Kojonazarov, B.; et al. Cystathionine γ Lyase Sulfhydrates the RNA Binding Protein Human Antigen R to Preserve Endothelial Cell Function and Delay Atherogenesis. Circulation 2019, 139, 101–114. [Google Scholar] [CrossRef]
- Bibli, S.-I.; Hu, J.; Leisegang, M.S.; Wittig, J.; Zukunft, S.; Kapasakalidi, A.; Fisslthaler, B.; Tsilimigras, D.; Zografos, G.; Filis, K.; et al. Shear stress regulates cystathionine γ lyase expression to preserve endothelial redox balance and reduce membrane lipid peroxidation. Redox Biol. 2020, 28, 101379. [Google Scholar] [CrossRef]
- Yuan, G.; Vasavda, C.; Peng, Y.-J.; Makarenko, V.V.; Raghuraman, G.; Nanduri, J.; Gadalla, M.M.; Semenza, G.L.; Kumar, G.K.; Snyder, S.H.; et al. Protein kinase G–regulated production of H2 S governs oxygen sensing. Sci. Signal. 2015, 8, ra37. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, Z.-H.; Yang, H.-B.; Zhang, Y.; Zhao, X.-N.; Zhang, M.; Liu, Y.-B.; Xu, Y.-Y.; Lei, Q.-Y. Cullin 3 targets methionine adenosyltransferase IIα for ubiquitylation-mediated degradation and regulates colorectal cancer cell proliferation. FEBS J. 2016, 283, 2390–2402. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ou, Y.; Zhou, Q.; Liang, Y.; Li, W.; Chen, Y.; Chen, W.; Wu, S.; Chen, Y.; Dai, X.; et al. Methionine orchestrates the metabolism vulnerability in cisplatin resistant bladder cancer microenvironment. Cell Death Dis. 2023, 14, 525. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.B.; Xu, Y.Y.; Zhao, X.N.; Zou, S.W.; Zhang, Y.; Zhang, M.; Li, J.T.; Ren, F.; Wang, L.Y.; Lei, Q.Y. Acetylation of MAT IIalpha represses tumour cell growth and is decreased in human hepatocellular cancer. Nat. Commun. 2015, 6, 6973. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-T.; Yang, H.; Lei, M.-Z.; Zhu, W.-P.; Su, Y.; Li, K.-Y.; Zhu, W.-Y.; Wang, J.; Zhang, L.; Qu, J.; et al. Dietary folate drives methionine metabolism to promote cancer development by stabilizing MAT IIA. Signal Transduct. Target. Ther. 2022, 7, 192. [Google Scholar] [CrossRef]
- Chen, Y.; Feng, X.; Wu, Z.; Yang, Y.; Rao, X.; Meng, R.; Zhang, S.; Dong, X.; Xu, S.; Wu, G.; et al. USP9X-mediated REV1 deubiquitination promotes lung cancer radioresistance via the action of REV1 as a Rad18 molecular scaffold for cystathionine γ-lyase. J. Biomed. Sci. 2024, 31, 55. [Google Scholar] [CrossRef]
- Bai, L.; Qi, Y.; Chen, S.; Wang, J.; Tang, C.; Du, J.; Jin, H.; Huang, Y. Angiotensin II downregulates vascular endothelial cell hydrogen sulfide production by enhancing cystathionine γ-lyase degradation through ROS-activated ubiquitination pathway. Biochem. Biophys. Res. Commun. 2019, 514, 907–912. [Google Scholar] [CrossRef]
- Chi, Z.; Le, T.P.H.; Lee, S.K.; Guo, E.; Kim, D.; Lee, S.; Seo, S.; Lee, S.Y.; Kim, J.H.; Lee, S.Y. Honokiol ameliorates angiotensin II-induced hypertension and endothelial dysfunction by inhibiting HDAC6-mediated cystathionine γ-lyase degradation. J. Cell. Mol. Med. 2020, 24, 10663–10676. [Google Scholar] [CrossRef]
- Chi, Z.; Byeon, H.-E.; Seo, E.; Nguyen, Q.-A.T.; Lee, W.; Jeong, Y.; Choi, J.; Pandey, D.; Berkowitz, D.E.; Kim, J.H.; et al. Histone deacetylase 6 inhibitor tubastatin A attenuates angiotensin II-induced hypertension by preventing cystathionine γ-lyase protein degradation. Pharmacol. Res. 2019, 146, 104281. [Google Scholar] [CrossRef]
- Das, A.; Thapa, P.; Santiago, U.; Shanmugam, N.; Banasiak, K.; Dąbrowska, K.; Nolte, H.; Szulc, N.A.; Gathungu, R.M.; Cysewski, D.; et al. A heterotypic assembly mechanism regulates CHIP E3 ligase activity. EMBO J. 2022, 41, e109566. [Google Scholar] [CrossRef]
- Floris, A.; Chandla, S.; Lim, Y.; Barbier-Torres, L.; Seth, K.; Khangholi, A.; Li, T.W.H.; Robison, A.; Murray, B.J.; Lee, S.; et al. Sumoylation of methionine adenosyltransferase alpha 1 promotes mitochondrial dysfunction in alcohol-associated liver disease. Hepatology 2024, 80, 102–118. [Google Scholar] [CrossRef]
- Tomasi, M.L.; Ryoo, M.; Ramani, K.; Tomasi, I.; Giordano, P.; Mato, J.M.; Lu, S.C. Methionine adenosyltransferase α2 sumoylation positively regulate Bcl-2 expression in human colon and liver cancer cells. Oncotarget 2015, 6, 37706–37723. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Luo, J.; Zhang, Y.; Zheng, S.; Zhang, H.; Huang, Y.; Wong, J.; Li, J. SUMOylation is enriched in the nuclear matrix and required for chromosome segregation. J. Biol. Chem. 2024, 300, 105547. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Fang, J.; Xie, X.; Carrico, C.; Meyer, J.G.; Wei, L.; Bons, J.; Rose, J.; Riley, R.; Kwok, R.; et al. Regulation of urea cycle by reversible high-stoichiometry lysine succinylation. Nat. Metab. 2024, 6, 550–566. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Banerjee, R. Human Polycomb 2 Protein Is a SUMO E3 Ligase and Alleviates Substrate-Induced Inhibition of Cystathionine β-Synthase Sumoylation. PLoS ONE 2008, 3, e4032. [Google Scholar] [CrossRef]
- Wang, Y.; Kavran, J.M.; Chen, Z.; Karukurichi, K.R.; Leahy, D.J.; Cole, P.A. Regulation of S-adenosylhomocysteine hydrolase by lysine acetylation. J. Biol. Chem. 2014, 289, 31361–31372. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Guo, H.; Lu, G.; Gu, J.; Wang, X.; Zhang, X.-E.; Deng, J. Lysine acetylation regulates the activity of Escherichia coli S-adenosylmethionine synthase. Acta Biochim. Biophys. Sin. 2016, 48, 723–731. [Google Scholar] [CrossRef]
- Wan, X.; Zeng, W.; Fan, H.; Wang, C.; Han, S.; Sun, Z.; Tang, M.; Shao, J.; Liu, Y.; Fang, Y.; et al. MAT2B regulates the protein level of MAT2A to preserve RNA N6-methyladenosine. Cell Death Dis. 2024, 15, 714. [Google Scholar] [CrossRef]
- Shen, S.; Liu, R.; Huang, J.; Sun, Y.; Tan, Q.; Luo, Q.; Liu, R. MAT1A activation of glycolysis to promote NSCLC progression depends on stabilizing CCND1. Cell Death Dis. 2024, 15, 768. [Google Scholar] [CrossRef]
- Morita, S.; Takeuchi, A.; Kitagawa, S. Functional analysis of two isoforms of phosphatidylethanolamine N-methyltransferase. Biochem. J. 2010, 432, 387–398. [Google Scholar] [CrossRef]
- Zhu, Q.; Cheng, X.; Cheng, Y.; Chen, J.; Xu, H.; Gao, Y.; Duan, X.; Ji, J.; Li, X.; Yi, W. O-GlcNAcylation regulates the methionine cycle to promote pluripotency of stem cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7755–7763. [Google Scholar] [CrossRef]
- Ichikawa, A.; Ohashi, Y.; Terada, S.; Natsuka, S.; Ikura, K. In vitro modification of betaine-homocysteine S-methyltransferase by tissue-type transglutaminase. Int. J. Biochem. Cell Biol. 2004, 36, 1981–1992. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, A.; Ishizaki, J.; Morita, M.; Tanaka, K.; Ikura, K. Identification of new amine acceptor protein substrate candidates of transglutaminase in rat liver extract: Use of 5-(biotinamido) pentylamine as a probe. Biosci. Biotechnol. Biochem. 2008, 72, 1056–1062. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gonzalez, B.; Pajares, M.A.; Martinez-Ripoll, M.; Blundell, T.L.; Sanz-Aparicio, J. Crystal structure of rat liver betaine homocysteine s-methyltransferase reveals new oligomerization features and conformational changes upon substrate binding. J. Mol. Biol. 2004, 338, 771–782. [Google Scholar] [CrossRef]
- Garrido, F.; Pacheco, M.; Vargas-Martinez, R.; Velasco-Garcia, R.; Jorge, I.; Serrano, H.; Portillo, F.; Vazquez, J.; Pajares, M.A. Identification of hepatic protein-protein interaction targets for betaine homocysteine S-methyltransferase. PLoS ONE 2018, 13, e0199472. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Su, H.; Liu, X.; Wang, H.; Feng, Y.; Wang, Y.; Chen, H.; Dai, L.; Lai, S.; Xu, S.; et al. mTORC1-c-Myc pathway rewires methionine metabolism for HCC progression through suppressing SIRT4 mediated ADP ribosylation of MAT2A. Cell Biosci. 2022, 12, 183. [Google Scholar] [CrossRef]
- Yang, H.; Liu, T.; Wang, J.; Li, T.W.H.; Fan, W.; Peng, H.; Krishnan, A.; Gores, G.J.; Mato, J.M.; Lu, S.C. Deregulated methionine adenosyltransferase α1, c-Myc, and Maf proteins together promote cholangiocarcinoma growth in mice and humans‡. Hepatology 2016, 64, 439–455. [Google Scholar] [CrossRef]
- Murray, B.; Antonyuk, S.V.; Marina, A.; Lu, S.C.; Mato, J.M.; Hasnain, S.S.; Rojas, A.L. Crystallography captures catalytic steps in human methionine adenosyltransferase enzymes. Proc. Natl. Acad. Sci. USA 2016, 113, 2104–2109. [Google Scholar] [CrossRef]
- Cabrero, C.; Duce, A.M.; Ortiz, P.; Alemany, S.; Mato, J.M. Specific loss of the high-molecular-weight form of S-adenosyl-L-methionine synthetase in human liver cirrhosis. Hepatology 1988, 8, 1530–1534. [Google Scholar] [CrossRef]
- Corrales, F.; Ochoa, P.; Rivas, C.; Martin-Lomas, M.; Mato, J.M.; Pajares, M.A. Inhibition of glutathione synthesis in the liver leads to S-adenosyl-L-methionine synthetase reduction. Hepatology 1991, 14, 528–533. [Google Scholar]
- Corrales, F.; Gimenez, A.; Alvarez, L.; Caballeria, J.; Pajares, M.A.; Andreu, H.; Pares, A.; Mato, J.M.; Rodes, J. S-adenosylmethionine treatment prevents carbon tetrachloride-induced S-adenosylmethionine synthetase inactivation and attenuates liver injury. Hepatology 1992, 16, 1022–1027. [Google Scholar] [CrossRef]
- Avila, M.A.; Mingorance, J.; Martinez-Chantar, M.L.; Casado, M.; Martin-Sanz, P.; Bosca, L.; Mato, J.M. Regulation of rat liver S-adenosylmethionine synthetase during septic shock: Role of nitric oxide. Hepatology 1997, 25, 391–396. [Google Scholar] [PubMed]
- Sanchez-Gongora, E.; Ruiz, F.; Mingorance, J.; An, W.; Corrales, F.J.; Mato, J.M. Interaction of liver methionine adenosyltransferase with hydroxyl radical. FASEB J. 1997, 11, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Pajares, M.A.; Corrales, F.J.; Ochoa, P.; Mato, J.M. The role of cysteine-150 in the structure and activity of rat liver S-adenosyl-L-methionine synthetase. Biochem. J. 1991, 274 Pt 1, 225–229. [Google Scholar] [CrossRef]
- Ruiz, F.; Corrales, F.J.; Miqueo, C.; Mato, J.M. Nitric oxide inactivates rat hepatic methionine adenosyltransferase In vivo by S-nitrosylation. Hepatology 1998, 28, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- Perez-Mato, I.; Castro, C.; Ruiz, F.A.; Corrales, F.J.; Mato, J.M. Methionine adenosyltransferase S-nitrosylation is regulated by the basic and acidic amino acids surrounding the target thiol. J. Biol. Chem. 1999, 274, 17075–17079. [Google Scholar] [CrossRef]
- Pajares, M.A.; Duran, C.; Corrales, F.; Pliego, M.M.; Mato, J.M. Modulation of rat liver S-adenosylmethionine synthetase activity by glutathione. J. Biol. Chem. 1992, 267, 17598–17605. [Google Scholar] [CrossRef]
- Martinez-Chantar, M.L.; Pajares, M.A. Role of thioltransferases on the modulation of rat liver S-adenosylmethionine synthetase activity by glutathione. FEBS Lett. 1996, 397, 293–297. [Google Scholar] [CrossRef]
- Wang, J.; Jia, G.; Li, H.; Yan, S.; Qian, J.; Guo, X.; Li, G.; Qi, H.; Zhu, Z.; Wu, Y.; et al. H2O2-Mediated Oxidative Stress Enhances Cystathionine γ-Lyase-Derived H2S Synthesis via a Sulfenic Acid Intermediate. Antioxidants 2021, 10, 1488. [Google Scholar] [CrossRef]
- Brown, J.M.; Kuhlman, C.; Terneus, M.V.; Labenski, M.T.; Lamyaithong, A.B.; Ball, J.G.; Lau, S.S.; Valentovic, M.A. S-adenosyl-l-methionine protection of acetaminophen mediated oxidative stress and identification of hepatic 4-hydroxynonenal protein adducts by mass spectrometry. Toxicol. Appl. Pharmacol. 2014, 281, 174–184. [Google Scholar] [CrossRef]
- Martinez-Chantar, M.L.; Pajares, M.A. Assignment of a single disulfide bridge in rat liver methionine adenosyltransferase. Eur. J. Biochem. FEBS 2000, 267, 132–137. [Google Scholar] [CrossRef]
- Sanchez-Perez, G.F.; Gasset, M.; Calvete, J.J.; Pajares, M.A. Role of an intrasubunit disulfide in the association state of the cytosolic homo-oligomer methionine adenosyltransferase. J. Biol. Chem. 2003, 278, 7285–7293. [Google Scholar] [CrossRef] [PubMed]
- Corrales, F.; Cabrero, C.; Pajares, M.A.; Ortiz, P.; Martin-Duce, A.; Mato, J.M. Inactivation and dissociation of S-adenosylmethionine synthetase by modification of sulfhydryl groups and its possible occurrence in cirrhosis. Hepatology 1990, 11, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Mingorance, J.; Alvarez, L.; Sanchez-Gongora, E.; Mato, J.M.; Pajares, M.A. Site-directed mutagenesis of rat liver S-adenosylmethionine synthetase. Identification of a cysteine residue critical for the oligomeric state. Biochem. J. 1996, 315 Pt 3, 761–766. [Google Scholar] [CrossRef]
- Niu, W.; Wang, J.; Qian, J.; Wang, M.; Wu, P.; Chen, F.; Yan, S. Allosteric control of human cystathionine β-synthase activity by a redox active disulfide bond. J. Biol. Chem. 2018, 293, 2523–2533. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.-N.; Yadav, P.K.; Adamec, J.; Banerjee, R. S-Glutathionylation Enhances Human Cystathionine β-Synthase Activity Under Oxidative Stress Conditions. Antioxid. Redox Signal. 2015, 22, 350–361. [Google Scholar] [CrossRef]
- Celano, L.; Gil, M.; Carballal, S.; Durán, R.; Denicola, A.; Banerjee, R.; Alvarez, B. Inactivation of cystathionine β-synthase with peroxynitrite. Arch. Biochem. Biophys. 2009, 491, 96–105. [Google Scholar] [CrossRef]
- Wang, H.; Sun, Q.; Zhou, Y.; Zhang, H.; Luo, C.; Xu, J.; Dong, Y.; Wu, Y.; Liu, H.; Wang, W. Nitration-mediated deficiency of cystathionine β-synthase activity accelerates the progression of hyperhomocysteinemia. Free. Radic. Biol. Med. 2017, 113, 519–529. [Google Scholar] [CrossRef]
- Rius-Pérez, S.; Pérez, S.; Torres-Cuevas, I.; Martí-Andrés, P.; Taléns-Visconti, R.; Paradela, A.; Guerrero, L.; Franco, L.; López-Rodas, G.; Torres, L.; et al. Blockade of the trans-sulfuration pathway in acute pancreatitis due to nitration of cystathionine β-synthase. Redox Biol. 2020, 28, 101324. [Google Scholar] [CrossRef]
- Luo, C.; Ji, D.; Li, Y.; Cao, Y.; Zhang, S.; Yan, W.; Xue, K.; Chai, J.; Wu, Y.; Liu, H.; et al. Abnormal nitration and S-sulfhydration modification of Sp1-CSE-H2S pathway trap the progress of hyperhomocysteinemia into a vicious cycle. Free. Radic. Biol. Med. 2021, 164, 20–33. [Google Scholar] [CrossRef]
- Mustafa, A.K.; Gadalla, M.M.; Sen, N.; Kim, S.; Mu, W.; Gazi, S.K.; Barrow, R.K.; Yang, G.; Wang, R.; Snyder, S.H. H2S signals through protein S-sulfhydration. Sci. Signal. 2009, 2, ra72. [Google Scholar] [CrossRef]
- Zheng, H.; Chen, H.; Cai, Y.; Shen, M.; Li, X.; Han, Y.; Deng, X.; Cao, H.; Liu, J.; Li, H.; et al. Hydrogen sulfide-mediated persulfidation regulates homocysteine metabolism and enhances ferroptosis in non-small cell lung cancer. Mol. Cell 2024, 84, 4016–4030.e6. [Google Scholar] [CrossRef]
| PTM | Digestion | Separation/Enrichment | Sample [Ref] |
|---|---|---|---|
| Ubiquitylation 1 | Lys-C/trypsin | UBISite antibody 2 | HepG2 [51], Jurkat [51] |
| Anti-Gly-Gly | HEK293 [52], HEK293T [53], MV4-11 [53] | ||
| trypsin | Anti-Gly-Gly | Jurkat [54], Jurkat E6-1 [55], HEK293T [56], HCT116 [56], HCT116 [57], HeLa [57], U2OS [58] | |
| Sumoylation | WaLP 3 | Anti-Gly-Gly | HeLa [57], HCT116 [57] |
| Acetylation | Lys-C/trypsin | UBISite antibody | HepG2 [51], Jurkat 4 [51] |
| NS 5 | HeLa [59] | ||
| trypsin | Anti-acetyl-Lys | Jurkat [54], U2OS [60], Jurkat [61], MV4-11 [61], A549 [61] | |
| SCX | A2780 [62] | ||
| Succinylation | Lys-C/trypsin | Anti-succinyl-Lys | HeLa [59] |
| trypsin | Anti-succinyl-Lys | MEFs [63] | |
| Monomethylation | Lys-C/trypsin | Anti-me1Arg 6 | HEK293T [64] |
| PTMScan me1Arg motif | HEK293 [65] | ||
| Anti-me1Lys | KYSE-150 [66] | ||
| Methylglyoxal | trypsin | Streptavidin beads | HEK293 [67], HCT116 [67], HeLa [67] |
| Phosphorylation | trypsin | TiO2-beads | U2OS [60], HEK293 [52], HEK293T [68], HeLa [69], HeLa S3 [70], B-cell NHL (11 lines) [71], WM239A [72], A549 [73] |
| Fe-IMAC or TiO2-beads | HeLa [74] | ||
| IMAC/Anti-phosphoTyr | HeLa S3 [75], GP293 11 [76] | ||
| Fe-IMAC | Jurkat [54], HCT116 [77], SW480 [77], SW620 [77], hESC [78], NIE-115 12 neurites [79] | ||
| IMAC | Jurkat [80], PC9 [81], PC9/gef 10 [81], RAW264.7 12 [82] | ||
| Anti-phosphoTyr | Jurkat [83], MV(4;11) [84], Nomo-1 [84], KY821 [84], Molm 14 [84], hESC [85], RAW264.7 12 [86], MEFs 8 [87] | ||
| Anti-phosphoSQ/ anti-phosphoTQ | HEK293T [88] | ||
| Anti-CK 7 motif | NIH3T3 [89] | ||
| SCX | HL-1 [90] | ||
| Lys-N | SCX | HEK293 [91] | |
| Lys-C/trypsin | IMAC | HeLa [92], NSCLC 8 (16 lines) [93], breast cancer (six lines) [93] | |
| TiO2-beads | HeLa [94], HeLa S3 [95], 3T3-L1 adipocytes [96,97], K562 [94], hESC [98] 13 | ||
| Glu-C | Anti-11OB7 9 | MKN-45 [99] | |
| Chymotrypsin | Anti-phosphoTyr | MEFs 9 [87] |
| PTM | Digestion | Separation/Enrichment | Sample 2 [Ref] |
|---|---|---|---|
| Ubiquitylation 1 | Lys-C/trypsin | Anti-Gly-Gly | MouseBLHKSkM [100] |
| N-glycosylation | Trypsin/Glu-C | Lectins (concanavalin A, wheat germ agglutinin, agglutinin RCA120) | MouseBLHKPl [101] |
| Acetylation | Lys-C/trypsin | Anti-Acetyl-Lys | RatLSPSkMSkThKPfBfBIHLuStTTf [102], HumanSkM [102] |
| NS 3 | MouseL [59] | ||
| trypsin | Anti-Acetyl-Lys | HumanL [103], MouseLC [104] | |
| Succinylation | Lys-C/trypsin | Anti-succinyl-Lys | MouseL [59] |
| Hydroxybutyrylation | trypsin | Pan-β-hydroxybutyryl-Lys | MouseL [105] |
| Lactylation | trypsin | Pan-lactyl-Lys | HumanL [106] |
| Malonylation | trypsin | Anti-malonyl-Lys | MouseL [107] |
| Formaldehyde | trypsin | MouseL [108] | |
| Phosphorylation | Glu-C/trypsin | Ti4+-IMAC | HumanL [109] |
| trypsin | Anti-phosphoTyr | Human cholangiocarcinoma [110] | |
| Fe-IMAC | Ovarian cancer [111], luminal breast cancer [111], breast cancer xenografts [111], colorectal cancer [77] | ||
| SCX/SAX | MouseL [112] | ||
| IMAC/Anti-phosphoTyr | MouseL [113] | ||
| IMAC | MouseL [114], MouseB [115], MouseBBfHLLuKPST [116] | ||
| TiO2-beads | MouseS [117], RatL [118] | ||
| TiO2-beads/Anti-phosphoTyr | MouseB [119] | ||
| Lys-C/trypsin | Fe3+-IMAC | 115 Breast tumors [120] | |
| TiO2-beads | MouseL [121], RatPBThHKPfLuSBlTSkMStIL [122] | ||
| IMAC | MouseL mitochondria [123] |
| Gene Name | Modification Site 1,2 [ref] |
|---|---|
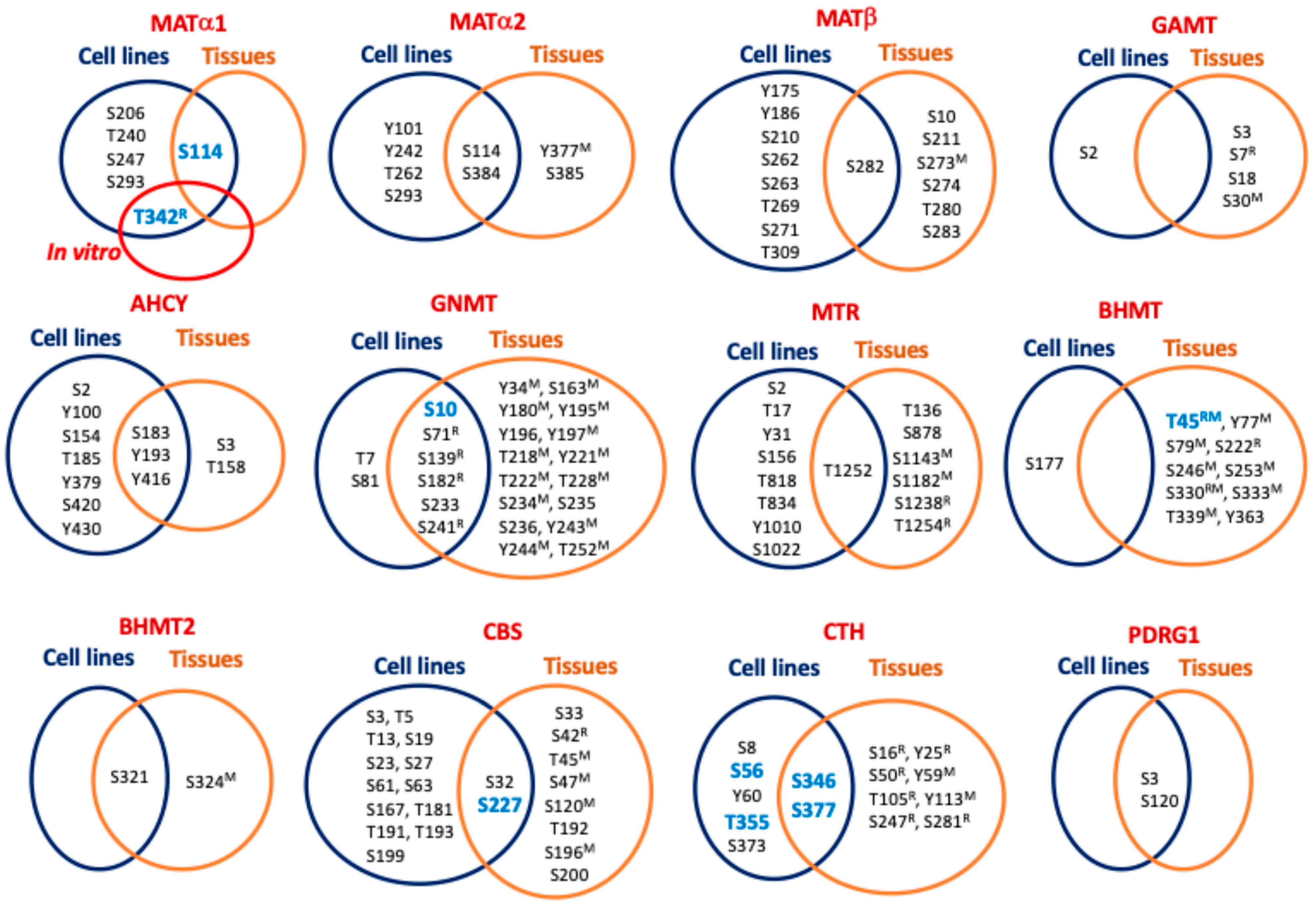
| MAT1A | S206 [72], T240 [68], S247 [68], S293 [70], S115 [121], S115 (mouseL) [114] |
| MAT2A | Y101 [99], S114 [54,69,74,75,96], S115 (humanBr) [120], Y242 [81], T262 [95], S293 [70], Y377 (mouseL) [116], S384 [54,75,77,78,92,94,111], S384 (NSCLC) [93], S385 (humanBr) [120] |
| MAT2B | S10 (humanBr) [120], Y175 [98], Y186 [98], S210 [75], S211 (humanBr) [120], S262 [96], S263 [96], T269 [96], S271 [72,78,96], S273 (mouseLK) [116], S273 (mouse) [111], S274 (mouseS) [116], T280 (mouseP) [116], T280 (NSCLC) [93], S282 (mouseBKP) [116], S282 [54,74,75,77,80,97], S282 (mouseB) [115,119], S282 (mouseL) [114], S282 (NSCLC, Breast carcinoma) [93], S282 (ratSTh) [122], S282 (mouseS) [117], S282 (human) [111], S282 (humanC) [77], S283 (humanBr) [120], T309 [75,94] |
| AHCY | S2 [121], S3 (humanBr) [120], Y100 [81], S154 [54], T158 (humanC) [77], S183 [75,77,94,121], S183 (mouseL) [114], S183 (ratLIKPS) [122], S183 (ratL) [118], T185 [75], Y193 [54,75,76,83,84,85,87], Y193 (mouse) [86], Y193 (mouseL) [113,114,116], Y379 [98], Y416 [98], Y416 (mouseL) [113], S420 [98], Y430 [98] |
| GNMT | S10 [121], S10 (ratL) [118], S10 (mouseL) [113,114,116], Y34 (mouseL) [113,114,116], S81 [109], S163 (mouseL) [114], Y180 (mouseL) [113], Y195 (mouseL) [113], Y196 (human cholangiocarcinoma) [110], S197 (mouseL) [113], T218 (mouseL) [113], Y221 (mouseL)mit [123] 3, Y221 (mouseL) [114], T222 (mouseL) [113], T228 (mouseL)mit [123] 3, T228 (mouseL) [116], S233 [121], S233 (mouseL) [114], S233 (mouseLP) [116], S234 (mouseL)mit [123] 3, S235 (human) [111], S236 (humanBr) [120], Y243 (mouseL) [113], Y244 (mouse) [111], Y244 (mouseL) [114], T252 (mouse) [111] |
| GAMT | S2 [54], S3 (humanBr) [120], S7 (ratLKI) [122], S18 (humanBr) [120], S30 (mouseL)mit [123] 3 |
| MTR | S2 [75], T17 [95], Y31 [90], T136 (human) [111], S156 [75], T818 [109], T834 [79], S878 (humanBr) [120], Y1010 [77], S1022 [77], S1143 (mouse) [111], S1182 (mouse) [82], S1238 (ratLHIKLuPSTTh) [122], T1252 [89], T1252 (ratL) [118], T1254 (ratLHIKLuPSTTh) [122] |
| BHMT | T45 (ratL) [118], T45 (mouseL) [113,114], Y77 (mouseL)mit [123], S79 (mouseL)mit [123], S79 (mouseL) [113,114,116], S177 [109], S222 (ratL) [118], S246 (mouseL) [114], S253 (mouseL) [113], S330 (mouseL)mit [123], S330 (mouseL) [113,114,116], S330 (ratLBlHIKPT) [122], S330 (ratL) [118], S333 (mouseL)mit [123], S333 (mouseL) [116], T339 (mouseL) [114], Y363 (mouseL) [113], Y363 (human cholangiocarcinoma) [110] |
| BHMT2 | S321 [109], S321 (mouseL)mit [123], S321 (mouseL) [113], S321 (mouseLK) [116], S324 (mouseK) [116] |
| CBS | S3 [75,91], T5 [75,91], T13 [91], S19 [91], S23 [91], S27 [91,98,109], S32 [52,72,95,98], S32 (human) [111], S33 (humanBr) [120], S42 (ratLIKLuPTh) [122], T45 (mouseL)mit [123], T45 (mouseLKP) [116], T45 (mouseL) [114], S47 (mouseL)mit [123], S61 [75], S63 [75], S120 (mouseP) [116], S167 [71], T181 [71], T191 [70,73], T192 (humanBr) [120], T193 [70,73], S196 (mouseP) [116], S199 [75,94], S200 (humanBr) [120] |
| CTH | S8 [88], S16 (ratL) [118], Y25 (ratL) [118], S50 (ratLHIKPSt) [122], S50 (ratL) [118], Y59 (mouseL) [113], Y60 [75], T105 (ratL) [118], Y113 (mouseL) [113], S247 (ratL) [118], S281 (ratLHIKPSt) [122], S373 [109], S376 [121], S376 (mouseL) [114], S377 (mouseL) [112] |
| PDRG1 | S3 [60,75,95], S3 (humanBr) [120], S120 [90], S120 (mouseB) [119], S120 (mouse) [82] |
| Gene Name | Modification Site 1 [ref] |
|---|---|
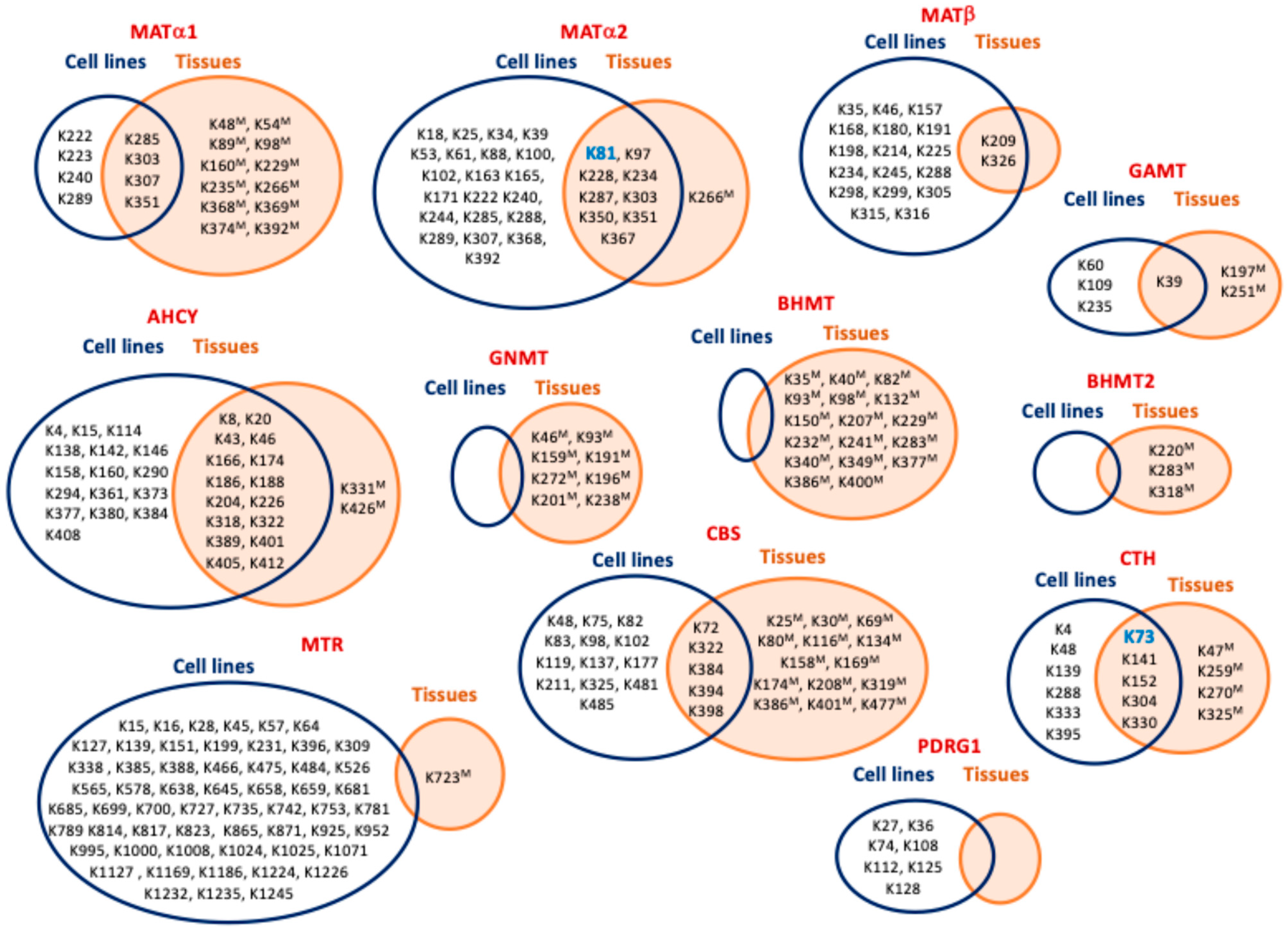
| MAT1A | K48 (mouseLH) [100], K54 (mouseLH) [100], K89 (mouseLH) [100], K98 (mouseLH) [100], K160 (mouseL) [100], K222 [51], K223 [55], K229 (mouseL) [100], K235 (mouseLH) [100], K240 [51], K266 (mouseL) [100], K285 [51,55], K286 (mouseL) [100], K289 [52,54,55], K303 [51,52,55], K304 (mouseL) [100], K307 [52], K308 (mouseL) [100], K351 [52,53,58], K352 (mouseL) [100], K368 (mouseL) [100], K369 (mouseL) [100], K374 (mouseL) [100], K392 (mouseL) [100] |
| MAT2A | K18 [51], K25 [51], K34 [51], K39 [51], K53 [51], K61 [54,55], K81 [51,54,55,56,58], K81 (mouseK) [100], K88 [51,54,55,56], K97 [51,53,54,55,56], K97 (mouseLKB) [100], K100 [51], K102 [51,55], K163 [51,54,55], K165 [51], K171 [51], K222 [51], K228 [51,54,55,56], K228 (mouseLK) [100], K234 [51,53,54,55,56,57,58], K234 (mouseLKBSkM) [100], K240 [51], K244 [51], K266 (mouseL) [100], K285 [51,55], K286 (mouseL) [100], K287 [51], K288 [51], K289 [52,54,55,56], K303 [51,52,55], K304 (mouseL) [100], K307 [51,52,53,54,55], K350 [51,53,54,55,56], K350 (mouseLKB) [100], K351 [51,52,53,54,55,57,58], K351 (mouseLKBHSkM) [100], K367 [51,53,54,55], K367 (mouseK) [100], K368 [55], K392 [55] |
| MAT2B | K35 [51], K46 [51,54,55], K157 [51], K168 [51], K180 [51], K191 [51], K198 [51], K209 [51,52,53,54,55,58], K209 (mouseLKB) [100], K214 [51], K225 [51,53,54], K234 [51], K245 [51,53], K288 [51], K298 [51], K299 [51,55], K305 [51], K315 [51], K316 [51,53,54,55], K326 [51,53,54,55,56,57,58], K326 (mouseLKB) [100] |
| AHCY | K4 [55], K8 [53,55], K8 (mouseL) [100], K15 [51], K20 [55,58], K20 (mouseLKBH) [100], K43 [51,52,54,55], K43 (mouseL) [100], K46 [52,54,55], K46 (mouseLKH) [100], K114 [51], K138 [51], K142 [51], K146 [51], K158 [51], K160 [51], K166 [51,53,54,55,56], K166 (mouseLKBHSkM) [100], K174 [51,55,56], K174 (mouseLK) [100], K186 [51,52,54,55,56,57,58], K186 (mouseLKBH) [100], K188 [51,52,53,54,55,56,58], K188 (mouseLKBH) [100], K204 [52,54,55], K204 (mouseLKH) [100], K226 [52,53,54,55,58], K226 (mouseLK) [100], K290 [51], K294 [51], K318 [51,55], K318 (mouseL) [100], K322 [51,54,55], K322 (mouseL) [100], K331 (mouseLKH) [100], K361 [51], K373 [51], K377 [51], K380 [51], K384 [51], K389 [51,55], K389 (mouseLKH) [100], K401 [51,55], K401 (mouseL) [100], K405 [51,52,54,55,56], K405 (mouseLKB) [100], K408 [51,52], K412 [51,53], K412 (mouseLK) [100], K426 (mouseL) [100] |
| GNMT | K46 (mouseL) [100], K93 (mouseL) [100], K159 (mouseL) [100], K191 (mouseL) [100], K272 (mouseL) [100], K196 (mouseLH) [100], K201 (mouseLH) [100], K238 (mouseLH) [100] |
| GAMT | K39 [51], K39 [55], K39 (mouseLB) [100], K60 [55], K109 [51], K197 (mouseL) [100], K235 [55,56], K251 (mouseL) [100] |
| MTR | K15 [55], K16 [51,53,55], K28 [51,53,54,55,57], K45 [51,54,55], K57 [51,55], K64 [51,55], K127 [51,55], K139 [51,52,55], K151 [51,52,54,55], K199 [56], K231 [53,55,57], K306 [55], K309 [51,55], K339 [51,55], K385 [55], K388 [55], K466 [55], K475 [51,54,55], K484 [51,54,55], K526 [55], K565 [54,55], K578 [56], K638 [51], K645 [51], K658 [57], K659 [51,55], K681 [51,55], K685 [51,53,55], K699 [51], K700 [55], K723 (mouseL) [100], K727 [51,53,54,55], K735 [52,54,55], K742 [51,55], K753 [51,55], K781 [55], K789 [53,54,55], K814 [52,53,55], K817 [51,52,53,54,55], K823 [55], K865 [51,52,53,54,55], K871 [51,54,55], K925 [55], K952 [51], K995 [51,55], K1000 [51,52,53,54,55], K1008 [51], K1024 [51], K1025 [55], K1071 [55], K1127 [51,55], K1169 [51,55], K1186 [51,55], K1224 [55], K1226 [55], K1232 [55], K1235 [51,55], K1245 [51,55] |
| BHMT | K35 (mouseL) [100], K40 (mouseLH) [100], K82 (mouseL) [100], K93 (mouseLH) [100], K98 (mouseLH) [100], K132 (mouseL) [100], K150 (mouseL) [100], K207 (mouseL) [100], K229 (mouseLH) [100], K232 (mouseLH) [100], K241 (mouseLH) [100], K283 (mouseLKH) [100], K340 (mouseL) [100], K349 (mouseL) [100], K377 (mouseLH) [100], K386 (mouseLH) [100], K400 (mouseL) [100] |
| BHMT2 | K220 (mouseL) [100], K283 (mouseLKH) [100], K318 (mouseL) [100] |
| CBS | K25 (mouseL) [100], K30 (mouseH) [100], K48 [51], K69 (mouseLKH) [100], K72 [51], K72 (mouseL) [100], K75 [51], K80 (mouseL) [100], K82 [51,55], K83 [51], K98 [51], K102 [55], K116 (mouseL) [100], K119 [55], K134 (mouseL) [100], K137 [55], K158 (mouseL) [100], K169 (mouseL) [100], K174 (mouseL) [100], K177 [52,55], K208 (mouseL) [100], K211 [51,55], K319 (mouseL) [100], K322 [51,55], K322 (mouseL) [100], K325 [55], K381 (mouseL) [100], K384 [55], K386 (mouseLKH) [100], K391 (mouseL) [100], K394 [53], K395 (mouseL) [100], K398 [51], K401 (mouseL) [100], K477 (mouseL) [100], K481 [52,54,55], K485 [52,55] |
| CTH | K4 [51], K47 (mouseL) [58], K48 [51,55], K72 (mouseLKBH) [58], K73 [51,52,53,55], K139 [51,55], K140 (mouseLKH) [58], K141 [51], K151 (mouseLKB) [58], K152 [51,56], K259, (mouseLK) [58], K270 (mouseLK) [58], K288 [51], K303 (mouseL) [58], K304 [55], K325 (mouseL) [58], K329 (mouseL) [58], K330 [51], K333 [55], K395 [51] |
| PDRG1 | K27 [51,55,56], K36 [51,52,55,56], K74 [56], K108 [51,55], K112 [51,54,55], K125 [51,54,55], K128 [55] |
| Gene Name | Modification Site 1 [ref] |
|---|---|
| MAT1A | N-term [51], K89 (ratL) [102], K89 (mouseL) [59], K98 (mouseL) [59], K229 (ratL) [102], K235 (ratL) [102], K235 (mouseL) [59,104], K286 (mouseL) [59], K352 (mouseL) [59], K353 (ratL) [102], K393 (ratL) [102] |
| MAT2A | N-term [51], K88 [54], K88 (ratTh) [102], K163 (ratK) [102], K234 [54,59], K234 (ratL) [102], K286 (mouseL) [59] |
| AHCY | N-term [51], K4 [54], K4 (ratLuSkMPSTTh) [102], K4 (mouseL) [104], K8 (ratLuBK) [102], K8 (mouseL) [59], K43 (ratLKLuTTh) [102], K43 (mouseL) [59], K142 (mouseL) [59], K166 (ratLKLuStTTh) [102], K174 (mouseL) [59], K186 (mouseL) [59], K204 (ratLB) [102], K318 (ratLK) [102], K322 (ratTh) [102], K322 (mouseL) [59], K389 (ratK) [102], K401 [54,59,60,61], K401 (ratL) [102], K401 (mouseL) [59], K405 (ratK) [102], K408 [54,59,60,61], K408 (ratLBKLuPSStTh) [102], K408 (humanSkM) [102], K412 (ratLu) [102], K412 (mouseL) [59], K426 (mouseL) [59] |
| GNMT | K46 (mouseL) [59], K93 (ratL) [102], K159ratL) [102], K159 (mouseL) [59], K191 (mouseL) [104], K196 (ratL) [102], K201 (ratK) [102], K201 (mouseL) [59,104], K238 (ratSk) [102], K238 (mouseL) [59] |
| GAMT | N-term [51], K105 (ratK) [102], K109 (ratT) [102] |
| MTR | N-term [51] |
| BHMT | K7 (ratL) [102], K40 (ratL) [102], K82 (ratL) [102], K82 (mouseL) [59], K93 (ratL) [102], K93 (mouseL) [59], K132 (mouseL) [104], K139 (ratL) [102], K150 (ratL) [102], K207 (mouseL) [104], K232 (ratLK) [102], K232 (mouseL) [59,104], K241 (mouseL) [104], K283 (ratL) [102], K283 (mouseL) [59,104], K327 (ratL) [102], K340 (ratL) [102], K349 (ratL) [102], K369 (ratLK) [102], K369 (humanL) [103], K386 (ratL) [102], K386 (mouseL) [59], K400 (ratL) [102] |
| BHMT2 | K11 (ratSt) [102], K123 (ratL) [102], K129 (ratL) [102], K220 (ratL) [102], K223 (ratL) [102], K274 (ratL) [102], K274 (mouseL) [59,104], K331 (ratL) [102] |
| CBS | K72 (ratBK) [102], K208 (ratBK) [102], K381 (mouseL) [59], K386 (ratK) [102], K386 (mouseL) [59], K481 [59] |
| CTH | K47 (mouseL) [59], K72 (mouseL) [104], K140 (mouseL) [59,104], K140 (ratLKPTh) [102], K151 (mouseL) [59], K164 (ratL) [102], K259 (ratLP) [102], K287 (ratLK) [102], K303 (ratLKPSkSTTh) [102], K329 (ratLKSkTh) [102], K361 (mouseL) [59], K383 (ratLBKT) [102], K394 (ratKT) [102] |
| PDRG1 | N-term [51] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pajares, M.Á. Posttranslational Regulation of Mammalian Sulfur Amino Acid Metabolism. Int. J. Mol. Sci. 2025, 26, 2488. https://doi.org/10.3390/ijms26062488
Pajares MÁ. Posttranslational Regulation of Mammalian Sulfur Amino Acid Metabolism. International Journal of Molecular Sciences. 2025; 26(6):2488. https://doi.org/10.3390/ijms26062488
Chicago/Turabian StylePajares, María Ángeles. 2025. "Posttranslational Regulation of Mammalian Sulfur Amino Acid Metabolism" International Journal of Molecular Sciences 26, no. 6: 2488. https://doi.org/10.3390/ijms26062488
APA StylePajares, M. Á. (2025). Posttranslational Regulation of Mammalian Sulfur Amino Acid Metabolism. International Journal of Molecular Sciences, 26(6), 2488. https://doi.org/10.3390/ijms26062488






