Regulatory Mechanisms of Yili Horses During an 80 km Race Based on Transcriptomics and Metabolomics Analyses
Abstract
1. Introduction
2. Results
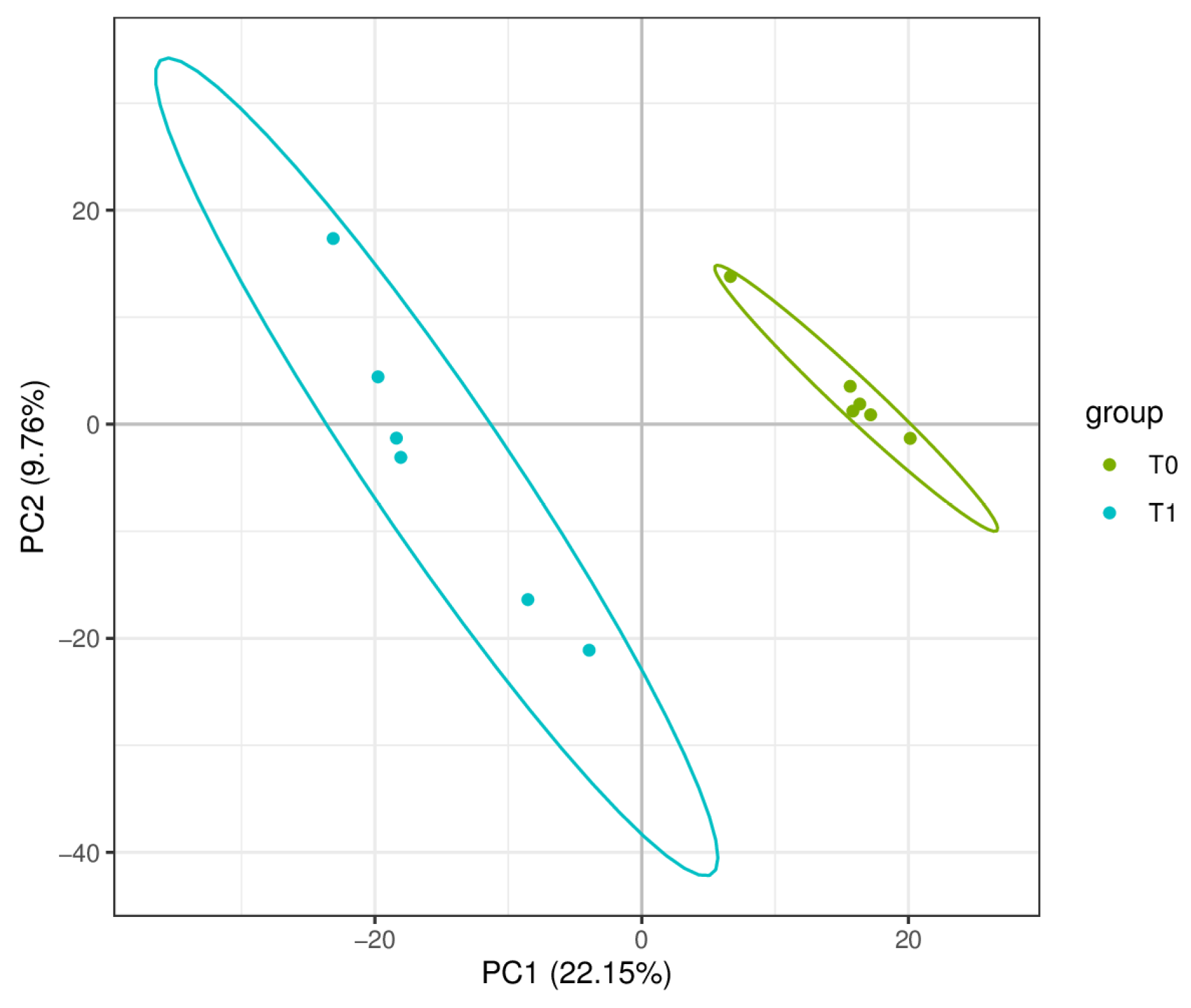
2.1. Transcriptome Sequencing Quality Control
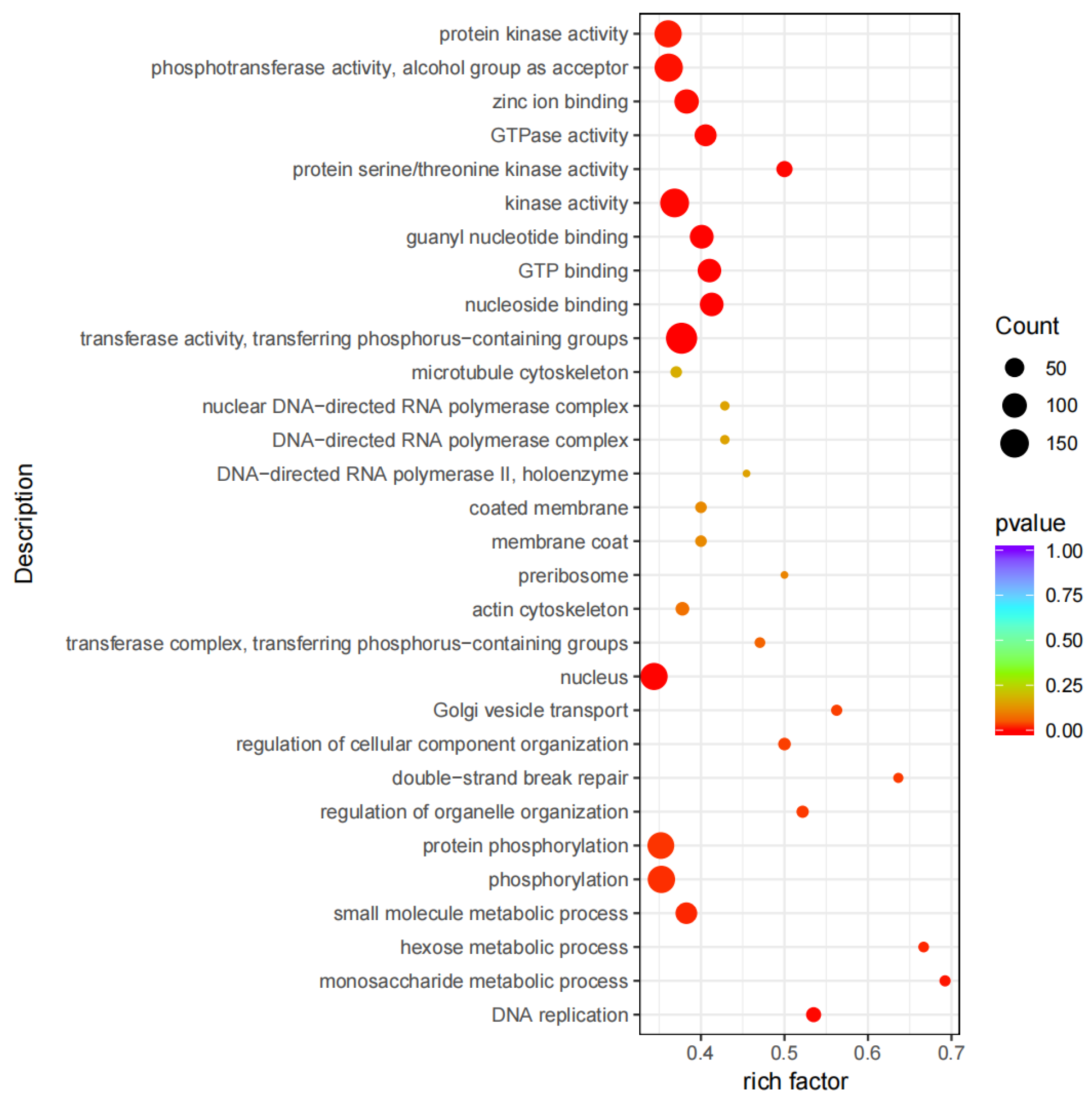
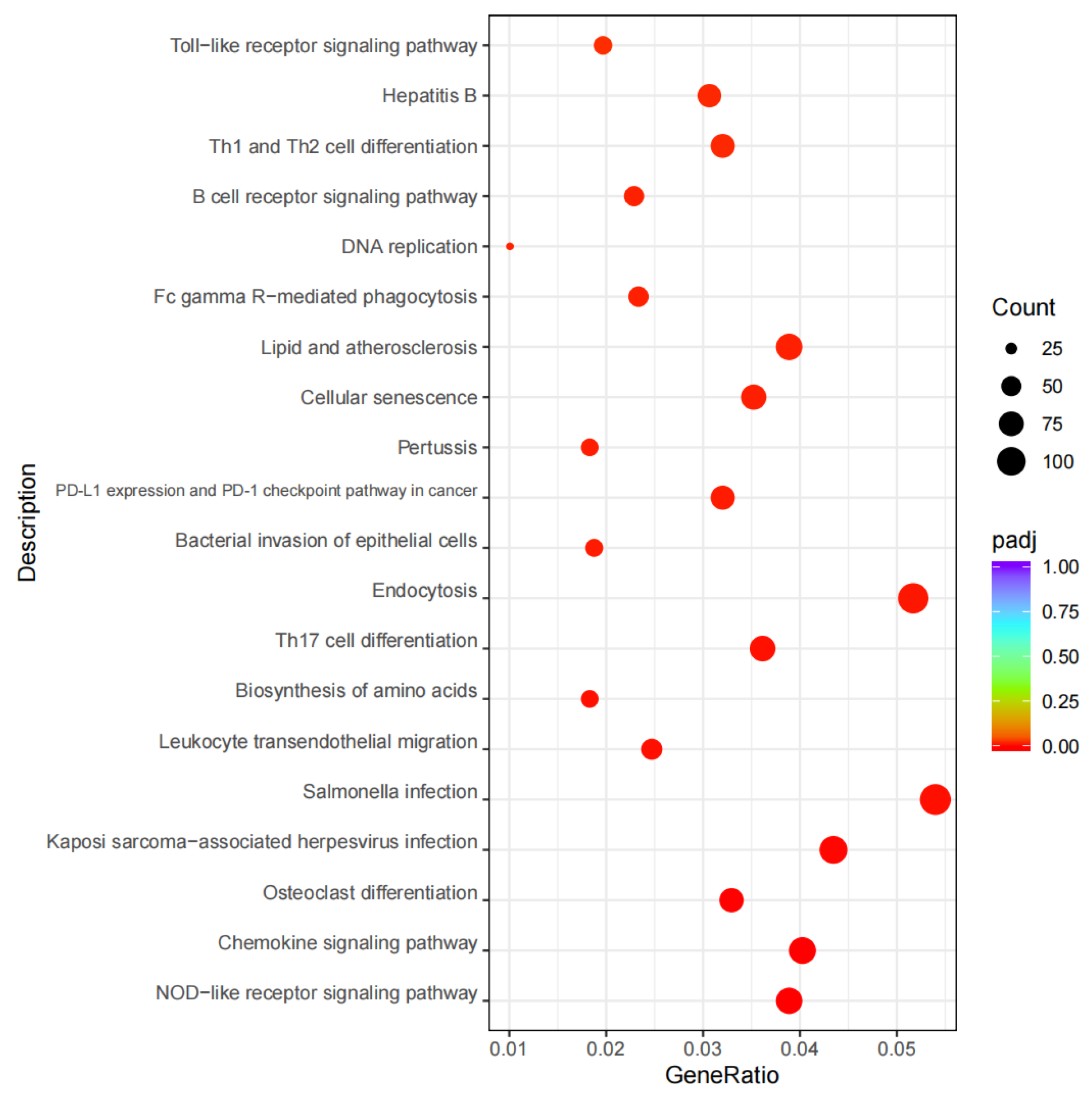
2.2. DEG Screening and Expression Analysis
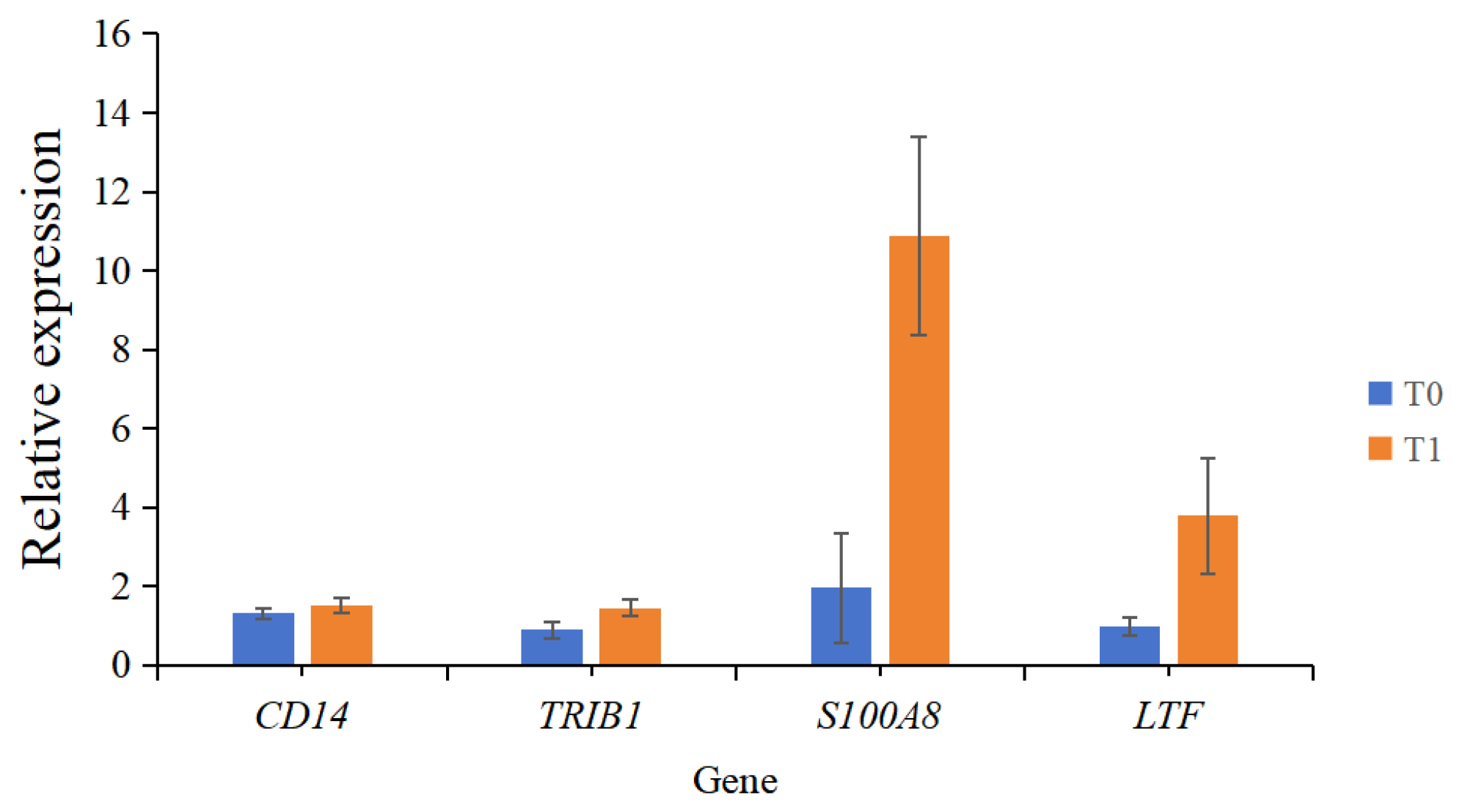
2.3. Real-Time Fluorescence Quantitative PCR Validation
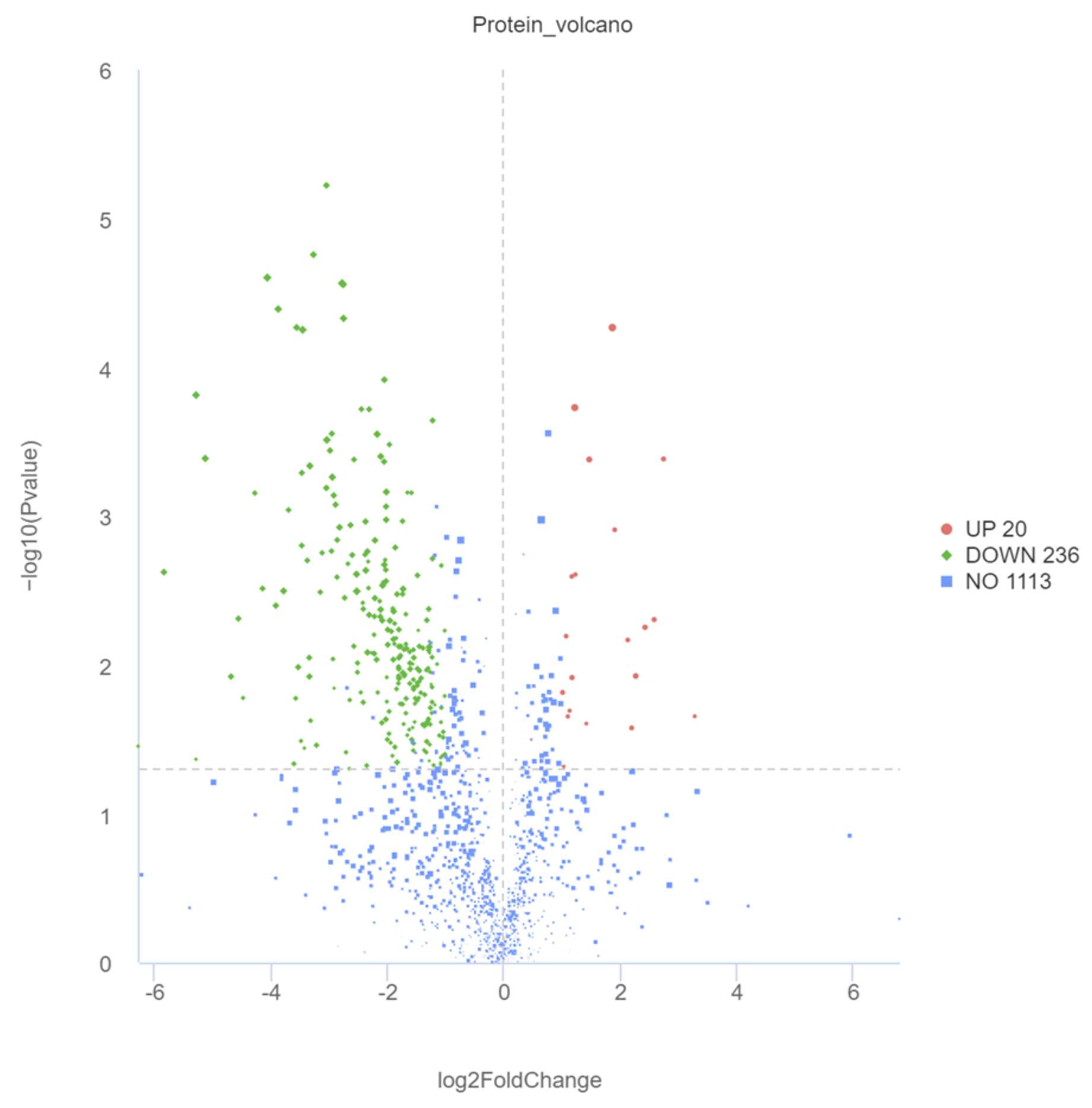
2.4. DM Screening and Analysis
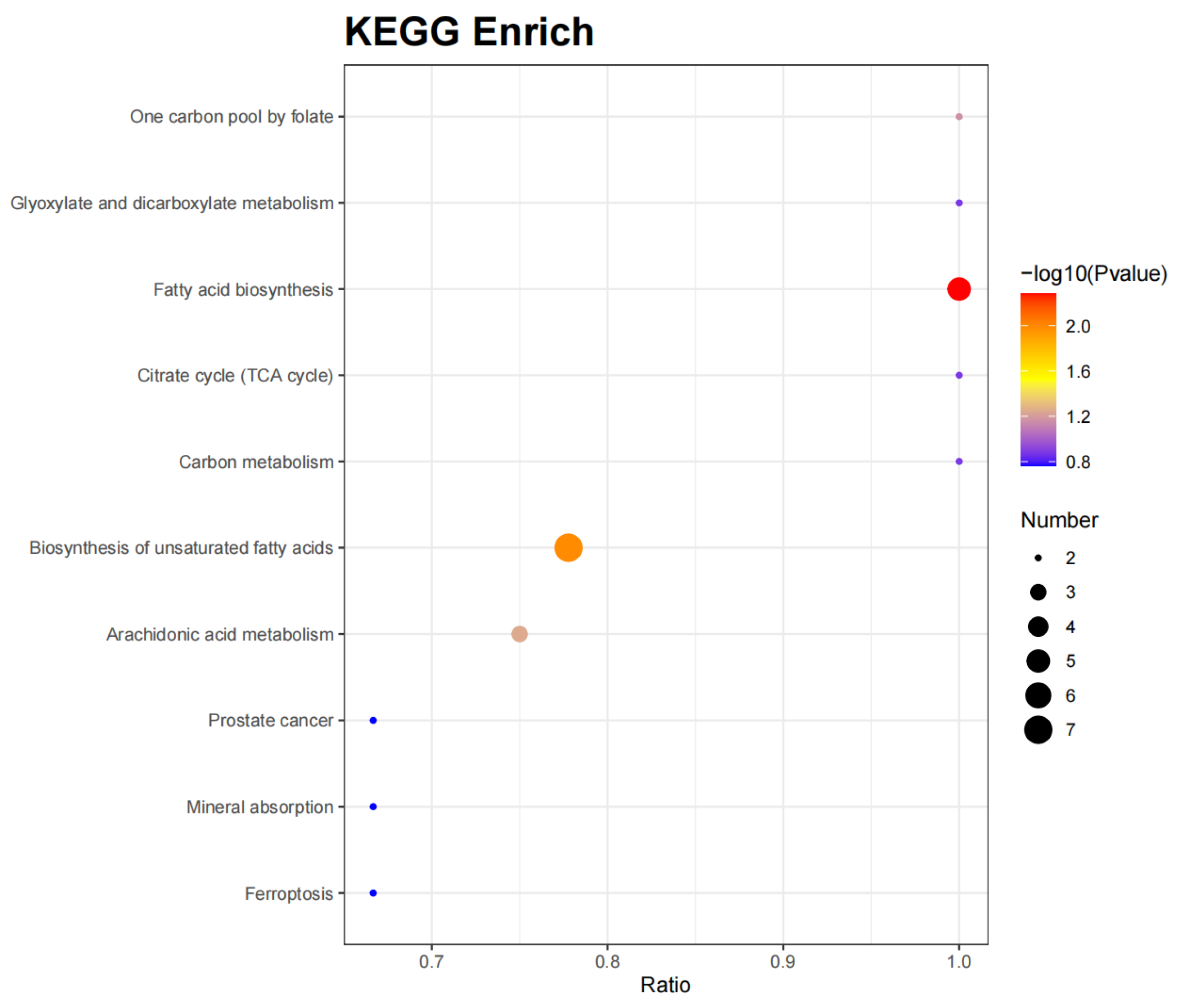
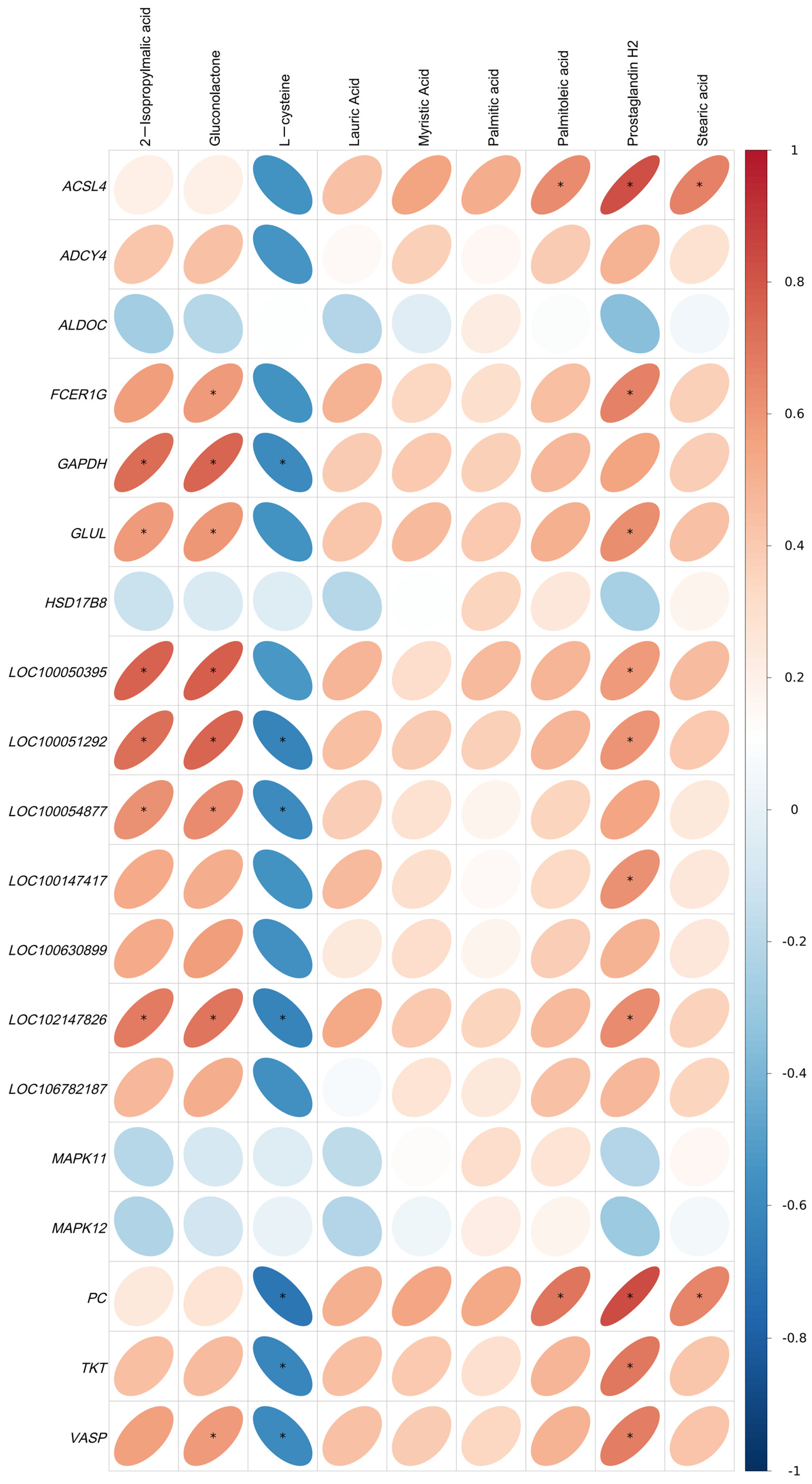
2.5. Correlation Analysis
3. Discussion
4. Materials and Methods
4.1. Animals and Sample Collection
4.2. Total RNA Extraction and Library Construction
4.3. RNA-Seq Data Processing and Analysis
4.4. Real-Time Quantitative PCR Validation
4.5. Metabolite Extraction
4.6. LC-MS Analysis
4.7. Metabolite Identification and Screening
4.8. Integrated Analysis of Transcriptomics and Metabolomics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bennet, E.D.; Parkin, T.D.H. The impact of the mandatory rest period in Federation Equestre Internationale endurance events. J. Equine Vet. 2020, 52, 268–272. [Google Scholar] [CrossRef]
- Bou, T.; Ding, W.Q.; Ren, X.J.; Liu, H.Y.; Gong, W.D.; Jia, Z.J.; Zhang, X.Z.; Dugarjaviin, M.; Bai, D.Y. Muscle fibre transition and transcriptional changes of horse skeletal muscles during traditional Mongolian endurance training. J. Equine Vet. 2024, 56, 178–192. [Google Scholar] [CrossRef]
- Pan, J.; Purev, C.; Zhao, H.W.; Zhang, Z.P.; Wang, F.; Wendoule, N.; Qi, G.C.; Liu, Y.B.; Zhou, H.M. Discovery of exercise-related genes and pathway analysis based on comparative genomes of Mongolian originated Abaga and Wushen horse. Open Life Sci. 2022, 17, 1269–1281. [Google Scholar] [CrossRef]
- Verdegaal, E.L.J.M.M.; Howarth, G.S.; McWhorter, T.J.; Boshuizen, B.; Franklin, S.H.; de Vega, C.V.M.; Jonas, S.E.; Folwell, L.E.; Delesalle, C.J.G. Continuous monitoring of the thermoregulatory response in endurance horses and trotter horses during field exercise: Baselining for future hot weather studies. Front. Physiol. 2021, 12, 708737. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Zukowski, K.; Piórkowska, K.; Gurgul, A.; Bugno-Poniewierska, M. Transcriptome profiling of Arabian horse blood during training regimens. BMC Genet. 2017, 18, 31. [Google Scholar] [CrossRef][Green Version]
- Farries, G.; Bryan, K.; McGivney, C.L.; McGettigan, P.A.; Gough, K.F.; Browne, J.A.; MacHugh, D.E.; Katz, L.M.; Hill, E.W. Expression quantitative trait loci in equine skeletal muscle reveals heritable variation in metabolism and the training responsive transcriptome. Front. Genet. 2019, 10, 1215. [Google Scholar] [CrossRef]
- Xing, J.Y.; Xie, L.; Qi, X.Z.; Liu, G.Q.; Akhtar, M.F.; Li, X.Y.; Bou, G.; Bai, D.Y.; Zhao, Y.P.; Dugarjaviin, M.; et al. Integrated analysis of transcriptome and proteome for exploring mechanism of promoting proliferation of equine satellite cells associated with leucine. Comp. Biochem. Phys. D 2023, 48, 101118. [Google Scholar] [CrossRef]
- Mycka, G.; Ropka-Molik, K.; Cywinska, A.; Szmatola, T.; Stefaniuk-Szmukier, M. The modifications of longevity regulating pathway resulting from endurance effort in Arabian horses. Ann. Anim. Sci. 2024, 24, 1161–1170. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Musial, A.D.; Velie, B.D. The genetics of racing performance in Arabian Horses. Int. J. Genom. 2019, 2019, 9013239. [Google Scholar] [CrossRef] [PubMed]
- Mach, N.; Moroldo, M.; Rau, A.; Lecardonnel, J.M.; Le Moyec, L.; Robert, C.; Barrey, E. Understanding the holobiont: Crosstalk between gut microbiota and mitochondria during long exercise in horse. Front. Mol. Biosci. 2021, 8, 656204. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Shi, X.Y.; Gao, Z.Z.; Li, R.; Tian, J.M.; Cao, X.D.; Yang, B.; Zhao, S.H.; Yang, Y. Acupoint catgut embedding improves lipid metabolism in exercise-induced fatigue rats via the PPAR signaling pathway. Animals 2023, 13, 558. [Google Scholar] [CrossRef]
- Ghosh, M.; Cho, H.W.; Park, J.W.; Choi, J.Y.; Chung, Y.H.; Sharma, N.; Singh, A.K.; Kim, N.E.; Mongre, R.K.; Huynh, D.; et al. Comparative transcriptomic analyses by RNA-seq to elucidate differentially expressed genes in the muscle of Korean Thoroughbred Horses. Appl. Biochem. Biotechnol. 2016, 180, 588–608. [Google Scholar] [CrossRef] [PubMed]
- Klobucar, K.; Vrbanac, Z.; Gotic, J.; Bojanic, K.; Bures, T.; Brkljaca Bottegaro, N. Changes in biochemical parameters in horses during 40 km and 80 km endurance races. Acta Vet.-Beogr. 2019, 69, 73–87. [Google Scholar] [CrossRef]
- de Siqueira, R.F.; Silva, B.O.; Fernandes, M.L.; Fernandes, W.R. Evaluation basal lipid metabolism components and adiposity in trained Arabian horses for Endurance and Racing. Cienc. Rural 2024, 54, e20230373. [Google Scholar] [CrossRef]
- Masko, M.; Domino, M.; Jasinski, T.; Witkowska-Pilaszewicz, O. The physical activity-dependent hematological and biochemical changes in school horses in comparison to blood profiles in endurance and race horses. Animals 2021, 11, 1128. [Google Scholar] [CrossRef]
- Buckley, P.; Buckley, D.J.; Freire, R.; Hughes, K.J. Pre-race and race management impacts serum muscle enzyme activity in Australian endurance horses. J. Equine Vet. 2022, 54, 895–904. [Google Scholar] [CrossRef]
- Halama, A.; Oliveira, J.M.; Filho, S.A.; Qasim, M.; Achkar, I.W.; Johnson, S.; Suhre, K.; Vinardell, T. Metabolic predictors of equine performance in endurance racing. Metabolites 2021, 11, 82. [Google Scholar] [CrossRef]
- van der Kolk, J.H.; Mach, N.; Ramseyer, A.; Burger, D.; Gerber, V.; Nuoffer, J.M. Serum acylcarnitine profile in endurance horses with and without metabolic dysfunction. Vet. J. 2020, 255, 105419. [Google Scholar] [CrossRef]
- Davis, T.Z.; Stegelmeier, B.L.; Lee, S.T.; Green, B.T.; Hall, J.O. Experimental rayless goldenrod (Isocoma pluriflora) toxicosis in horses. Toxicon 2013, 73, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Luck, M.M.; Le Moyec, L.; Barrey, E.; Triba, M.N.; Bouchemal, N.; Savarin, P.; Robert, C. Energetics of endurance exercise in young horses determined by nuclear magnetic resonance metabolomics. Front. Physiol. 2015, 6, 198. [Google Scholar] [CrossRef][Green Version]
- Mukai, K.; Ohmura, H.; Takahashi, Y.; Ebisuda, Y.; Yoneda, K.; Miyata, H. Physiological and skeletal muscle responses to high-intensity interval exercise in Thoroughbred horses. Front. Vet. Sci. 2023, 10, 1241266. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, J.D.; Hawley, J.A.; Morton, J.P. Carbohydrate availability and exercise training adaptation: Too much of a good thing? Eur. J. Sport. Sci. 2015, 15, 3–12. [Google Scholar] [CrossRef]
- Siqueira, R.F.; Weigel, R.A.; Nunes, G.R.; Mori, C.S.; Fernandes, W.R. Oxidative profiles of endurance horses racing different distances. Arq. Bras. Med. Vet. Zootec. 2014, 66, 455–461. [Google Scholar] [CrossRef]
- Park, K.D.; Park, J.; Ko, J.; Kim, B.C.; Kim, H.S.; Ahn, K.; Do, K.T.; Choi, H.; Kim, H.M.; Song, S.; et al. Whole transcriptome analyses of six thoroughbred horses before and after exercise using RNA-seq. BMC Genet. 2012, 13, 473. [Google Scholar] [CrossRef] [PubMed]
- Capomaccio, S.; Vitulo, N.; Verini-Supplizi, A.; Barcaccia, G.; Albiero, A.; D’Angelo, M.; Campagna, D.; Valle, G.; Felicetti, M.; Silvestrelli, M.; et al. RNA sequencing of the exercise transcriptome in equine athletes. PLoS ONE 2013, 8, e83504. [Google Scholar] [CrossRef]
- Clauss, M.; Gérard, P.; Mosca, A.; Leclerc, M. Interplay between exercise and gut microbiome in the context of human health and performance. Front. Nutr. 2021, 8, 637010. [Google Scholar] [CrossRef]
- Walker, J.B. Creatine: Biosynthesis, regulation, and function. Adv. Enzymol. Relat. Areas Mol. Biol. 1979, 50, 177–242. [Google Scholar] [CrossRef]
- Castro, A.; Duft, R.G.; Ferreira, M.L.V.; de Andrade, A.L.L.; Gáspari, A.F.; Silva, L.D.; de Oliveira-Nunes, S.G.; Cavaglieri, C.R.; Ghosh, S.; Bouchard, C.; et al. Association of skeletal muscle and serum metabolites with maximum power output gains in response to continuous endurance or high-intensity interval training programs: The TIMES study-A randomized controlled trial. PLoS ONE 2019, 14, e0212115. [Google Scholar] [CrossRef]
- Al-Khelaifi, F.; Diboun, I.; Donati, F.; Botre, F.; Alsayrafi, M.; Georgakopoulos, C.; Suhre, K.; Yousri, N.A.; Elrayess, M.A. A pilot study comparing the metabolic profiles of elite-level athletes from different sporting disciplines. Sports Med.-Open 2018, 4, 2. [Google Scholar] [CrossRef]
- James, L.A.; Lunn, P.G.; Elia, M. Glutamine metabolism in the gastrointestinal tract of the rat assess by the relative activities of glutaminase (EC 3.5.1.2) and glutamine synthetase (EC 6.3.1.2). Br. J. Nutr. 1998, 79, 365–372. [Google Scholar] [CrossRef]
- Perry, M.C.; Dufour, C.R.; Tam, I.S.; B’chir, W.; Giguère, V. Estrogen-related receptor-α coordinates transcriptional programs essential for exercise tolerance and muscle fitness. Mol. Endocrinol. 2014, 28, 2060–2071. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.T.; Wang, W.P.; Peng, S.C.; Lu, Y.; Du, J.; Cai, W. Farnesoid X receptor agonist tropifexor detoxifies ammonia by regulating the glutamine metabolism and urea cycles in cholestatic livers. Eur. J. Pharmacol. 2024, 966, 176334. [Google Scholar] [CrossRef]
- Adammek, F.; Chirino, T.Y.W.; Walzik, D.; Trebing, S.; Belen, S.; Renpening, D.; Zimmer, P.; Joisten, N. Kinetics of immune cell mobilization during acute aerobic exercise in healthy adults. Int. J. Sports Med. 2024, 45, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Adkins, J.N.; Bodine, S.C. Endurance exercise causes a multi-organ full-body molecular reaction. Nature 2024, 629, 174–183. [Google Scholar] [CrossRef]
- Casella, G.; Munk, R.; Kim, K.M.; Piao, Y.L.; De, S.; Abdelmohsen, K.; Gorospe, M. Transcriptome signature of cellular senescence. Nucleic Acids Res. 2019, 47, 11476. [Google Scholar] [CrossRef]
- Arsenis, N.C.; You, T.J.; Ogawa, E.F.; Tinsley, G.M.; Zuo, L. Physical activity and telomere length: Impact of aging and potential mechanisms of action. Oncotarget 2017, 8, 45008–45019. [Google Scholar] [CrossRef] [PubMed]
- Bolotta, A.; Filardo, G.; Abruzzo, P.M.; Astolfi, A.; De Sanctis, P.; Di Martino, A.; Hofer, C.; Indio, V.; Kern, H.; Löfler, S.; et al. Skeletal muscle gene expression in long-term endurance and resistance trained elderly. Int. J. Mol. Sci. 2020, 21, 3988. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, J.H.; Lee, H.Y.; Min, K.J. Sirtuin signaling in cellular senescence and aging. BMB Rep. 2019, 52, 24–34. [Google Scholar] [CrossRef]
- Marcotte, G.R.; Miller, M.J.; Kunz, H.E.; Ryan, Z.C.; Strub, M.D.; Vanderboom, P.M.; Heppelmann, C.J.; Chau, S.; Von Ruff, Z.D.; Kilroe, S.P.; et al. GADD45A is a mediator of mitochondrial loss, atrophy, and weakness in skeletal muscle. JCI Insight 2023, 8, e171772. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, G.Q.; Mao, P.; Zhang, J.; Zhang, L.; Liu, L.K.; Wang, J.; Owusu, L.; Ren, B.Y.; Tang, Y.W.; et al. Down-regulation of GADD45A enhances chemosensitivity in melanoma. Sci. Rep. 2018, 8, 4111. [Google Scholar] [CrossRef]
- Wang, X.; Zhan, Q.; Coursen, J.D.; Khan, M.A.; Kontny, H.U.; Yu, L.; Hollander, M.C.; O’Connor, P.M.; Fornace, A.J., Jr.; Harris, C.C. GADD45 induction of a G2/M cell cycle checkpoint. Proc. Biol. Sci. 1999, 96, 3706–3711. [Google Scholar] [CrossRef]
- Pilat, C.; Krüger, K.; Frech, T.; Mooren, F.C. Exercise-induced cytokine changes in antigen stimulated whole-blood cultures compared to serum. J. Immunol. Methods 2017, 440, 58–66. [Google Scholar] [CrossRef]
- Reichel, T.; Held, S.; Schwarz, A.; Hacker, S.; Wesemann, F.; Donath, L.; Krüger, K. Acute response of biomarkers in plasma from capillary blood after a strenuous endurance exercise bout. Eur. J. Appl. Physiol. 2022, 123, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, H.H.; Urhausen, A.; Valet, G.; Heidelbach, U.; Kindermann, W. Overtraining and immune system: A prospective longitudinal study in endurance athletes. Med. Sci. Sports Exerc. 1998, 30, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Laughlin, M.H.; Padilla, J.; Jenkins, N.T.; Thorne, P.K.; Martin, J.S.; Rector, R.S.; Akter, S.; Davis, J.W. Exercise-induced differential changes in gene expression among arterioles of skeletal muscles of obese rats. J. Appl. Physiol. 2015, 119, 583–603. [Google Scholar] [CrossRef]
- Ducharme, A.; Frantz, S.; Aikawa, M.; Rabkin, E.; Lindsey, M.; Rohde, L.E.; Schoen, F.J.; Kelly, R.A.; Werb, Z.; Libby, P.; et al. Targeted deletion of matrix metalloproteinase-9 attenuates left ventricular enlargement and collagen accumulation after experimental myocardial infarction. J. Clin. Invest. 2000, 106, 55–62. [Google Scholar] [CrossRef]
- Regouski, M.; Galenko, O.; Doleac, J.; Olsen, A.L.; Jacobs, V.; Liechty, D.; White, K.L.; Bunch, T.J.; Lee, P.M.; Rutigliano, H.M.; et al. Spontaneous atrial fibrillation in transgenic goats with TGF (transforming growth factor)-β1 induced atrial myopathy with endurance exercise. Circ. Arrhythmia Electrophysiol. 2019, 12, e007499. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.L.; Hsieh, P.L.; Hung, C.H.; Cheng, H.C.; Chou, W.C.; Chu, P.M.; Chang, Y.C.; Tsai, K.L. Early moderate intensity aerobic exercise intervention prevents doxorubicin-caused cardiac dysfunction through inhibition of cardiac fibrosis and inflammation. Cancers 2020, 12, 1102. [Google Scholar] [CrossRef]
- Goj, T.; Hoene, M.; Fritsche, L.; Schneeweiss, P.; Machann, J.; Petrera, A.; Hauck, S.M.; Fritsche, A.; Birkenfeld, A.L.; Peter, A.; et al. The acute cytokine response to 30-minute exercise bouts before and after 8-week endurance training in individuals with obesity. J. Clin. Endocr. Metab. 2023, 108, 865–875. [Google Scholar] [CrossRef]
- Mycka, G.; Ropka-Molik, K.; Cywinska, A.; Szmatola, T.; Stefaniuk-Szmukier, M. Molecular insights into the lipid-carbohydrates metabolism switch under the endurance effort in Arabian horses. Equine Vet. J. 2024, 56, 586–597. [Google Scholar] [CrossRef]
- Le Moyec, L.; Robert, C.; Triba, M.N.; Bouchemal, N.; Mach, N.; Rivière, J.; Zalachas-Rebours, E.; Barrey, E. A first step toward unraveling the energy metabolism in endurance horses: Comparison of plasma nuclear magnetic resonance metabolomic profiles before and after different endurance race distances. Front. Mol. Biosci. 2019, 6, 45. [Google Scholar] [CrossRef] [PubMed]
- Edwin, R.P.; Christopher, G.G. The effect of muscle phospholipid fatty acid composition on exercise performance: A direct test in the migratory white-throated sparrow (Zonotrichia albicollis). Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 297, R775–R782. [Google Scholar] [CrossRef]
- Lyudinina, A.Y.; Ivankova, G.E.; Bojko, E.R. Priority use of medium-chain fatty acids during high-intensity exercise in cross-country skiers. J. Int. Soc. Sports Nutr. 2018, 15, 57. [Google Scholar] [CrossRef] [PubMed]
- Mougios, V.; Ring, S.; Petridou, A.; Nikolaidis, M.G. Duration of coffee- and exercise-induced changes in the fatty acid profile of human serum. J. Appl. Physiol. 2003, 94, 476–484. [Google Scholar] [CrossRef]
- Gemmink, A.; Schrauwen, P.; Hesselink, M.K.C. Exercising your fat (metabolism) into shape: A muscle-centred view. Diabetologia 2020, 63, 1453–1463. [Google Scholar] [CrossRef]
- Behrend, A.M.; Harding, C.O.; Shoemaker, J.D.; Matern, D.; Elliot, D.L.; Gillingham, M.B. Substrate oxidation and cardiac performance during exercise in disorders of long chain fatty acid oxidation. Mol. Genet. Metab. 2012, 105, 110–115. [Google Scholar] [CrossRef]

| Group | Sample | Raw Date | Clean Date | Clean Ratio | Mapped Reads | Q20% | Q30% | GC Content% | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Read | Base (G) | Read | Base (G) | |||||||
| T0 | 1 | 42,199,338 | 6.33 | 41,398,488 | 6.21 | 98.10 | 38,532,112 (93.08%) | 98.05 | 94.62 | 56.36 |
| 2 | 46,136,320 | 6.92 | 45,499,530 | 6.82 | 98.62 | 42,987,209 (94.48%) | 98.20 | 94.92 | 55.25 | |
| 3 | 42,628,206 | 6.39 | 42,202,070 | 6.33 | 99.00 | 40,298,138 (95.49%) | 98.03 | 94.42 | 54.3 | |
| 4 | 48,256,328 | 7.24 | 47,745,646 | 7.16 | 98.94 | 43,232,922 (90.55%) | 98.03 | 94.55 | 58.74 | |
| 5 | 48,678,986 | 7.3 | 48,039,608 | 7.21 | 98.69 | 43,347,330 (90.23%) | 97.82 | 94.10 | 60.34 | |
| 6 | 46,770,976 | 7.02 | 45,942,990 | 6.89 | 98.23 | 43,698,460 (95.11%) | 98.10 | 94.64 | 53.21 | |
| T1 | 1 | 44,806,188 | 6.72 | 44,262,422 | 6.64 | 98.79 | 41,365,321 (93.45%) | 98.04 | 94.55 | 56.69 |
| 2 | 47,663,710 | 7.15 | 47,111,612 | 7.07 | 98.84 | 44,558,797 (94.58%) | 98.08 | 94.67 | 57.95 | |
| 3 | 48,090,328 | 7.21 | 47,441,622 | 7.12 | 98.65 | 44,392,457 (93.57%) | 97.85 | 94.21 | 58.2 | |
| 4 | 41,936,500 | 6.29 | 41,013,050 | 6.15 | 97.80 | 38,099,695 (92.9%) | 97.91 | 94.40 | 57.29 | |
| 5 | 49,041,564 | 7.36 | 48,512,654 | 7.28 | 98.92 | 45,381,440 (93.55%) | 97.84 | 94.16 | 56.55 | |
| 6 | 47,552,158 | 7.13 | 46,749,566 | 7.01 | 98.31 | 42,493,180 (90.9%) | 98.09 | 94.78 | 57.69 | |
| Gene Name | Forward Primer Sequence (5′→3′) | Reverse Primer Sequence (5′→3′) | Size (bp) |
|---|---|---|---|
| CD14 | GGAGCAGGTGCCTAAAGGACTA | AGTTCTGGTCTTGCTGCTTGGA | 167 |
| TRIB1 | AAATCCGACGTGGACAGTTCTG | GCTTCCAAGACGGACTCAAAC | 148 |
| S100A8 | GCCATCTATAGGGACGACTTGAA | GATGAGGAACTCCTGGAAGTTAACA | 138 |
| LTF | GATGGCGGTTTGGTGTATGA | GCTGCCCTTCTTCACTACGG | 129 |
| GAPDH | ATGGTGAAGGTCGGAGTAAACG | CATGGGTGGAATCATACTGAAACA | 154 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Ren, W.; Li, Z.; Li, L.; Wang, R.; Ma, S.; Zeng, Y.; Meng, J.; Yao, X. Regulatory Mechanisms of Yili Horses During an 80 km Race Based on Transcriptomics and Metabolomics Analyses. Int. J. Mol. Sci. 2025, 26, 2426. https://doi.org/10.3390/ijms26062426
Wang J, Ren W, Li Z, Li L, Wang R, Ma S, Zeng Y, Meng J, Yao X. Regulatory Mechanisms of Yili Horses During an 80 km Race Based on Transcriptomics and Metabolomics Analyses. International Journal of Molecular Sciences. 2025; 26(6):2426. https://doi.org/10.3390/ijms26062426
Chicago/Turabian StyleWang, Jianwen, Wanlu Ren, Zexu Li, Luling Li, Ran Wang, Shikun Ma, Yaqi Zeng, Jun Meng, and Xinkui Yao. 2025. "Regulatory Mechanisms of Yili Horses During an 80 km Race Based on Transcriptomics and Metabolomics Analyses" International Journal of Molecular Sciences 26, no. 6: 2426. https://doi.org/10.3390/ijms26062426
APA StyleWang, J., Ren, W., Li, Z., Li, L., Wang, R., Ma, S., Zeng, Y., Meng, J., & Yao, X. (2025). Regulatory Mechanisms of Yili Horses During an 80 km Race Based on Transcriptomics and Metabolomics Analyses. International Journal of Molecular Sciences, 26(6), 2426. https://doi.org/10.3390/ijms26062426






