Investigating the Mechanisms of Adventitious Root Formation in Semi-Tender Cuttings of Prunus mume: Phenotypic, Phytohormone, and Transcriptomic Insights
Abstract
1. Introduction
2. Results
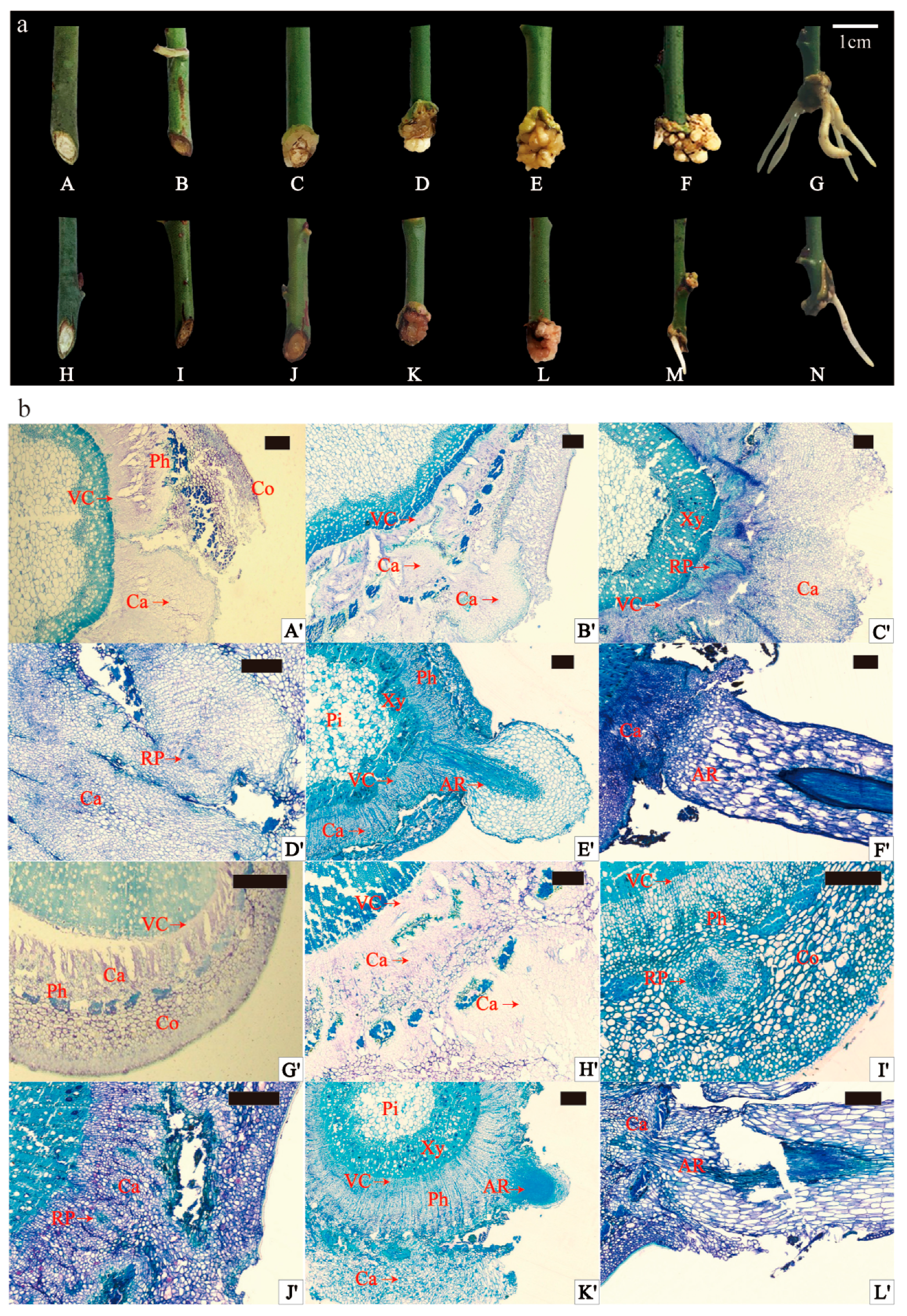
2.1. External Morphology and Anatomical Observation of Adventitious Rooting of Scion
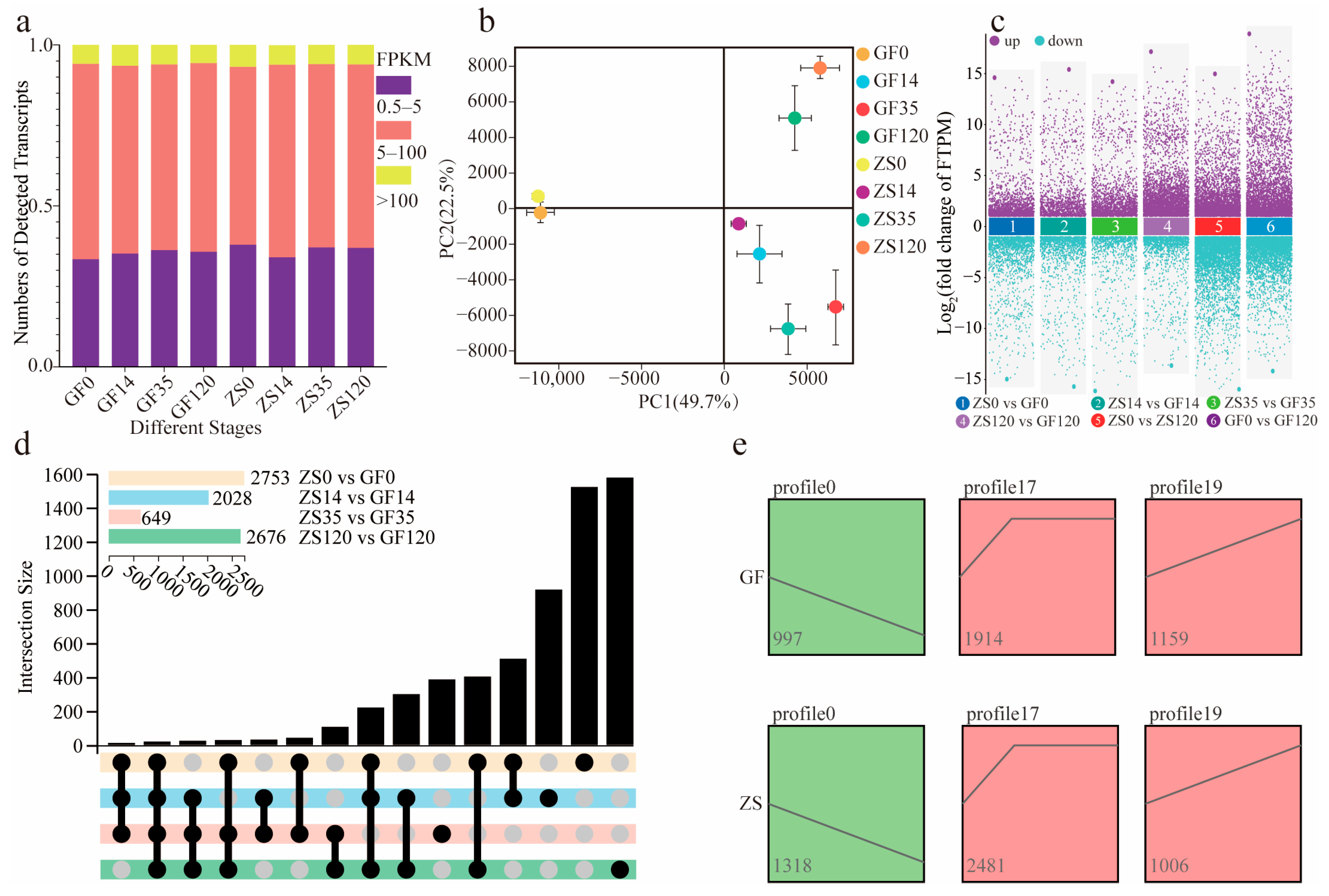
2.2. Transcriptome Sequencing Overview and Differential Expression Analysis
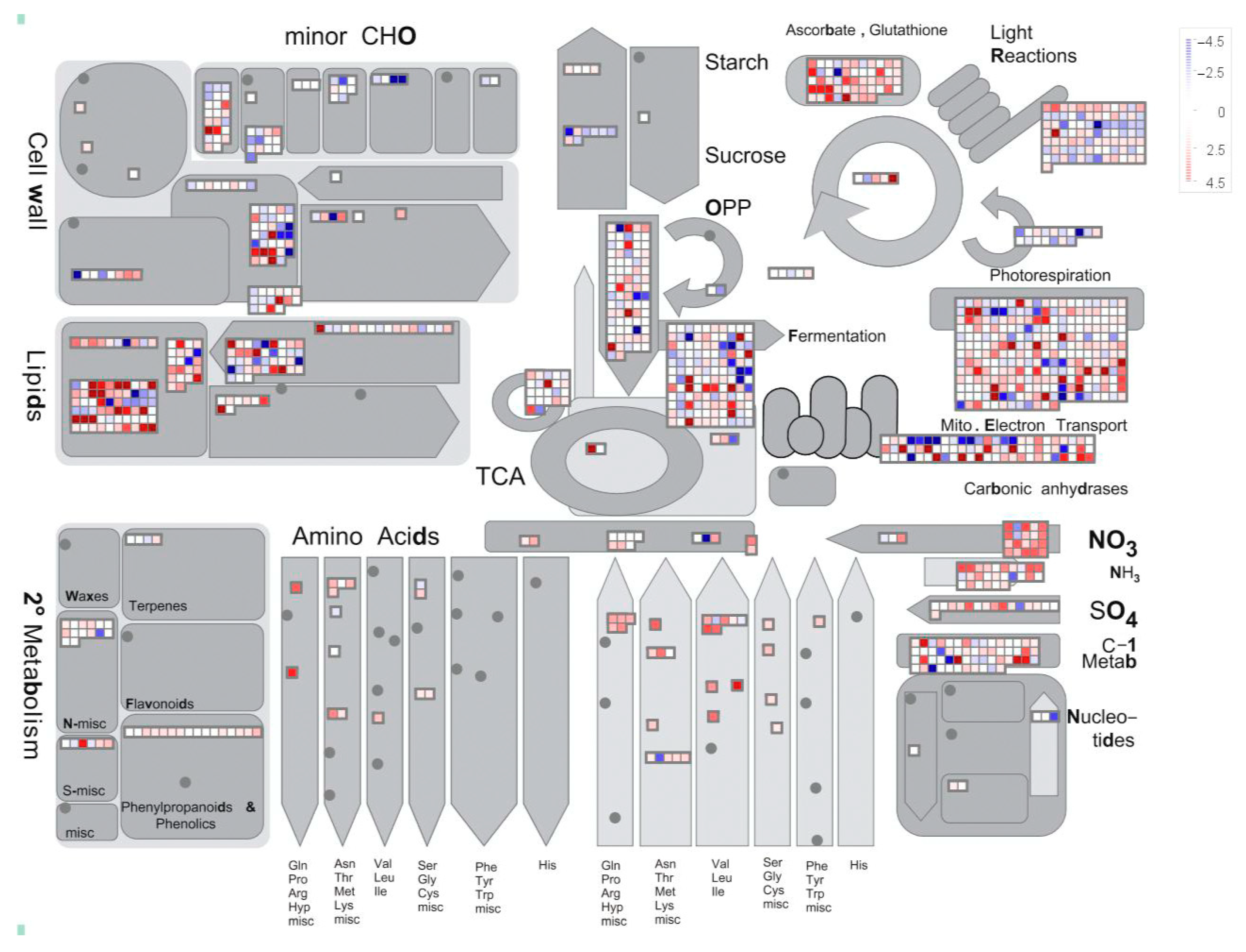
2.3. Annotation and Classification of Differentially Expressed Genes
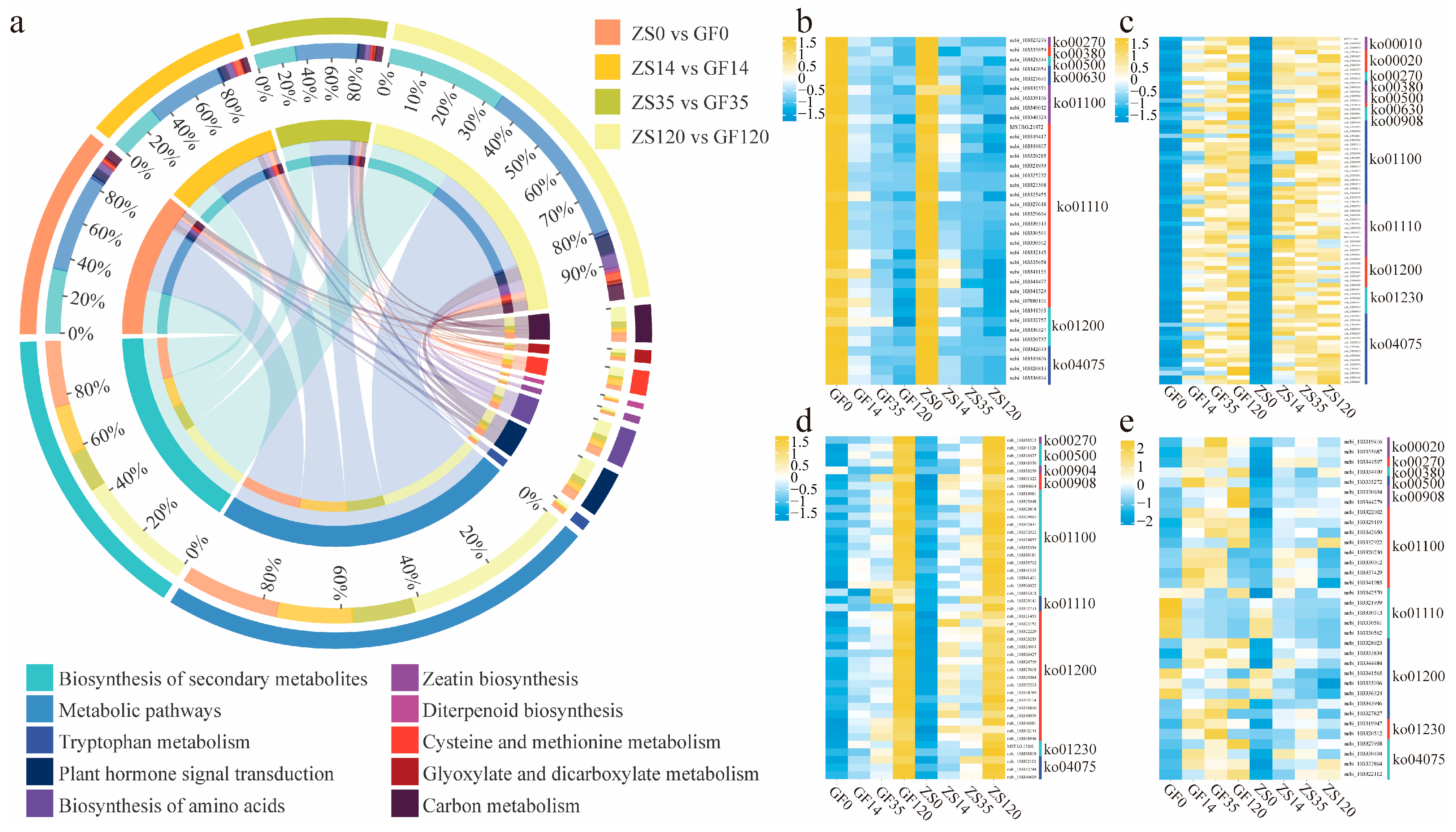
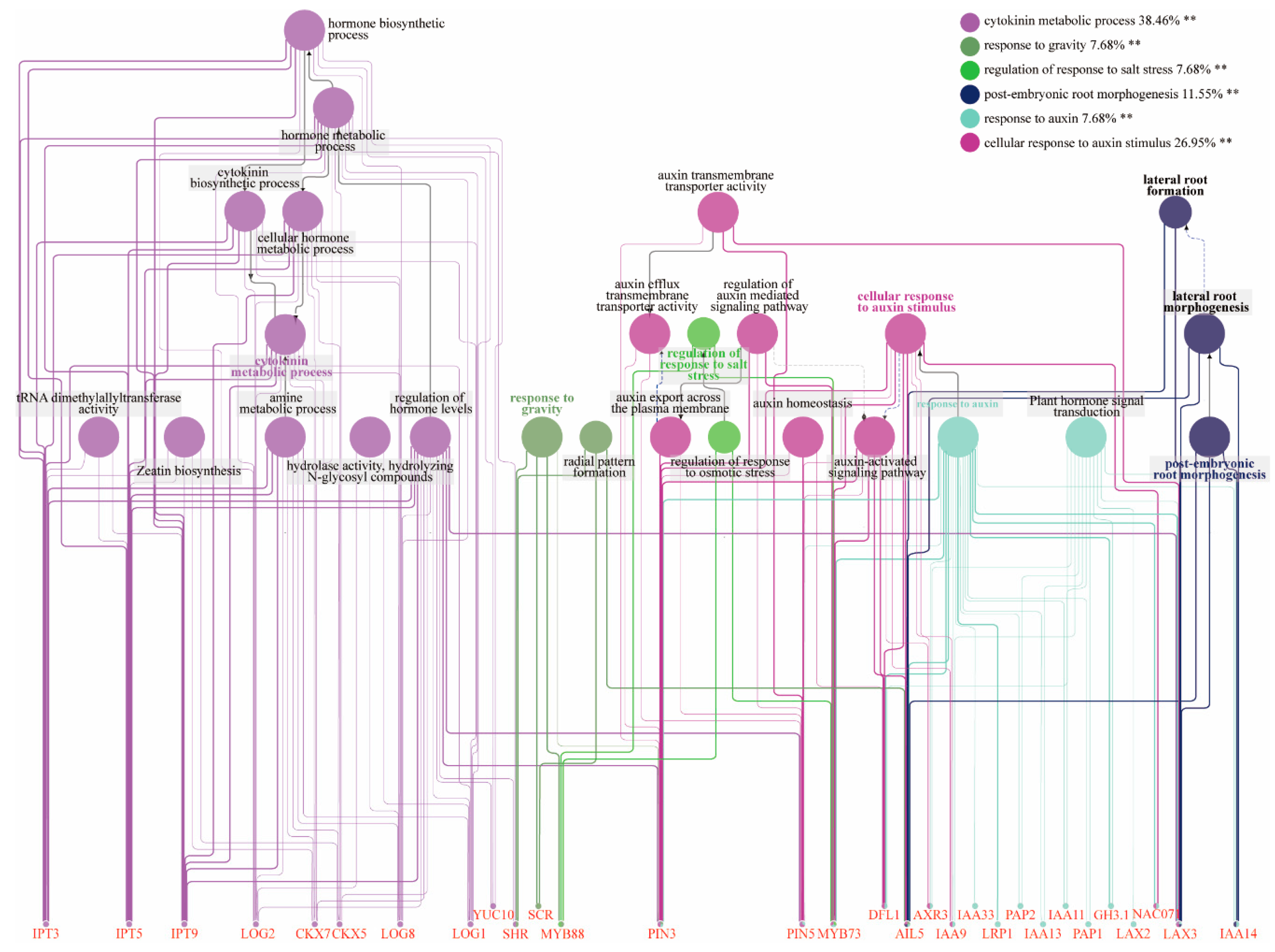
2.4. Identification of DEGs Related to Root Developmental Regulation Across Different Stages
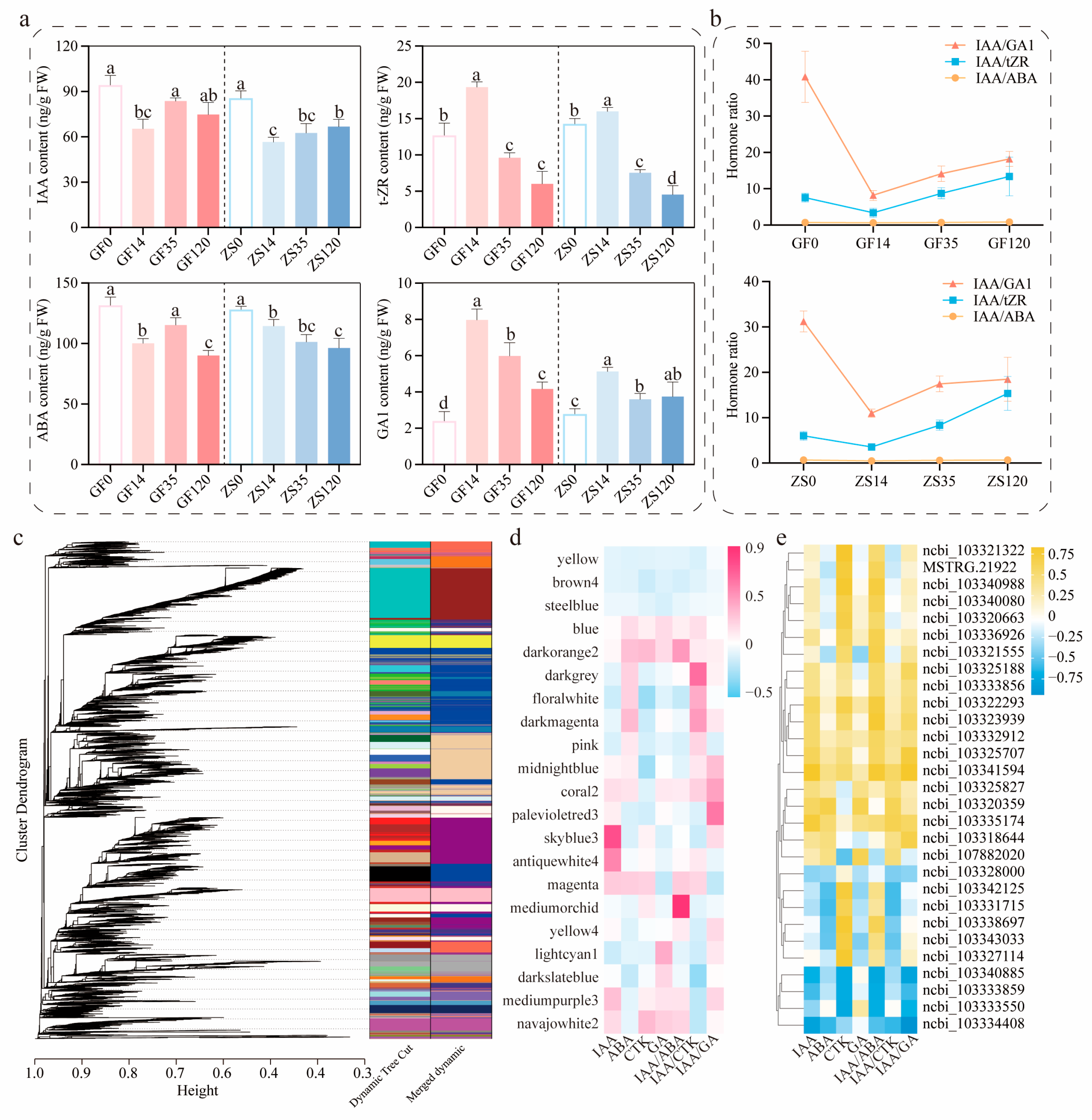
2.5. Co-Expression Analysis and Validation of Key Hormone Levels and Related Genes
2.6. Construction of Gene Co-Expression Networks
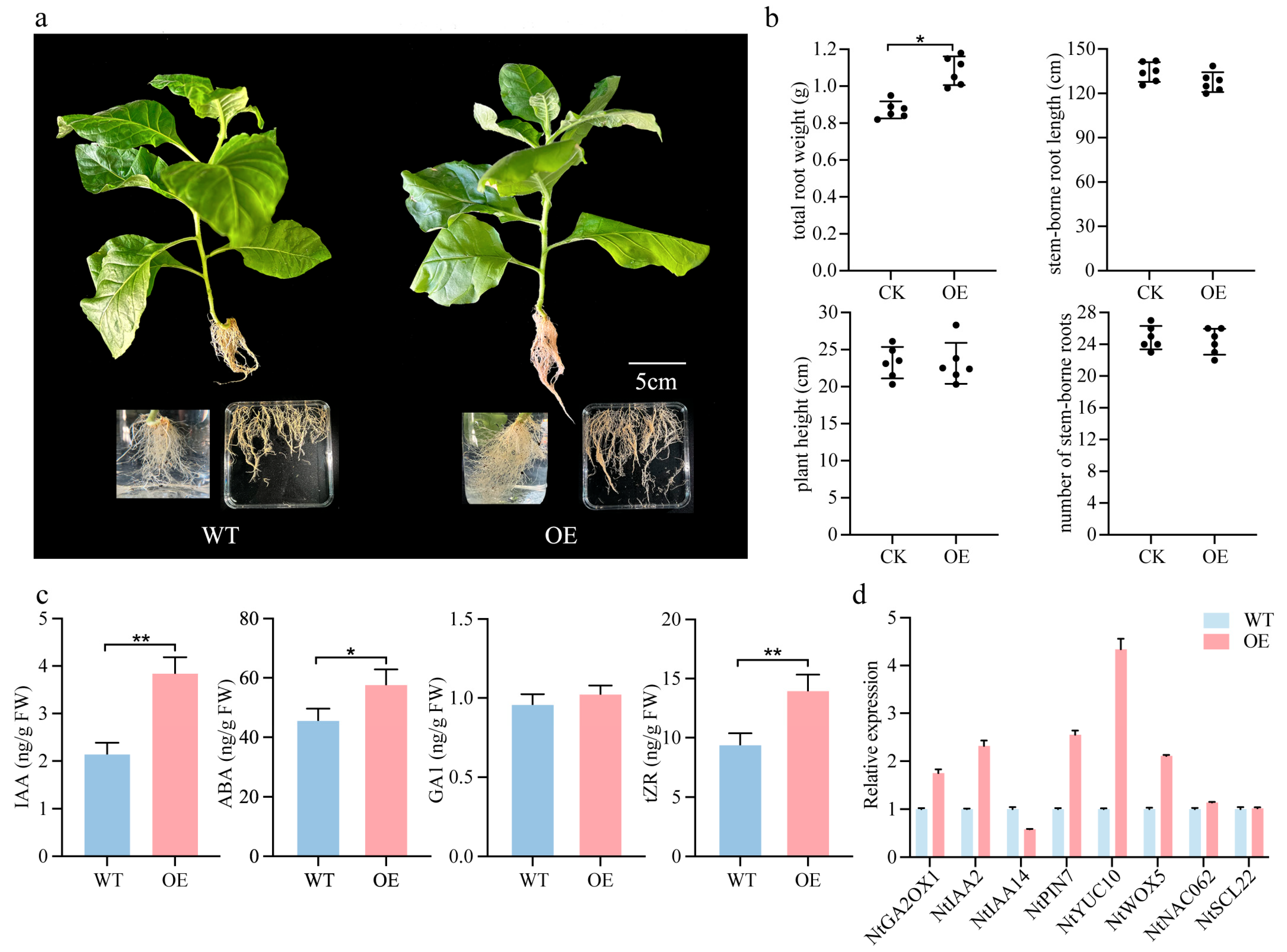
2.7. Overexpression of PmSHR Increases the Number of Lateral Roots in Tobacco
3. Discussion
3.1. Anatomical Structures and Rooting Patterns in Adventitious Root Formation of P. mume Cuttings
3.2. Endogenous Hormonal Regulation and Gene Expression in Adventitious Root Formation of P. mume
3.3. Hormonal Ratios and Gene Interactions in the Regulation of Adventitious Root Formation in P. mume
3.4. Role of the SHR Gene in Adventitious Root Formation
4. Materials and Methods
4.1. Experimental Materials
4.2. Cutting Conditions
4.3. Morphological and Anatomical Analysis of Adventitious Root Formation in P. mume Cuttings
4.4. Quantitative Analysis of Endogenous Hormones
4.5. RNA Isolation, Transcriptome Sequencing, and Differential Gene Expression Analysis
4.6. Tobacco Transformation
4.7. Quantitative Real-Time PCR Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IAA | indole-3-acetic acid |
| PIN | puroindoline |
| LOG | cytokinin-activating enzyme; Lonely Guy |
| CKX | cytokinin oxidase |
| LAX | lax panicle |
| GA2OX | GA2-oxidase |
| WOX | WUSCHEL-related homeobox |
| SHR | SHORT-ROOT |
| NAC | no apical meristem |
| SCL | SCARECROW-like |
| ABA | abscisic acid |
| tZR | trans-zeatin nucleoside |
| YUC | YUCCA |
References
- Darras, A.I. Implementation of sustainable practices to ornamental plant cultivation worldwide: A critical review. Agronomy 2020, 10, 1570. [Google Scholar] [CrossRef]
- Wang, J.; Kan, J.; Wang, J.; Yan, X.; Li, Y.; Soe, T.; Tembrock, L.R.; Xing, G.; Li, S.; Wu, Z.; et al. The pan-plastome of Prunus mume: Insights into Prunus diversity, phylogeny, and domestication history. Front. Plant Sci. 2024, 15, 1404071. [Google Scholar] [CrossRef] [PubMed]
- Chen, J. China Mei Flower (Prunus mume) Cultivars in Colour; China Forestry Publishing House: Beijing, China, 2017; pp. 1–8. [Google Scholar]
- Fan, D.; Miao, R.; Lv, W.; Wen, Z.; Meng, J.; Liu, X.; Cheng, T.; Zhang, Q.; Sun, L. Prunus mume genome research: Current status and prospects. Ornam. Plant Res. 2024, 4, e006. [Google Scholar] [CrossRef]
- Ătefančič, M.; Ătampar, F.; Osterc, G. Influence of endogenous IAA levels and exogenous IBA on rooting and quality of leafy cuttings of Prunus ‘GiSelA 5’. J. Hortic. Sci. Biotechnol. 2006, 81, 508–512. [Google Scholar] [CrossRef]
- Anderson, N.O.; Hoover, E.; Kostick, S.A.; Tepe, E.; Tillman, J. Cutting type and time-of-year affect rooting ability of hardy minnesota Prunus species. J. Amer. Pomolog. Soc. 2016, 70, 114–123. [Google Scholar]
- Dong, R.R.; Chen, R.D. Effects of different cutting date on softwood-cutting rooting and related physiological and biochemical changes during rooting of Prunus mume Meiren (Chinese). Acta Agric. Zhejiangensis 2016, 28, 1522–1529. [Google Scholar]
- Bellini, C.; Păcurar, D.I.; Perrone, I. Adventitious roots and lateral roots: Similarities and differences. Annu. Rev. Plant Biol. 2014, 65, 639–666. [Google Scholar] [CrossRef]
- Wei, P.; Lv, Y.; Guang, Q.; Han, J.; Wang, Y.; Wang, X.; Song, L. Chifnα regulates adventitious root development in Lotus Japonicus via an auxin-mediated pathway. Plant Signal. Behav. 2023, 18, 2218670. [Google Scholar] [CrossRef]
- Păcurar, D.I.; Perrone, I.; Bellini, C. Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiol. Plant. 2014, 151, 83–96. [Google Scholar] [CrossRef]
- Druege, U.; Franken, P.; Hajirezaei, M.-R. Plant hormone homeostasis, signaling, and function during adventitious root formation in cuttings. Front. Plant Sci. 2016, 7, 381. [Google Scholar] [CrossRef]
- Manokari, M.; Raj, M.C.; Dey, A.; Faisal, M.; Alatar, A.A.; Joshee, N.; Shekhawat, M.S. Improvements in morpho-anatomical traits of adventitious roots of Hedyotis Biflora (L.) Lam. using silicon nanoparticles. Silicon 2023, 15, 5747–5755. [Google Scholar] [CrossRef]
- Da Costa, C.T.; de Almeida, M.R.; Ruedell, C.M.; Schwambach, J.; Maraschin, F.D.S.; Fett-Neto, A.G. When Stress and development go hand in hand: Main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 2013, 4, 133. [Google Scholar] [CrossRef] [PubMed]
- Zhi, L.; Hu, X. Adventitious root regeneration: Molecular basis and influencing factors. Phyton-Int. J. Exp. Bot. 2023, 92, 2825–2840. [Google Scholar] [CrossRef]
- Ahkami, A.; Scholz, U.; Steuernagel, B.; Strickert, M.; Haensch, K.-T.; Druege, U.; Reinhardt, D.; Nouri, E.; von Wiren, N.; Franken, P.; et al. Comprehensive transcriptome analysis unravels the existence of crucial genes regulating primary metabolism during adventitious root formation in Petunia hybrida. PLoS ONE 2014, 9, e100997. [Google Scholar] [CrossRef]
- Li, S.-W.; Shi, R.-F.; Leng, Y. De novo characterization of the mung bean transcriptome and transcriptomic analysis of adventitious rooting in seedlings using RNA-Seq. PLoS ONE 2015, 10, e0132969. [Google Scholar] [CrossRef]
- Chen, L.; Tong, J.; Xiao, L.; Ruan, Y.; Liu, J.; Zeng, M.; Huang, H.; Wang, J.-W.; Xu, L. Yucca-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 2016, 67, 4273–4284. [Google Scholar] [CrossRef]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. Wox11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Sun, P.; Zhang, J.; Xia, Y.; Hu, J.; Wang, L.; Lu, M. The Wuschela (Ptowusa) is involved in developmental plasticity of adventitious root in poplar. Genes 2020, 11, 176. [Google Scholar] [CrossRef]
- Li, M.; Fan, D.; Wen, Z.; Meng, J.; Li, P.; Cheng, T.; Zhang, Q.; Sun, L. Genome-wide identification of the Dof gene family: How it plays a part in mediating cold stress response in Prunus mume. Plant Physiol. Biochem. 2024, 217, 109215. [Google Scholar] [CrossRef]
- Park, S.-H.; Elhiti, M.; Wang, H.; Xu, A.; Brown, D.; Wang, A. Adventitious root formation of in vitro peach shoots is regulated by auxin and ethylene. Sci. Hortic. 2017, 226, 250–260. [Google Scholar] [CrossRef]
- Chen, H.; Lei, Y.; Sun, J.; Ma, M.; Deng, P.; Quan, J.E.; Bi, H. Effects of different growth hormones on rooting and endogenous hormone content of two Morus alba L. cuttings. Horticulturae 2023, 9, 552. [Google Scholar] [CrossRef]
- Mu, H.; Jin, X.; Ma, X.; Zhao, A.; Gao, Y.; Lin, L. Ortet age effect, anatomy and physiology of adventitious rooting in Tilia mandshurica softwood cuttings. Forests 2022, 13, 1427. [Google Scholar] [CrossRef]
- Sosnowski, J.; Truba, M.; Vasileva, V. The impact of auxin and cytokinin on the growth and development of selected crops. Agriculture 2023, 13, 724. [Google Scholar] [CrossRef]
- Mao, J.; Zhang, D.; Meng, Y.; Li, K.; Wang, H.; Han, M. Inhibition of adventitious root development in apple rootstocks by cytokinin is based on its suppression of adventitious root primordia formation. Physiol. Plant. 2019, 166, 663–676. [Google Scholar] [CrossRef]
- Mauriat, M.; Petterle, A.; Bellini, C.; Moritz, T. Gibberellins Inhibit Adventitious Rooting in Hybrid Aspen and Arabidopsis by Affecting Auxin Transport. Plant J. 2014, 78, 372–384. [Google Scholar] [CrossRef]
- Zhao, M.; Lei, Y.; Wu, L.; Qi, H.; Song, Z.; Xu, M. The Mir159a-Pemyb33 module regulates poplar adventitious rooting through the abscisic acid signal pathway. Plant J. 2024, 118, 879–891. [Google Scholar] [CrossRef]
- Chagas, E.A.; Pio, R.; Bettiol Neto, J.E.; Sobierajski, G.D.R.; Campo Dall’Orto, F.A.; Signorini, G. Rooting of peach and clones of japanese apricot cutting treated with iba. Cienc. Agrotecnol. 2008, 32, 986–991. [Google Scholar] [CrossRef]
- Sándor, G.; Rabnecz, G.; Hajagos, A.; Nehiba, B. Iba uptake and metabolism of different type of plum rootstocks hardwood cuttings. Acta Biol. Szeged. 2008, 52, 237–240. [Google Scholar]
- Wamhoff, D.; Guendel, A.; Wagner, S.; Ortleb, S.; Borisjuk, L.; Winkelmann, T. Anatomical limitations in adventitious root formation revealed by magnetic resonance imaging, infrared spectroscopy, and histology of rose genotypes with contrasting rooting phenotypes. J. Exp. Bot. 2024, 75, 4784–4801. [Google Scholar] [CrossRef]
- Porfirio, S.; Gomes da Silva, M.D.R.; Cabrita, M.J.; Azadi, P.; Peixe, A. Reviewing current knowledge on olive (Olea europaea L.) adventitious root formation. Sci. Hortic. 2016, 198, 207–226. [Google Scholar] [CrossRef]
- Zhao, X.; Zheng, H.; Li, S.; Yang, C.; Jiang, J.; Liu, G. The rooting of poplar cuttings: A review. New For. 2014, 45, 21–34. [Google Scholar] [CrossRef]
- Abshahi, M.; Zarei, H.; Zahedi, B.; García-Morote, F.A.; Rezaei Nejad, A. Secondary metabolite changes in maymars juniper cuttings (Juniperus sabina) under different treatments of propagation (iba, substrate and harvest time of cutting). Adv. Hortic. Sci. 2022, 36, 163–174. [Google Scholar] [CrossRef]
- Maynard, B.K.; Bassuk, N.L. Effects of stock plant etiolation, shading, banding, and shoot development on histology and cutting propagation of Carpinus betulus L. fastigiata. J. Am. Soc. Hortic. Sci. 1996, 121, 853–860. [Google Scholar] [CrossRef]
- Liu, G.; Zhao, J.; Liao, T.; Wang, Y.; Guo, L.; Yao, Y.; Cao, J. Histological dissection of cutting-inducible adventitious rooting in Platycladus orientalis reveals developmental endogenous hormonal homeostasis. Ind. Crops Prod. 2021, 170, 113817. [CrossRef]
- Uddin, S.; Munir, M.Z.; Larriba, E.; Perez-Perez, J.M.; Gull, S.; Pervaiz, T.; Mahmood, U.; Mahmood, Z.; Sun, Y.; Li, Y. Temporal profiling of physiological, histological, and transcriptomic dissection during auxin-induced adventitious root formation in tetraploid Robinia pseudoacacia micro-cuttings. Planta 2024, 259, 66. [Google Scholar] [CrossRef]
- Zhu, H.; Li, H.; Yu, J.; Zhao, H.; Zhang, K.; Ge, W. Regulatory mechanisms of Araux/IAA13 and Araux/IAA16 in the rooting process of Acer rubrum. Genes 2023, 14, 1206. [Google Scholar] [CrossRef]
- Suchkova, S.A.; Yamburov, M.S.; Astafurova, T.P.; Sirotkina, E.E. Iron oxyhydroxide effect on rooting of cuttings of Ribes nigrum and Ribes rubrum. Int. J. Geomate 2019, 17, 169–173. [Google Scholar] [CrossRef]
- Dong, R. The Studies on Softwood Cutting Technique and Rooting Mechanism of Cold-Resistant Mei; Beijing Forestry University: Beijing, China, 2016. [Google Scholar]
- Lambolez, A.; Kawamura, A.; Takahashi, T.; Rymen, B.; Iwase, A.; Favero, D.S.; Ikeuchi, M.; Suzuki, T.; Cortijo, S.; Jaeger, K.E.; et al. Warm temperature promotes shoot regeneration in Arabidopsis thaliana. Plant Cell Physiol. 2022, 63, 618–634. [Google Scholar] [CrossRef]
- Shao, F.; Wang, S.; Huang, W.; Liu, Z. Effects of iba on the rooting of branch cuttings of Chinese jujube (Zizyphus jujuba Mill.) and changes to nutrients and endogenous hormones. J. For. Res. 2018, 29, 1557–1567. [Google Scholar] [CrossRef]
- Hayat, F.; Bai, Y.; Iqbal, S.; Ma, C.; Ali, M.M.; Shahid, M.A.; Ul Hasan, M.; Mosa, W.F.A.; Khan, U.; Xiao, H.; et al. Genome-wide analysis and expression profiling of Yucca gene family associated with plant vigor in japanese apricot (Prunus mume Sieb. Et Zucc). Hortic. Environ. Biotechnol. 2023, 64, 819–833. [Google Scholar] [CrossRef]
- Zeng, Y.; Verstraeten, I.; Trinh, H.K.; Heugebaert, T.; Stevens, C.V.; Garcia-Maquilon, I.; Geelen, D. Arabidopsis hypocotyl adventitious root formation is suppressed by ABA signaling. Genes 2021, 8, 1141. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, L.; Forde, B.G.; Davies, W.J. The biphasic root growth response to abscisic acid in Arabidopsis involves interaction with ethylene and auxin signalling pathways. Front. Plant Sci. 2017, 8, 1493. [Google Scholar] [CrossRef] [PubMed]
- Mironova, V.; Teale, W.; Shahriari, M.; Dawson, J.; Palme, K. The systems biology of auxin in development emoryos. Trends Plant Sci. 2017, 22, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Baluska, F.; Samaj, J.; Menzel, D. Polar transport of auxin: Carrier-mediated flux across the plasma membrane or neurotransmitter-like secretion? Trends Cell Biol. 2003, 13, 282–285. [Google Scholar] [CrossRef]
- Bai, Y.; Cai, M.; Dou, Y.; Xie, Y.; Zheng, H.; Gao, J. Phytohormone crosstalk of cytokinin biosynthesis and signaling family genes in moso bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 2023, 24, 10863. [Google Scholar] [CrossRef]
- Gonzalez-Rizzo, S.; Crespi, M.; Frugier, F. The Medicago truncatula cre1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell. 2006, 18, 2680–2693. [Google Scholar] [CrossRef]
- Hartmann, A.; Senning, M.; Hedden, P.; Sonnewald, U.; Sonnewald, S. Reactivation of meristem activity and sprout growth in potato tubers require both cytokinin and gibberellin1. Plant Physiol. 2010, 155, 776–796. [Google Scholar] [CrossRef]
- Mai, N.T.N.; Phong, T.H.; Khai, H.D.; Cuong, D.M.; Luan, V.Q.; Tung, H.T.; Thu, P.T.M.; Phuong, H.T.N.; Vinh, B.V.T.; Vinh, N.Q.; et al. Endogenous hormone alteration during callus and adventitious root formation through thin cell layer culture system in Phyllanthus amarus. Plant Cell Tissue Organ Cult. 2024, 159, 42. [Google Scholar] [CrossRef]
- Wang, S.T.; Sun, G.D.; Luo, Y.; Qian, W.J.; Fan, K.; Ding, Z.T.; Hu, J.H. Role of IAA and primary metabolites in two rounds of adventitious root formation in softwood cuttings of Camellia sinensis (L.). Agronomy 2022, 12, 2486. [Google Scholar] [CrossRef]
- Zhang, L.; Hu, J.; Han, X.; Li, J.; Gao, Y.; Richards, C.M.; Zhang, C.; Tian, Y.; Liu, G.; Gul, H.; et al. A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 2019, 10, 1494. [Google Scholar] [CrossRef]
- Saito, T.; Opio, P.; Wang, S.; Ohkawa, K.; Kondo, S.; Maejima, T.; Ohara, H. Association of auxin, cytokinin, abscisic acid, and plant peptide response genes during adventitious root formation in marubakaido apple rootstock (Malus prunifolia Borkh. Var. Ringo Asami). Acta Physiol. Plant. 2019, 41, 41. [Google Scholar] [CrossRef]
- Chen, R.; Zhao, D.; Huang, X. Transcriptome analysis of easy- and hard-to-root tea plants uncovers roles for CSGH3.2 and CSGH3.3 in adventitious root formation. Plant Cell Tissue Organ Cult. 2022, 150, 385–398. [Google Scholar] [CrossRef]
- Zhao, D.; Wang, Y.; Feng, C.; Wei, Y.; Peng, X.; Guo, X.; Guo, X.; Zhai, Z.; Li, J.; Shen, X.; et al. Overexpression of MSGH3.5 inhibits shoot and root development through the auxin and cytokinin pathways in apple plants. Plant J. 2020, 103, 166–183. [Google Scholar] [CrossRef]
- Li, J.; Xu, P.; Zhang, B.; Song, Y.; Wen, S.; Bai, Y.; Ji, L.; Lai, Y.; He, G.; Zhang, D. Paclobutrazol promotes root development of difficult-to-root plants by coordinating auxin and abscisic acid signaling pathways in Phoebe bournei. Int. J. Mol. Sci. 2023, 24, 3753. [Google Scholar] [CrossRef]
- Emenecker, R.J.; Strader, L.C. Auxin-abscisic acid interactions in plant growth and development. Biomolecules 2020, 10, 281. [Google Scholar] [CrossRef]
- Cui, H.; Kong, D.; Liu, X.; Hao, Y. Scarecrow, SCR-Like 23 and short-root control bundle sheath cell fate and function in Arabidopsis thaliana. Plant J. 2014, 78, 319–327. [Google Scholar] [CrossRef]
- Dhar, S.; Kim, J.; Yoon, E.K.; Jang, S.; Ko, K.; Lim, J. Short-root controls cell elongation in the etiolated Arabidopsis hypocotyl. Mol. Cells 2022, 45, 243–256. [Google Scholar] [CrossRef]
- Shaar-Moshe, L.; Brady, S.M.M. Short-Root and scarecrow homologs regulate patterning of diverse cell types within and between species. New Phytol. 2023, 237, 1542–1549. [Google Scholar] [CrossRef]
- Druege, U.; Hilo, A.; Manuel Perez-Perez, J.; Klopotek, Y.; Acosta, M.; Shahinnia, F.; Zerche, S.; Franken, P.; Hajirezaei, M.R. Molecular and physiological control of adventitious rooting in cuttings: Phytohormone action meets resource allocation. Ann. Bot. 2019, 123, 929–949. [Google Scholar] [CrossRef]
- Winter, C.M.; Szekely, P.; Popov, V.; Belcher, H.; Carter, R.; Jones, M.; Fraser, S.E.; Truong, T.V.; Benfey, P.N. SHR and SCR coordinate root patterning and growth early in the cell cycle. Nature 2024, 626, 611–616. [Google Scholar] [CrossRef]
- Henry, S.; Dievart, A.; Divol, F.; Pauluzzi, G.; Meynard, D.; Swarup, R.; Wu, S.; Gallagher, K.L.; Perin, C. SHR overexpression induces the formation of supernumerary cell layers with cortex cell identity in rice. Dev. Biol. 2017, 425, 1–7. [Google Scholar] [CrossRef]
- Curci, P.L.; Zhang, J.; Maehler, N.; Seyfferth, C.; Mannapperuma, C.; Diels, T.; Van Hautegem, T.; Jonsen, D.; Street, N.; Hvidsten, T.R.; et al. Identification of growth regulators using cross-species network analysis in plants. Plant Physiol. 2022, 190, 2350–2365. [Google Scholar] [CrossRef] [PubMed]
- Ljung, K.; Sandberg, G.; Moritz, T. Plant Hormones; Springer: Dordrecht, The Netherlands, 2010; pp. 717–718. [Google Scholar]
- Van Meulebroek, L.; Vanden Bussche, J.; Steppe, K.; Vanhaecke, L. Ultra-high performance liquid chromatography coupled to high resolution orbitrap mass spectrometry for metabolomic profiling of the endogenous phytohormonal status of the tomato plant. J. Chromatogr. A 2012, 1260, 67–80. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. Hisat: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. Stringtie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A guide to using mapman to visualize and compare omics data in plants: A case study in the crop species, maize. Plant Cell Environ. 2009, 32, 1211–1229. [Google Scholar] [CrossRef]
- Anwar, A.; Zhang, S.; Wang, L.; He, L.; Gao, J. BrCYP71A15 negatively regulates hg stress tolerance by modulating cell wall biosynthesis in yeast. Plants 2023, 12, 723. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Li, Y.; Li, Z.; Gu, X.; Wang, Z.; Qin, X.; Li, Q. Investigating the Mechanisms of Adventitious Root Formation in Semi-Tender Cuttings of Prunus mume: Phenotypic, Phytohormone, and Transcriptomic Insights. Int. J. Mol. Sci. 2025, 26, 2416. https://doi.org/10.3390/ijms26062416
Wang X, Li Y, Li Z, Gu X, Wang Z, Qin X, Li Q. Investigating the Mechanisms of Adventitious Root Formation in Semi-Tender Cuttings of Prunus mume: Phenotypic, Phytohormone, and Transcriptomic Insights. International Journal of Molecular Sciences. 2025; 26(6):2416. https://doi.org/10.3390/ijms26062416
Chicago/Turabian StyleWang, Xiujun, Yue Li, Zihang Li, Xiaowen Gu, Zixu Wang, Xiaotian Qin, and Qingwei Li. 2025. "Investigating the Mechanisms of Adventitious Root Formation in Semi-Tender Cuttings of Prunus mume: Phenotypic, Phytohormone, and Transcriptomic Insights" International Journal of Molecular Sciences 26, no. 6: 2416. https://doi.org/10.3390/ijms26062416
APA StyleWang, X., Li, Y., Li, Z., Gu, X., Wang, Z., Qin, X., & Li, Q. (2025). Investigating the Mechanisms of Adventitious Root Formation in Semi-Tender Cuttings of Prunus mume: Phenotypic, Phytohormone, and Transcriptomic Insights. International Journal of Molecular Sciences, 26(6), 2416. https://doi.org/10.3390/ijms26062416





