Construction of T-Cell-Related Prognostic Risk Models and Prediction of Tumor Immune Microenvironment Regulation in Pancreatic Adenocarcinoma via Integrated Analysis of Single-Cell RNA-Seq and Bulk RNA-Seq
Abstract
1. Introduction
2. Results
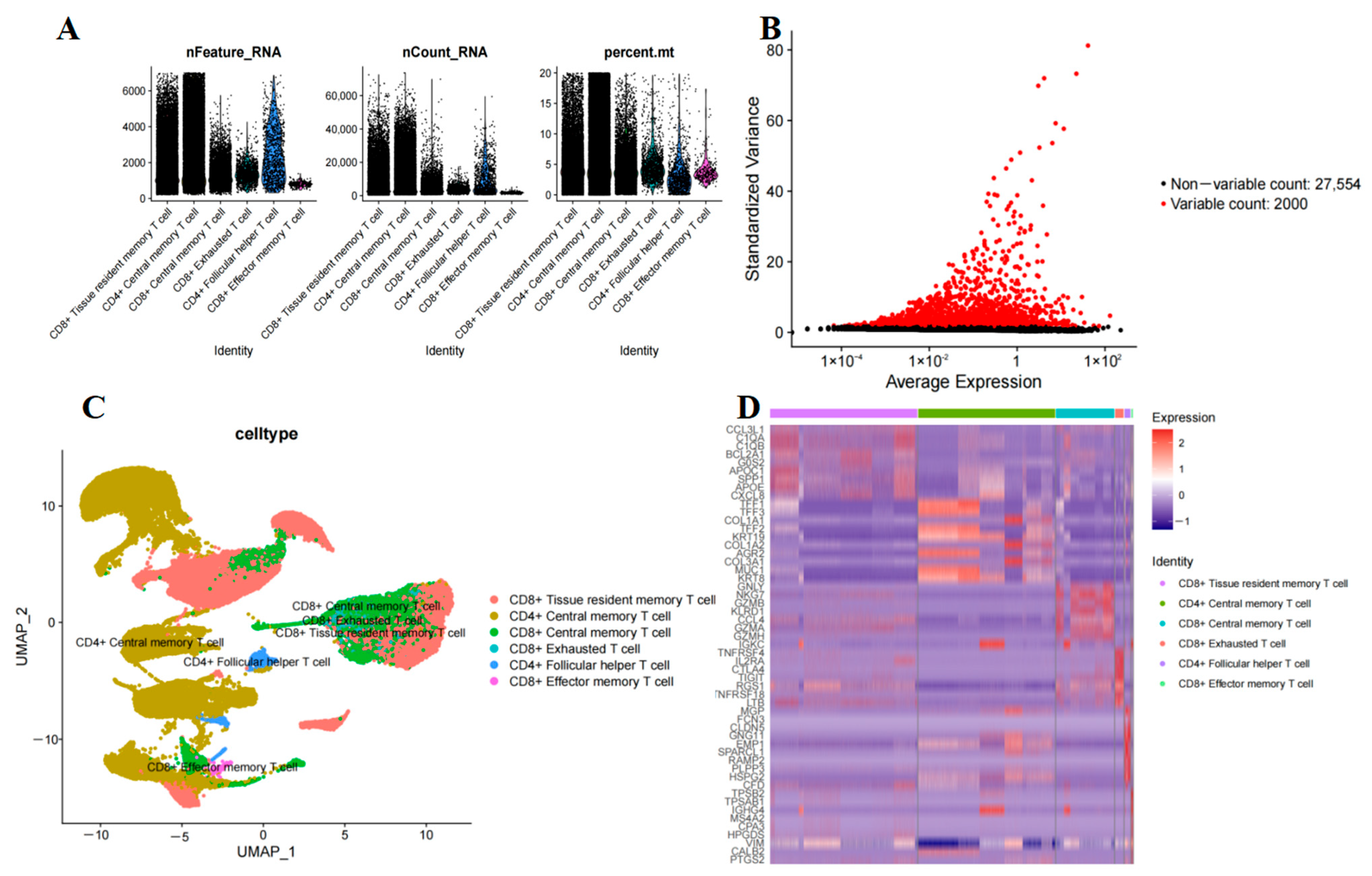
2.1. T-Cell Subset Analysis in PAAD
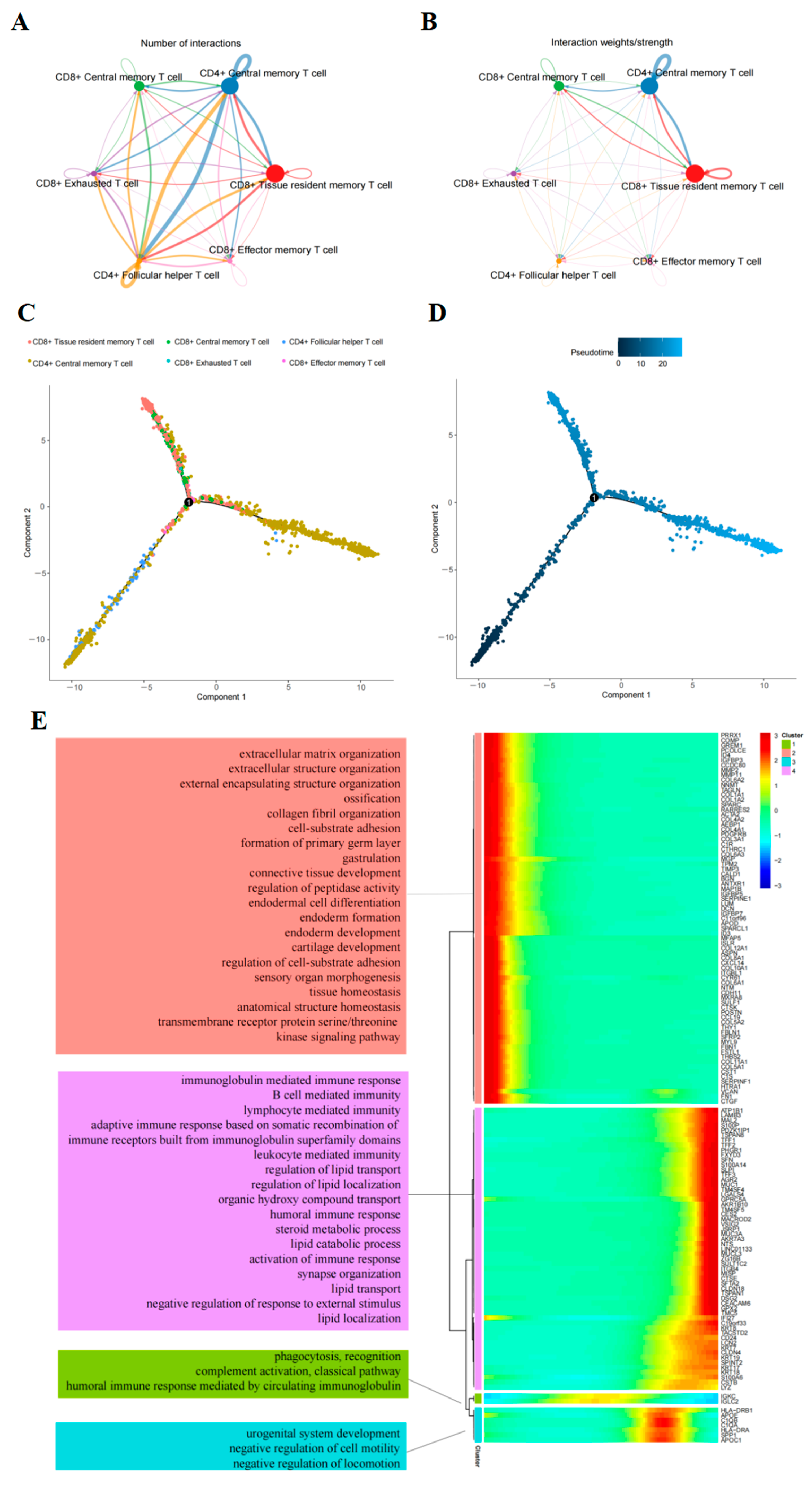
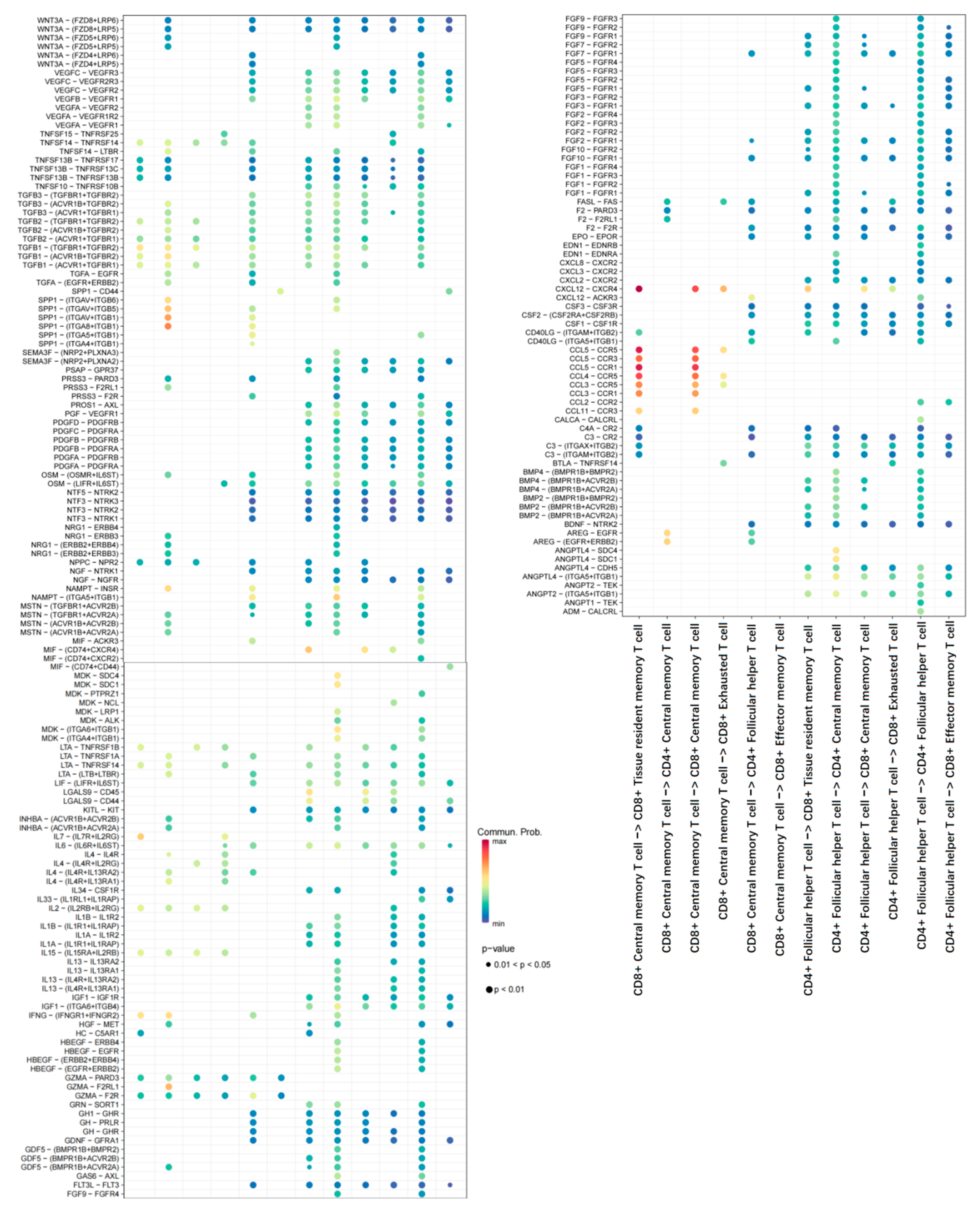
2.2. Cell–Cell Interaction Analysis Related to T-Cells
2.3. Trajectory of T-Cells Maturation in PAAD
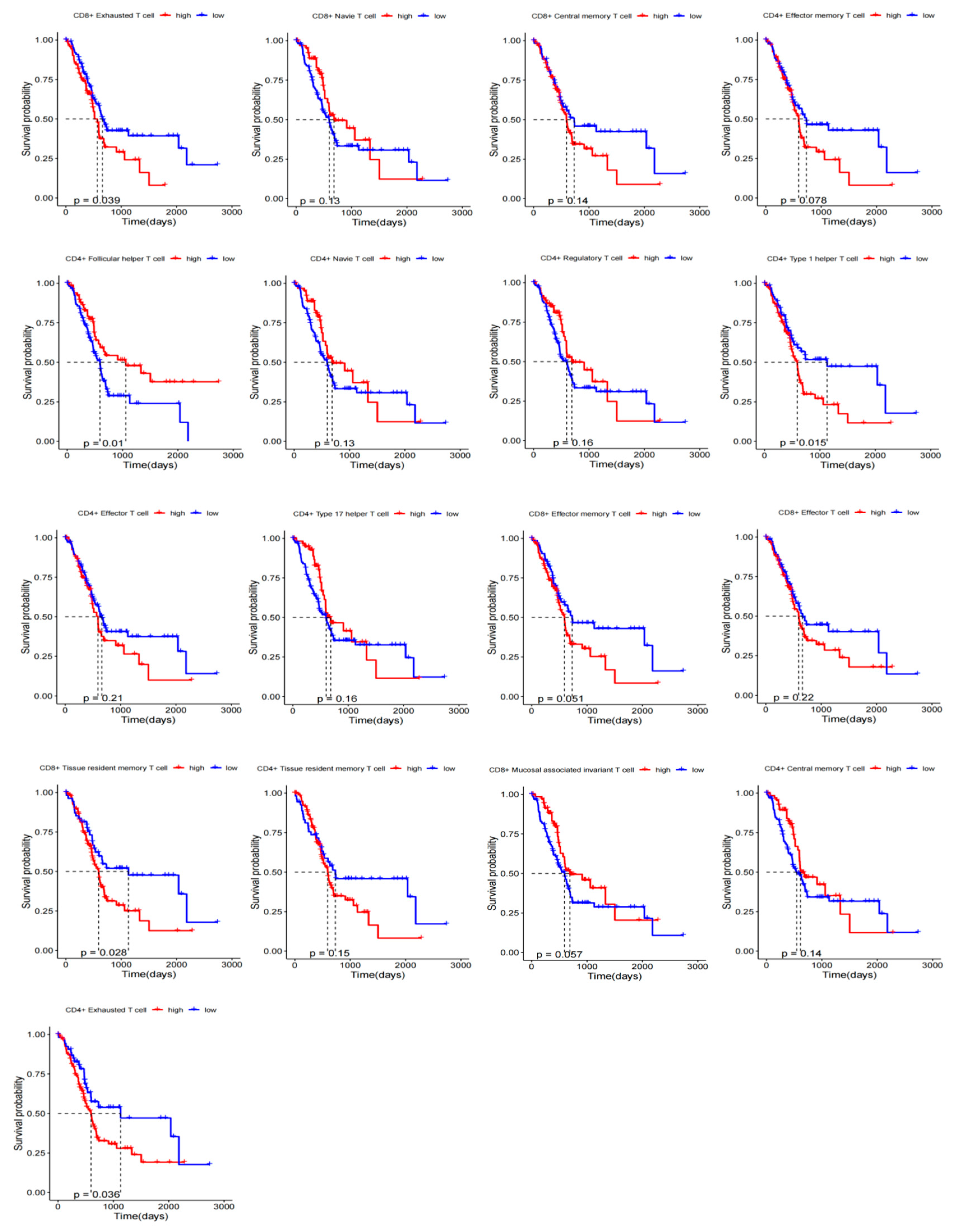
2.4. Prognostic KM Curves of T-Cell Subsets
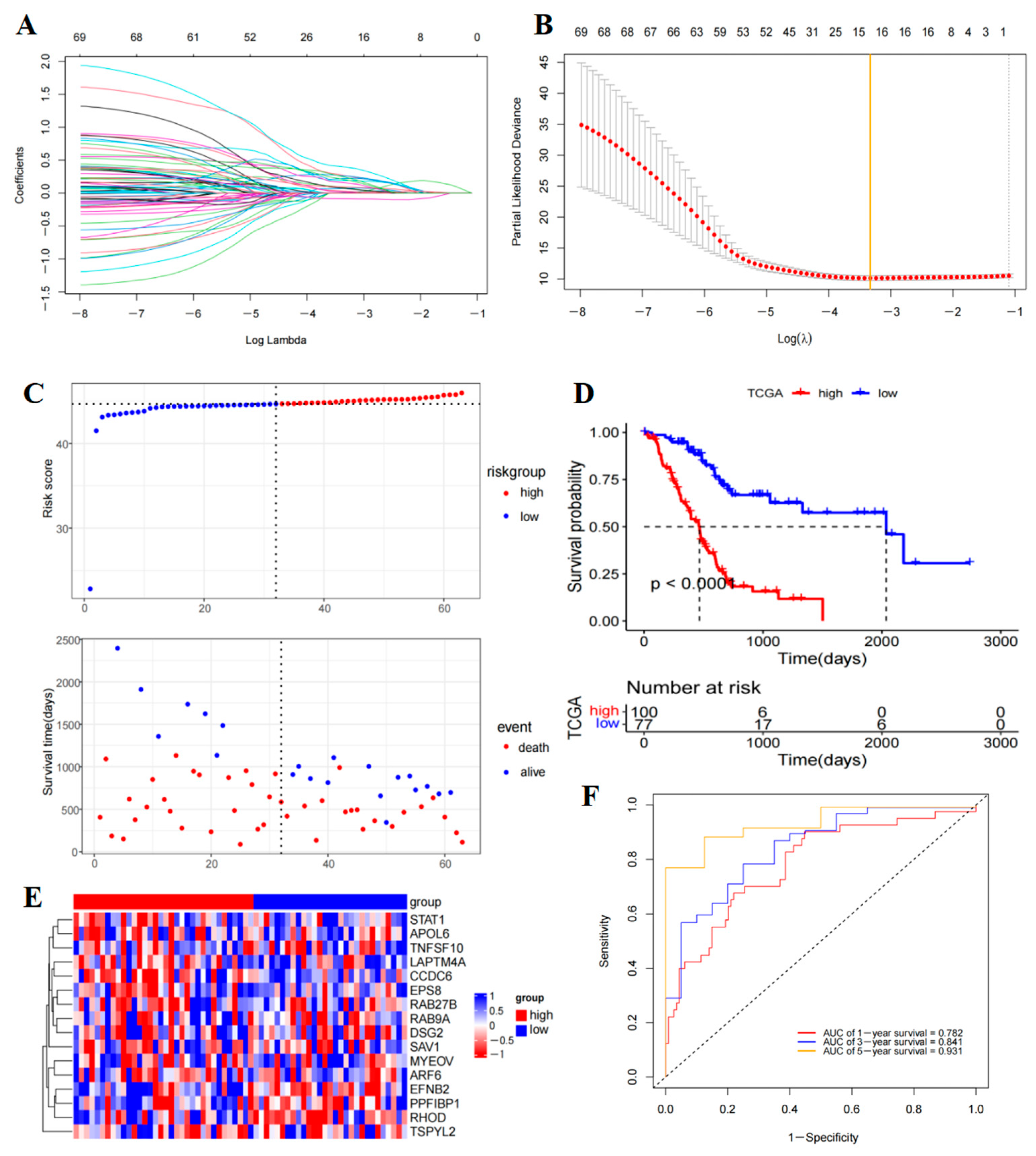
2.5. Prognostic Model Construction and Validation
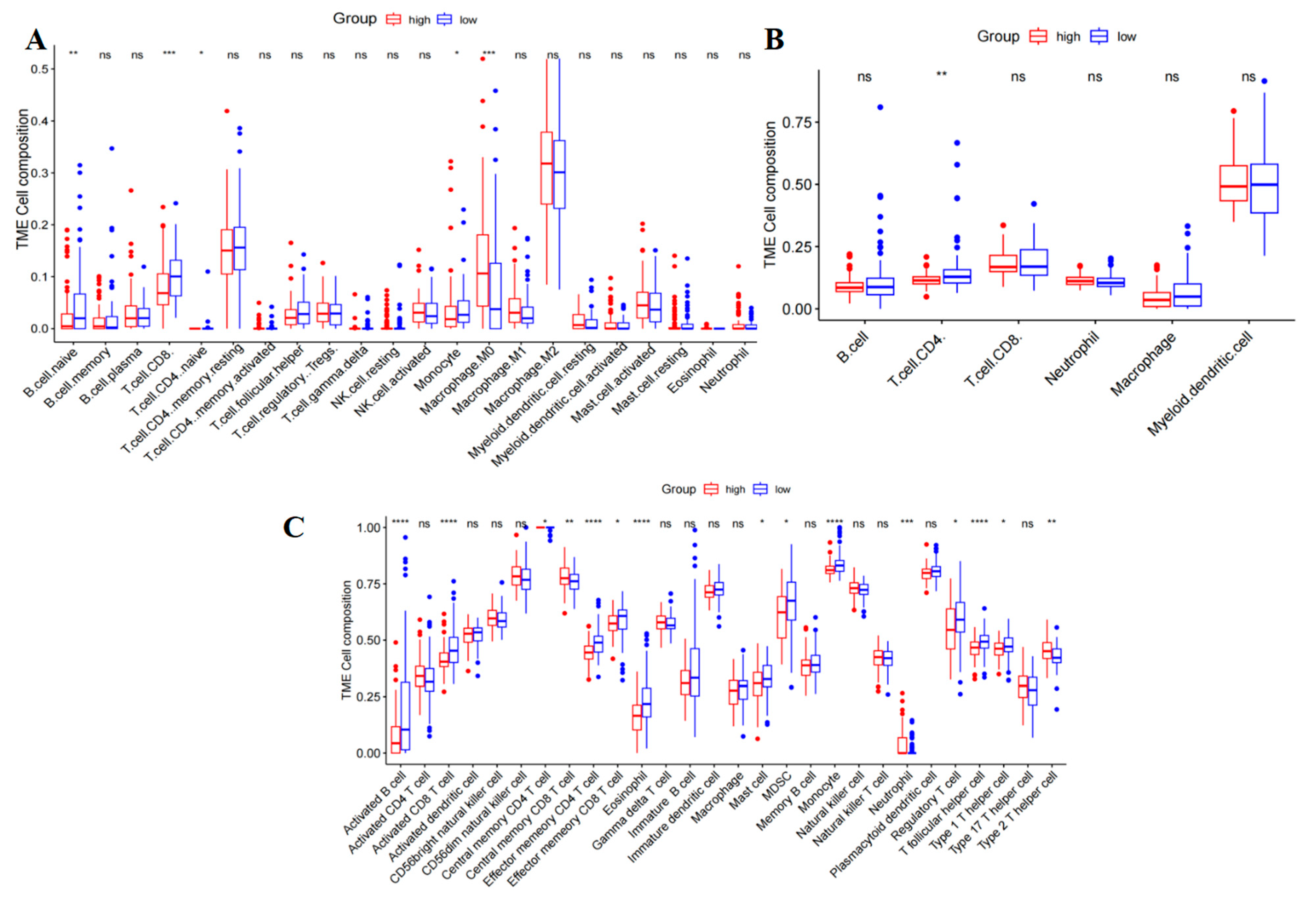
2.6. Correlation of the PAAD with Immune Cell Infiltration and Immune Checkpoint
2.7. Analyses of Tumor Mutation and Half-Inhibitory Centration
3. Discussion
4. Materials and Methods
4.1. Data Source and Preprocessing
4.2. T-Cell Subtypes and Differential Gene Expression Analysis
4.3. Cell–Cell Communication Analysis
4.4. Pseudo-Time Trajectories Analysis
4.5. Prognostic KM Curves of T-Cell Subsets
4.6. Prognostic Model Construction and Validation
4.7. Immune Cell Infiltration Analysis
4.8. Analysis of Drug Sensitivity and Immunotherapy Sensitivity
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klein, A.P. Pancreatic cancer epidemiology: Understanding the role of lifestyle and inherited risk factors. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Deng, T.; Cheng, H.; Liu, W.; Shi, D.; Yuan, J.; He, Z.; Wang, W.; Chen, B.; Ma, L.; et al. Macrophage-organoid co-culture model for identifying treatment strategies against macrophage-related gemcitabine resistance. J. Exp. Clin. Cancer Res. 2023, 42, 199. [Google Scholar] [CrossRef] [PubMed]
- Morrison, A.H.; Byrne, K.T.; Vonderheide, R.H. Immunotherapy and Prevention of Pancreatic Cancer. Trends Cancer 2018, 4, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M.H.; Beatty, G.L. Tumor Microenvironment in Pancreatic Cancer Pathogenesis and Therapeutic Resistance. Annu. Rev. Pathol. 2023, 18, 123–148. [Google Scholar] [CrossRef]
- O’Donnell, J.S.; Teng, M.W.L.; Smyth, M.J. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol. 2019, 16, 151–167. [Google Scholar] [CrossRef]
- Paijens, S.T.; Vledder, A.; de Bruyn, M.; Nijman, H.W. Tumor-infiltrating lymphocytes in the immunotherapy era. Cell. Mol. Immunol. 2021, 18, 842–859. [Google Scholar] [CrossRef]
- Han, L.; Gao, Q.L.; Zhou, X.M.; Shi, C.; Chen, G.Y.; Song, Y.P.; Yao, Y.J.; Zhao, Y.M.; Wen, X.Y.; Liu, S.L.; et al. Characterization of CD103(+) CD8(+) tissue-resident T cells in esophageal squamous cell carcinoma: May be tumor reactive and resurrected by anti-PD-1 blockade. Cancer Immunol. Immunother. CII 2020, 69, 1493–1504. [Google Scholar] [CrossRef]
- Djenidi, F.; Adam, J.; Goubar, A.; Durgeau, A.; Meurice, G.; de Montpreville, V.; Validire, P.; Besse, B.; Mami-Chouaib, F. CD8+CD103+ tumor-infiltrating lymphocytes are tumor-specific tissue-resident memory T cells and a prognostic factor for survival in lung cancer patients. J. Immunol. 2015, 194, 3475–3486. [Google Scholar] [CrossRef]
- Yi, Q.; Wang, J.; Liu, T.; Yao, Y.; Loveless, I.; Subedi, K.; Toor, J.; Adrianto, I.; Xiao, H.; Chen, B.; et al. scRNA-Seq and imaging mass cytometry analyses unveil iNKT cells-mediated anti-tumor immunity in pancreatic cancer liver metastasis. Cancer Lett. 2023, 561, 216149. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, X.; Zheng, L.; Zhang, Y.; Li, Y.; Fang, Q.; Gao, R.; Kang, B.; Zhang, Q.; Huang, J.Y.; et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018, 564, 268–272. [Google Scholar] [CrossRef]
- Zheng, L.; Qin, S.; Si, W.; Wang, A.; Xing, B.; Gao, R.; Ren, X.; Wang, L.; Wu, X.; Zhang, J.; et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 2021, 374, abe6474. [Google Scholar] [CrossRef] [PubMed]
- Blair, A.B.; Kim, V.M.; Muth, S.T.; Saung, M.T.; Lokker, N.; Blouw, B.; Armstrong, T.D.; Jaffee, E.M.; Tsujikawa, T.; Coussens, L.M.; et al. Dissecting the Stromal Signaling and Regulation of Myeloid Cells and Memory Effector T Cells in Pancreatic Cancer. Clin. Cancer Res. 2019, 25, 5351–5363. [Google Scholar] [CrossRef] [PubMed]
- Steele, M.M.; Jaiswal, A.; Delclaux, I.; Dryg, I.D.; Murugan, D.; Femel, J.; Son, S.; du Bois, H.; Hill, C.; Leachman, S.A.; et al. T cell egress via lymphatic vessels is tuned by antigen encounter and limits tumor control. Nat. Immunol. 2023, 24, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Sun, Z.; Chen, L. Memory T cells: Strategies for optimizing tumor immunotherapy. Protein Cell 2020, 11, 549–564. [Google Scholar] [CrossRef]
- Whitesell, J.C.; Lindsay, R.S.; Olivas-Corral, J.G.; Yannacone, S.F.; Schoenbach, M.H.; Lucas, E.D.; Friedman, R.S. Islet Lymphocytes Maintain a Stable Regulatory Phenotype Under Homeostatic Conditions and Metabolic Stress. Front. Immunol. 2022, 13, 814203. [Google Scholar] [CrossRef]
- Hu, Q.; Tong, Z.; Yalikong, A.; Ge, L.P.; Shi, Q.; Du, X.; Wang, P.; Liu, X.Y.; Zhan, W.; Gao, X.; et al. DNAzyme-based faithful probing and pulldown to identify candidate biomarkers of low abundance. Nat. Chem. 2024, 16, 122–131. [Google Scholar] [CrossRef]
- Li, R.; Peng, C.; Zhang, X.; Wu, Y.; Pan, S.; Xiao, Y. Roles of Arf6 in cancer cell invasion, metastasis and proliferation. Life Sci. 2017, 182, 80–84. [Google Scholar] [CrossRef]
- Ye, Z.; Hu, Q.; Zhuo, Q.; Zhu, Y.; Fan, G.; Liu, M.; Sun, Q.; Zhang, Z.; Liu, W.; Xu, W.; et al. Abrogation of ARF6 promotes RSL3-induced ferroptosis and mitigates gemcitabine resistance in pancreatic cancer cells. Am. J. Cancer Res. 2020, 10, 1182–1193. [Google Scholar]
- Hashimoto, A.; Handa, H.; Hata, S.; Tsutaho, A.; Yoshida, T.; Hirano, S.; Hashimoto, S.; Sabe, H. Inhibition of mutant KRAS-driven overexpression of ARF6 and MYC by an eIF4A inhibitor drug improves the effects of anti-PD-1 immunotherapy for pancreatic cancer. Cell Commun. Signal. 2021, 19, 54. [Google Scholar] [CrossRef]
- Cerrato, A.; Merolla, F.; Morra, F.; Celetti, A. CCDC6: The identity of a protein known to be partner in fusion. Int. J. Cancer 2018, 142, 1300–1308. [Google Scholar] [CrossRef]
- Yang, T.; Gu, X.; Jia, L.; Guo, J.; Tang, Q.; Zhu, J.; Zhao, W.; Feng, Z. DSG2 expression is low in colon cancer and correlates with poor survival. BMC Gastroenterol. 2021, 21, 7. [Google Scholar] [CrossRef] [PubMed]
- Hutz, K.; Zeiler, J.; Sachs, L.; Ormanns, S.; Spindler, V. Loss of desmoglein 2 promotes tumorigenic behavior in pancreatic cancer cells. Mol. Carcinog. 2017, 56, 1884–1895. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Dai, S.N.; Xu, D.L.; Hou, C.Q.; Liu, T.T.; Chen, Q.Y.; Wu, J.L.; Miao, Y. EFNB2 facilitates cell proliferation, migration, and invasion in pancreatic ductal adenocarcinoma via the p53/p21 pathway and EMT. Biomed. Pharmacother. 2020, 125, 109972. [Google Scholar] [CrossRef] [PubMed]
- Welsch, T.; Endlich, K.; Giese, T.; Buchler, M.W.; Schmidt, J. Eps8 is increased in pancreatic cancer and required for dynamic actin-based cell protrusions and intercellular cytoskeletal organization. Cancer Lett. 2007, 255, 205–218. [Google Scholar] [CrossRef]
- Chen, C.; Liang, Z.; Huang, W.; Li, X.; Zhou, F.; Hu, X.; Han, M.; Ding, X.; Xiang, S. Eps8 regulates cellular proliferation and migration of breast cancer. Int. J. Oncol. 2015, 46, 205–214. [Google Scholar] [CrossRef]
- Ding, Y.; Jiang, Y.; Zeng, H.; Zhou, M.; Zhou, X.; Yu, Z.; Pan, J.; Geng, X.; Zhu, Y.; Zheng, H.; et al. Identification of a robust biomarker LAPTM4A for glioma based on comprehensive computational biology and experimental verification. Aging 2024, 16, 6954–6989. [Google Scholar] [CrossRef]
- Li, W.; Wang, J.; Tang, C. A comprehensive analysis of the prognostic value and immune microenvironment of lysosome-dependent cell death in glioma: Including glioblastoma and low-grade glioma. Medicine 2024, 103, e36960. [Google Scholar] [CrossRef]
- Tange, S.; Hirano, T.; Idogawa, M.; Hirata, E.; Imoto, I.; Tokino, T. MYEOV overexpression induced by demethylation of its promoter contributes to pancreatic cancer progression via activation of the folate cycle/c-Myc/mTORC1 pathway. BMC Cancer 2023, 23, 85. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Guo, Q.; Li, X.; Zou, X. Identification of MYEOV-Associated Gene Network as a Potential Therapeutic Target in Pancreatic Cancer. Cancers 2022, 14, 5439. [Google Scholar] [CrossRef]
- Dong, C.; Li, X.; Yang, J.; Yuan, D.; Zhou, Y.; Zhang, Y.; Shi, G.; Zhang, R.; Liu, J.; Fu, P.; et al. PPFIBP1 induces glioma cell migration and invasion through FAK/Src/JNK signaling pathway. Cell Death Dis. 2021, 12, 827. [Google Scholar] [CrossRef]
- Li, J.; Jin, Q.; Huang, F.; Tang, Z.; Huang, J. Effects of Rab27A and Rab27B on Invasion, Proliferation, Apoptosis, and Chemoresistance in Human Pancreatic Cancer Cells. Pancreas 2017, 46, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Zhang, Z.; Qin, X.; Gao, Y.; Zhao, P.; Liu, J.; Zeng, W. Long noncoding RNA ZFAS1 promotes tumorigenesis through regulation of miR-150-5p/RAB9A in melanoma. Melanoma Res. 2019, 29, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Kazmi, N.; Robinson, T.; Zheng, J.; Kar, S.; Martin, R.M.; Ridley, A.J. Rho GTPase gene expression and breast cancer risk: A Mendelian randomization analysis. Sci. Rep. 2022, 12, 1463. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Chang, W.; Fu, Y.; Gao, Y.; Zhao, C.; Zhang, X.; Zhang, S. SAV1 represses the development of human colorectal cancer by regulating the Akt-mTOR pathway in a YAP-dependent manner. Cell. Prolif. 2017, 50, e12351. [Google Scholar] [CrossRef]
- Wang, L.; Wang, M.; Hu, C.; Li, P.; Qiao, Y.; Xia, Y.; Liu, L.; Jiang, X. Protein salvador homolog 1 acts as a tumor suppressor and is modulated by hypermethylation in pancreatic ductal adenocarcinoma. Oncotarget 2017, 8, 62953–62961. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, F.; Liu, F.; Xu, G. Predicting STAT1 as a prognostic marker in patients with solid cancer. Ther. Adv. Med. Oncol. 2020, 12, 1758835920917558. [Google Scholar] [CrossRef]
- Han, Y.J.; Zhang, J.; Hardeman, A.; Liu, M.; Karginova, O.; Romero, R.; Khramtsova, G.F.; Zheng, Y.; Huo, D.; Olopade, O.I. An enhancer variant associated with breast cancer susceptibility in Black women regulates TNFSF10 expression and antitumor immunity in triple-negative breast cancer. Hum. Mol. Genet. 2023, 32, 139–150. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, X.; Yao, W.; Wang, Y.H. A tumor-suppressing role of TSPYL2 in thyroid cancer: Through interacting with SIRT1 and repressing SIRT1/AKT pathway. Exp. Cell Res. 2023, 432, 113777. [Google Scholar] [CrossRef]
- Liu, Z.; Li, C.; Yu, C.; Chen, Z.; Zhao, C.; Ye, L. TSPYL2 reduced gefitinib resistance and DNA damage repair via suppressing SIRT1-mediated FOXO3 deacetylation. Future Med. Chem. 2022, 14, 407–419. [Google Scholar] [CrossRef]
- Li, M.; Yang, Y.; Xiong, L.; Jiang, P.; Wang, J.; Li, C. Metabolism, metabolites, and macrophages in cancer. J. Hematol. Oncol. 2023, 16, 80. [Google Scholar] [CrossRef]
- Yang, D.; Zhao, F.; Su, Y.; Zhou, Y.; Shen, J.; Zhao, K.; Ding, Y. Analysis of M2 macrophage-associated risk score signature in pancreatic cancer TME landscape and immunotherapy. Front. Mol. Biosci. 2023, 10, 1184708. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Z.; Chen, W.; Wang, X.; Cao, M.; Han, X.; Zhang, K.; Teng, B.; Cao, J.; Wu, W.; et al. M2 Macrophage-Derived Exosomes Promote Angiogenesis and Growth of Pancreatic Ductal Adenocarcinoma by Targeting E2F2. Mol. Ther. 2021, 29, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Postow, M.A.; Sidlow, R.; Hellmann, M.D. Immune-Related Adverse Events Associated with Immune Checkpoint Blockade. N. Engl. J. Med. 2018, 378, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Bannoura, S.F.; Uddin, M.H.; Nagasaka, M.; Fazili, F.; Al-Hallak, M.N.; Philip, P.A.; El-Rayes, B.; Azmi, A.S. Targeting KRAS in pancreatic cancer: New drugs on the horizon. Cancer Metastasis Rev. 2021, 40, 819–835. [Google Scholar] [CrossRef]
- Abe, K.; Kitago, M.; Kitagawa, Y.; Hirasawa, A. Hereditary pancreatic cancer. Int. J. Clin. Oncol. 2021, 26, 1784–1792. [Google Scholar] [CrossRef]
- Cicenas, J.; Kvederaviciute, K.; Meskinyte, I.; Meskinyte-Kausiliene, E.; Skeberdyte, A.; Cicenas, J. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 Mutations in Pancreatic Cancer. Cancers 2017, 9, 42. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets--update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Slovin, S.; Carissimo, A.; Panariello, F.; Grimaldi, A.; Bouche, V.; Gambardella, G.; Cacchiarelli, D. Single-Cell RNA Sequencing Analysis: A Step-by-Step Overview. Methods Mol. Biol. 2021, 2284, 343–365. [Google Scholar] [CrossRef]
- Fang, Z.; Tian, Y.; Sui, C.; Guo, Y.; Hu, X.; Lai, Y.; Liao, Z.; Li, J.; Feng, G.; Jin, L.; et al. Single-Cell Transcriptomics of Proliferative Phase Endometrium: Systems Analysis of Cell-Cell Communication Network Using CellChat. Front. Cell Dev. Biol. 2022, 10, 919731. [Google Scholar] [CrossRef]
- Ni, L.; Yang, H.; Wu, X.; Zhou, K.; Wang, S. The expression and prognostic value of disulfidptosis progress in lung adenocarcinoma. Aging 2023, 15, 7741–7759. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Bach, D.R.; Dayan, P. Algorithms for survival: A comparative perspective on emotions. Nat. Rev. Neurosci. 2017, 18, 311–319. [Google Scholar] [CrossRef]
- Rich, J.T.; Neely, J.G.; Paniello, R.C.; Voelker, C.C.; Nussenbaum, B.; Wang, E.W. A practical guide to understanding Kaplan-Meier curves. Otolaryngol. Head Neck Surg. 2010, 143, 331–336. [Google Scholar] [CrossRef]
- Ren, N.; Liang, B.; Li, Y. Identification of prognosis-related genes in the tumor microenvironment of stomach adenocarcinoma by TCGA and GEO datasets. Biosci. Rep. 2020, 40, BSR20200980. [Google Scholar] [CrossRef]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, Z.; He, D.; Zhu, Y.; Chen, X.; Cao, K. Identification of novel subtypes based on ssGSEA in immune-related prognostic signature for tongue squamous cell carcinoma. Cancer Med. 2021, 10, 8693–8707. [Google Scholar] [CrossRef]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Wang, W.; Lu, Z.; Wang, M.; Liu, Z.; Wu, B.; Yang, C.; Huan, H.; Gong, P. The cuproptosis-related signature associated with the tumor environment and prognosis of patients with glioma. Front. Immunol. 2022, 13, 998236. [Google Scholar] [CrossRef] [PubMed]



| Low-Risk Group | High-Risk Group | |
|---|---|---|
| A.443654 | Better | |
| A.770041 | Better | |
| AKT.inhibitor.VIII | Better | |
| AZD.0530 | Better | |
| BI.2536 | Better | |
| BIBW2992 | Better | |
| Bicalutamide | Better | |
| BMS.509744 | Better | |
| BMS.536924 | Better | |
| Bortezomib | Better | |
| Bryostatin.1 | Better | |
| CGP.60474 | Better | |
| CGP.082996 | Better | |
| CHIR.99021 | Better | |
| CI.1040 | Better | |
| CMK | Better | |
| Dasatinib | Better | |
| Epothilone.B | Better | |
| Erlotinib | Better | |
| FH535 | Better | |
| FTI.277 | Better | |
| GNF.2 | Better | |
| GSK.650394 | Better | |
| JW.7.52.1 | Better | |
| Lapatinib | Better | |
| LFM.A13 | Better | |
| NSC.87877 | Better | |
| NVP.TAE684 | Better | |
| Paclitaxel | Better | |
| PD.0325901 | Better | |
| PF.562271 | Better | |
| PHA.665752 | Better | |
| RDEA119 | Better | |
| Thapsigargin | Better | |
| VX.680 | Better | |
| WH.4.023 | Better | |
| WZ.1.84 | Better | |
| X17.AAG | Better | |
| Z.LLNle.CHO | Better | |
| AUY922 | Better | |
| AZ628 | Better | |
| AZD6244 | Better | |
| Bexarotene | Better | |
| Bleomycin | Better | |
| BMS.754807 | Better | |
| Docetaxel | Better | |
| GW843682X | Better | |
| KIN001.135 | Better | |
| MG.132 | Better | |
| Midostaurin | Better | |
| Rapamycin | Better | |
| S.Trityl.L.cysteine | Better | |
| Sorafenib | Better | |
| XMD8.85 | Better | |
| AP.24534 | Better | |
| BI.D1870 | Better | |
| BX.795 | Better | |
| Cytarabine | Better | |
| DMOG | Better | |
| Elesclomol | Better | |
| GDC.0449 | Better | |
| GDC0941 | Better | |
| JNK.9L | Better | |
| Mitomycin.C | Better | |
| ABT.263 | Better | |
| ABT.888 | Better | |
| AG.014699 | Better | |
| AMG.706 | Better | |
| ATRA | Better | |
| Axitinib | Better | |
| AZD.2281 | Better | |
| AZD8055 | Better | |
| BIRB.0796 | Better | |
| Camptothecin | Better | |
| CCT007093 | Better | |
| CEP.701 | Better | |
| EHT.1864 | Better | |
| IPA.3 | Better | |
| JNK.Inhibitor.VIII | Better | |
| Lenalidomide | Better | |
| Metformin | Better | |
| Methotrexate | Better | |
| MK.2206 | Better | |
| Nilotinib | Better | |
| NU.7441 | Better | |
| Nutlin.3a | Better | |
| NVP.BEZ235 | Better | |
| PD.173074 | Better | |
| PD.0332991 | Better | |
| PLX4720 | Better | |
| QS11 | Better | |
| Salubrinal | Better | |
| SB590885 | Better | |
| SL.0101.1 | Better | |
| Temsirolimus | Better | |
| TW.37 | Better | |
| Vorinostat | Better | |
| VX.702 | Better | |
| X681640 | Better | |
| ZM.447439 | Better |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, D.; Hu, Y.; Peng, J.; Wang, S. Construction of T-Cell-Related Prognostic Risk Models and Prediction of Tumor Immune Microenvironment Regulation in Pancreatic Adenocarcinoma via Integrated Analysis of Single-Cell RNA-Seq and Bulk RNA-Seq. Int. J. Mol. Sci. 2025, 26, 2384. https://doi.org/10.3390/ijms26062384
Sun D, Hu Y, Peng J, Wang S. Construction of T-Cell-Related Prognostic Risk Models and Prediction of Tumor Immune Microenvironment Regulation in Pancreatic Adenocarcinoma via Integrated Analysis of Single-Cell RNA-Seq and Bulk RNA-Seq. International Journal of Molecular Sciences. 2025; 26(6):2384. https://doi.org/10.3390/ijms26062384
Chicago/Turabian StyleSun, Dingya, Yijie Hu, Jun Peng, and Shan Wang. 2025. "Construction of T-Cell-Related Prognostic Risk Models and Prediction of Tumor Immune Microenvironment Regulation in Pancreatic Adenocarcinoma via Integrated Analysis of Single-Cell RNA-Seq and Bulk RNA-Seq" International Journal of Molecular Sciences 26, no. 6: 2384. https://doi.org/10.3390/ijms26062384
APA StyleSun, D., Hu, Y., Peng, J., & Wang, S. (2025). Construction of T-Cell-Related Prognostic Risk Models and Prediction of Tumor Immune Microenvironment Regulation in Pancreatic Adenocarcinoma via Integrated Analysis of Single-Cell RNA-Seq and Bulk RNA-Seq. International Journal of Molecular Sciences, 26(6), 2384. https://doi.org/10.3390/ijms26062384





