Identification of Aldehyde Dehydrogenase Gene Family in Glycyrrhiza uralensis and Analysis of Expression Pattern Under Drought Stress
Abstract
1. Introduction
2. Results
2.1. The Identification and Analysis of the ALDH Gene in the Whole Genome of Glycyrrhiza uralensis
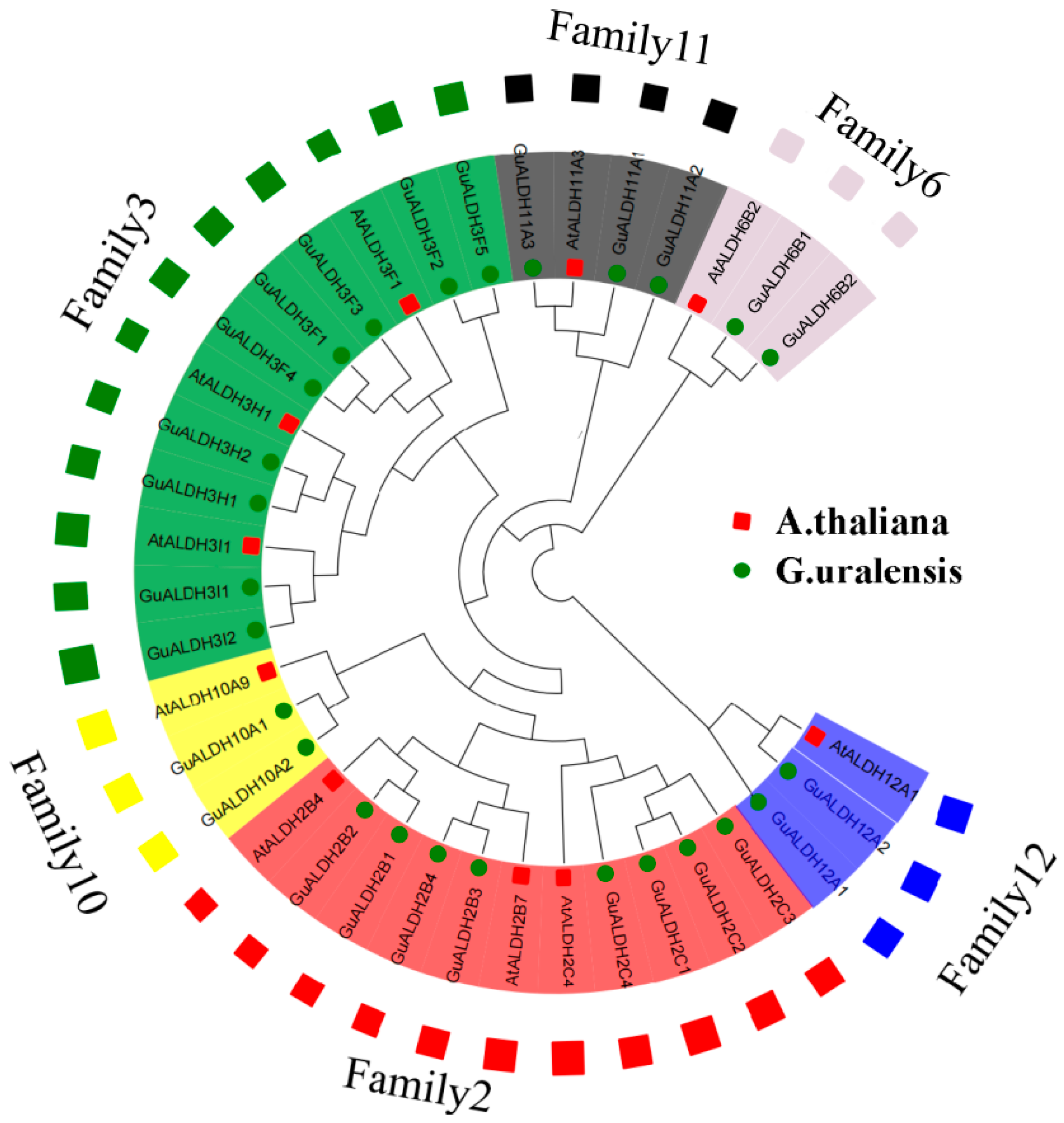
2.2. Phylogenetic Analysis of ALDHs
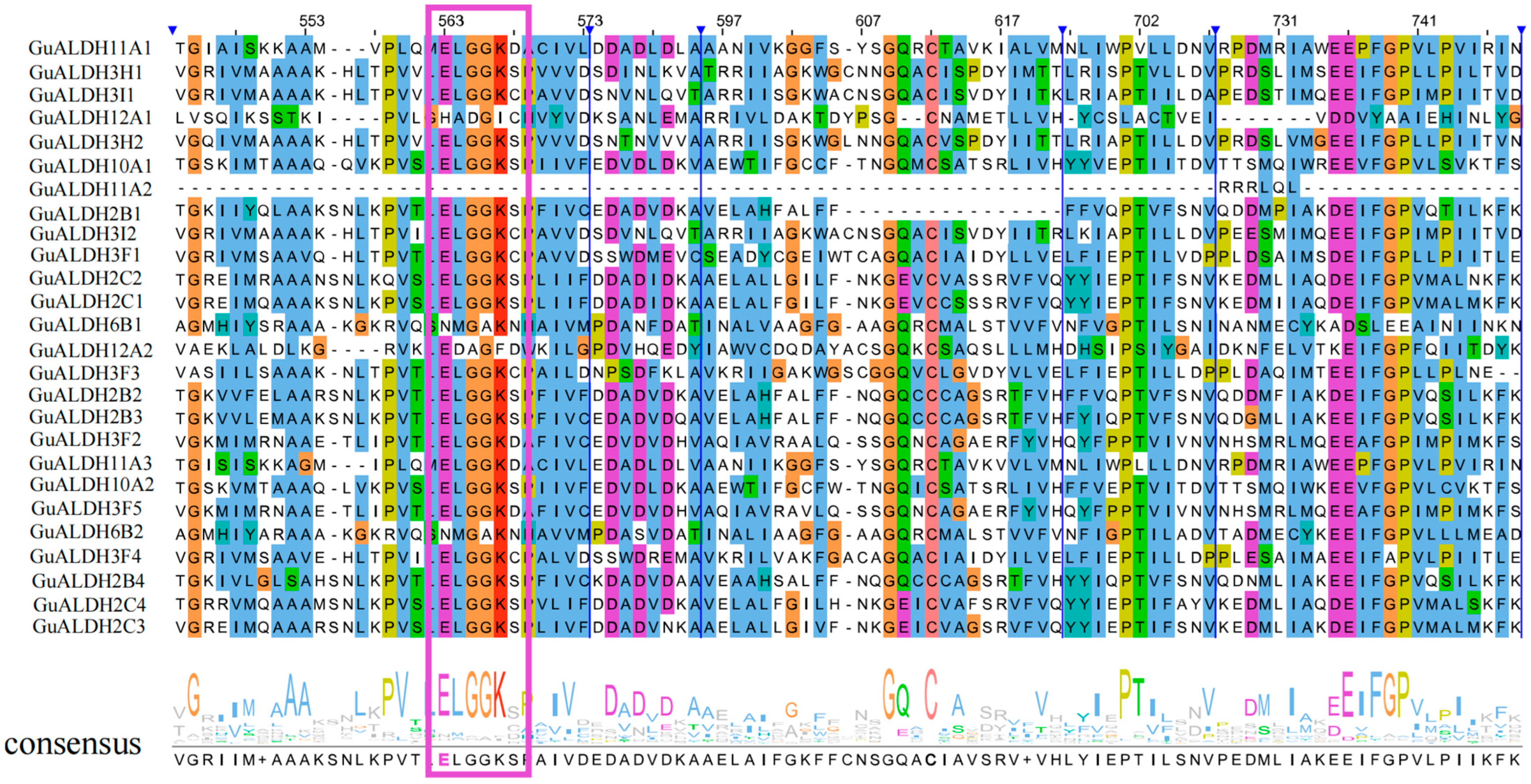
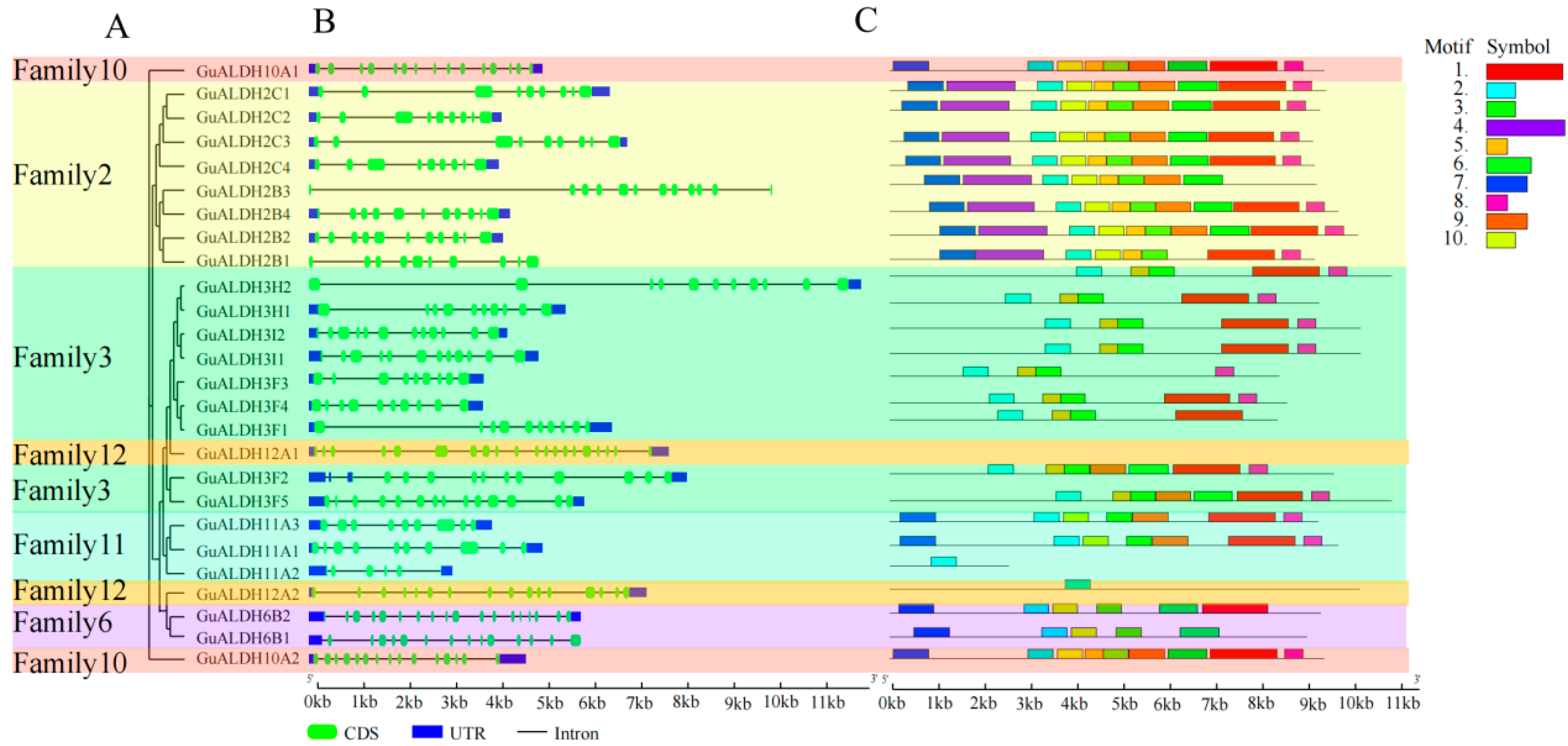
2.3. Analysis of Gene Structure and Conserved Protein Moift of GuALDH Subfamily Members in Glycyrrhiza uralensis
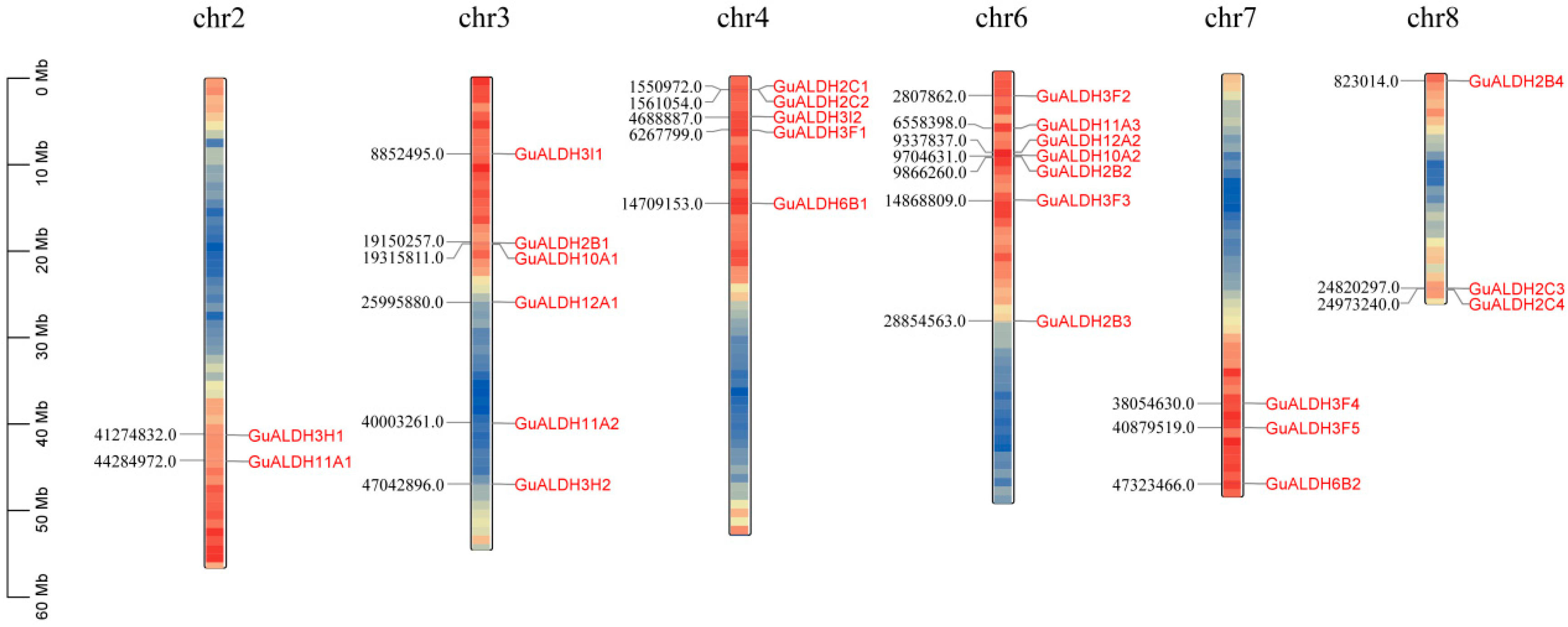
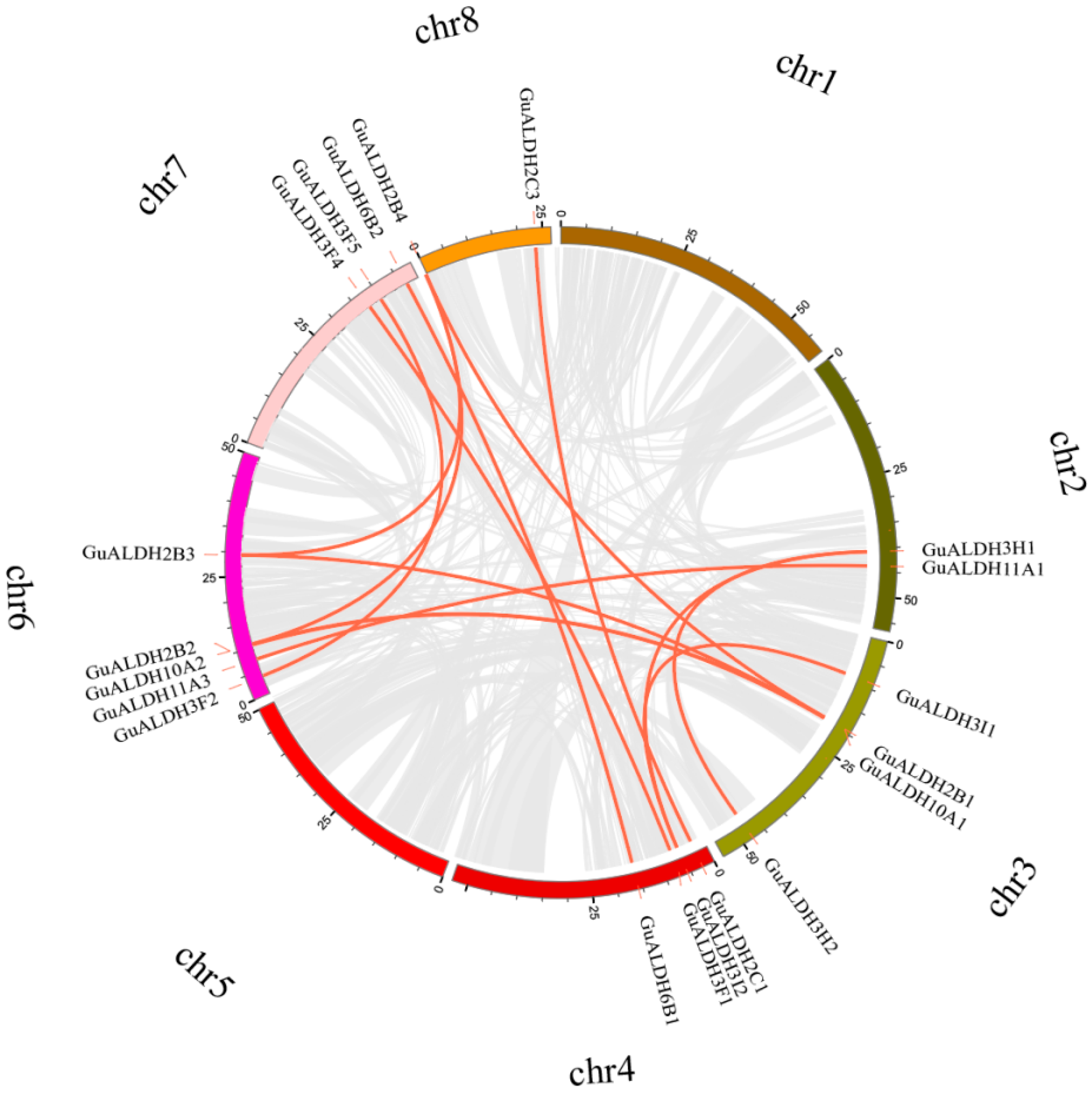
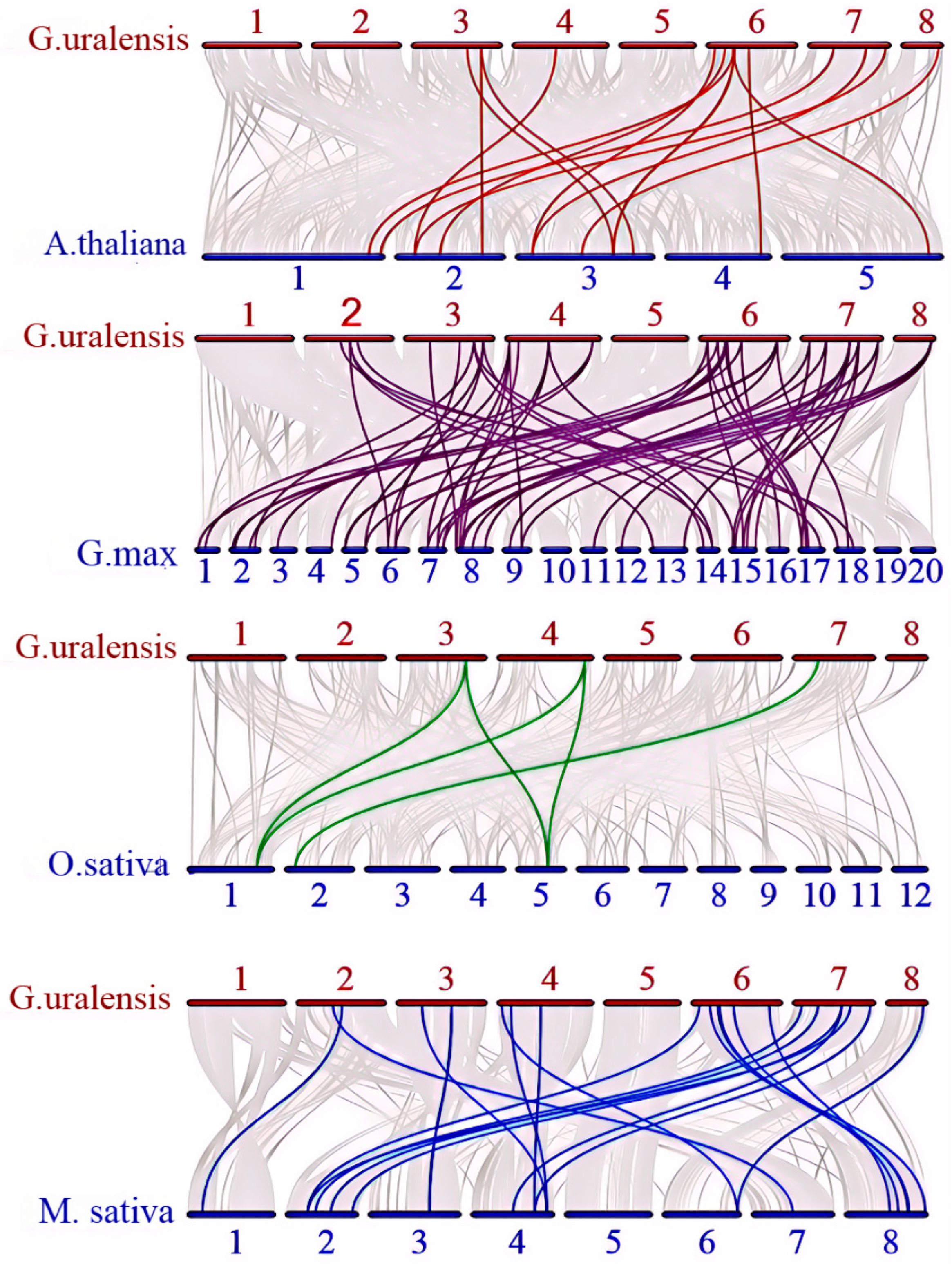
2.4. Genome Localisation, Homology, and Evolutionary Analysis of GuALDHs
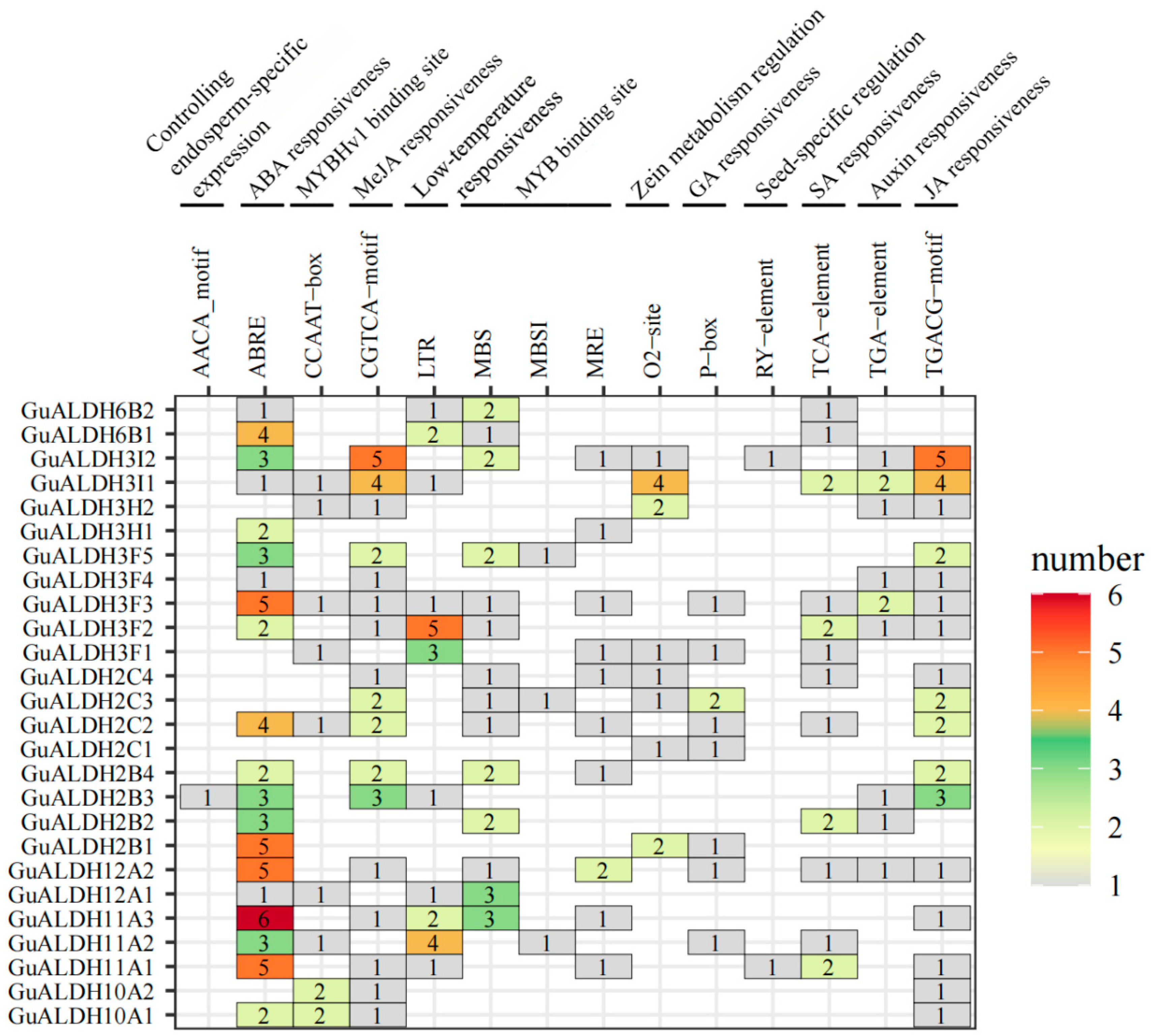
2.5. Identifying Cis-Regulatory Elements in the GuALDHs Promoter
2.6. Expression Patterns of GuALDH in Different Tissues of Glycyrrhiza uralensis
2.7. Co-Expression Analysis of GuALDHs
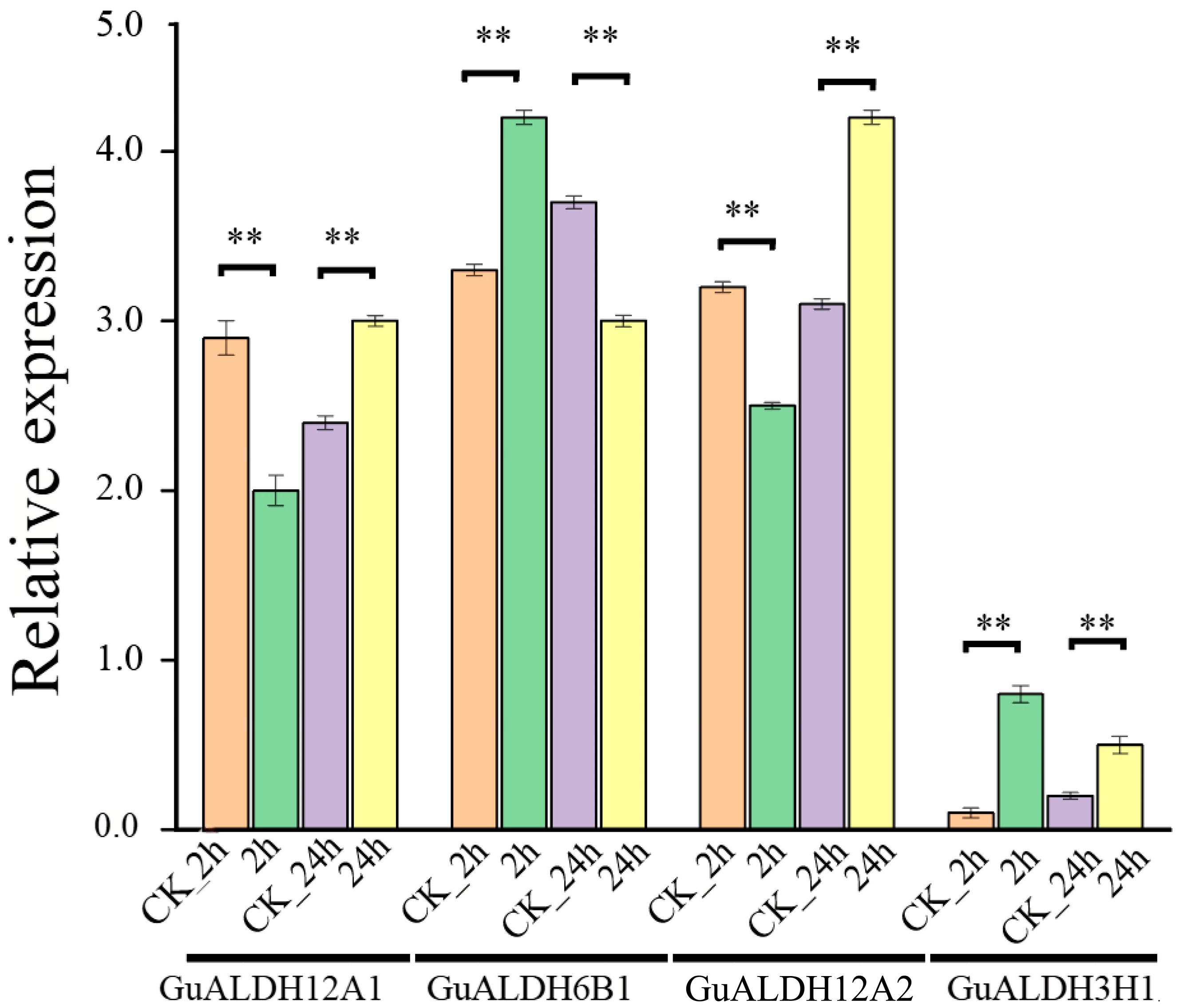
2.8. Response of GuALDH Genes to Drought Stress
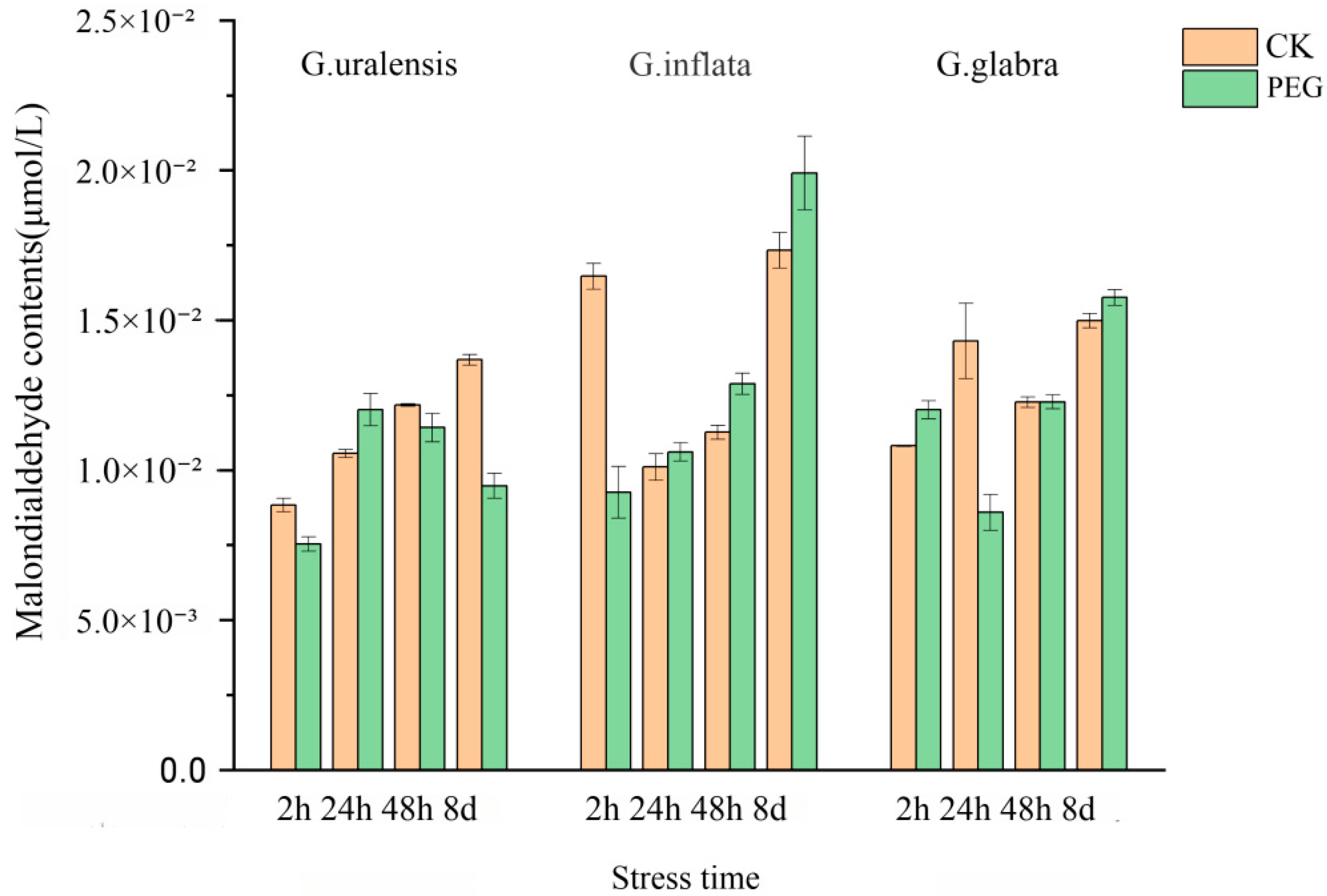
2.9. Changes in Malondialdehyde Content Under Drought Stress
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification of GuALDH Gene in Glycyrrhiza uralensis
4.2. Sequence Alignment and Construction of Phylogenetic Tree
4.3. Gene Structure, Motif Recognition, and Physical Location
4.4. Analysis of Cis-Regulatory Elements of Predicted Promoter Sequences
4.5. Homology and Gene Duplication Analysis
4.6. Gene Co-Expression Networks and Gene Annotation
4.7. Analysis of ALDH Gene Expression in Glycyrrhiza uralensis Based on RNA-Seq
4.8. Plant Materials, Drought Stress, RNA Extraction, and qRT-PCR Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Czarnocka, W.; Karpiński, S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 2018, 122, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Brocker, C.; Koppaka, V.; Chen, Y.; Jackson, B.C.; Matsumoto, A.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic. Biol. Med. 2013, 56, 89–101. [Google Scholar] [CrossRef]
- Stiti, N.; Missihoun, T.D.; Kotchoni, S.O.; Kirch, H.H.; Bartels, D. Aldehyde Dehydrogenases in Arabidopsis thaliana: Biochemical Requirements, Metabolic Pathways, and Functional Analysis. Front. Plant Sci. 2011, 2, 65. [Google Scholar] [CrossRef]
- Carmona-Molero, R.; Jimenez-Lopez, J.C.; Caballo, C.; Gil, J.; Millán, T.; Die, J.V. Aldehyde Dehydrogenase 3 Is an Expanded Gene Family with Potential Adaptive Roles in Chickpea. Plants 2021, 10, 2429. [Google Scholar] [CrossRef]
- Rodrigues, S.M.; Andrade, M.O.; Gomes, A.P.; Damatta, F.M.; Baracat-Pereira, M.C.; Fontes, E.P. Arabidopsis and tobacco plants ectopically expressing the soybean antiquitin-like ALDH7 gene display enhanced tolerance to drought, salinity, and oxidative stress. J. Exp. Bot. 2006, 57, 1909–1918. [Google Scholar] [CrossRef]
- Yoshida, A.; Rzhetsky, A.; Hsu, L.C.; Chang, C. Human Aldehyde dehydrogenase gene family. Eur. J. Biochem. 1998, 251, 549–557. [Google Scholar] [CrossRef]
- Vasiliou, V.; Bairoch, A.; Tipton, K.F.; Nebert, D.W. Eukaryotic Aldehyde dehydrogenase (ALDH) genes: Human polymorphisms, and recommended nomenclature based on divergent evolution and chromosomal mapping. Pharmacogenetics 1999, 9, 421–434. [Google Scholar] [PubMed]
- Sophos, N.A.; Vasiliou, V. Aldehyde dehydrogenase gene superfamily: The 2002 update. Chem. Biol. Interact. 2003, 143, 5–22. [Google Scholar] [CrossRef]
- Brocker, C.; Vasiliou, M.; Carpenter, S.; Carpenter, C.; Zhang, Y.; Wang, X.; Kotchoni, S.O.; Wood, A.J.; Kirch, H.H.; Kopečný, D.; et al. Aldehyde dehydrogenase (ALDH) superfamily in plants: Gene nomenclature and comparative genomics. Planta 2013, 237, 189–210. [Google Scholar] [CrossRef]
- Missihoun, T.D.; Schmitz, J.; Klug, R.; Kirch, H.H.; Bartels, D. Betaine Aldehyde dehydrogenase genes from Arabidopsis with different sub-cellular localization affect stress responses. Planta 2011, 233, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Kirch, H.H.; Bartels, D.; Wei, Y.; Schnable, P.S.; Wood, A.J. The ALDH gene superfamily of Arabidopsis. Trends Plant Sci. 2004, 9, 371–377. [Google Scholar] [CrossRef]
- Kirch, H.H.; Schlingensiepen, S.; Kotchoni, S.; Sunkar, R.; Bartels, D. Detailed expression analysis of selected genes of the Aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant Mol. Biol. 2005, 57, 315–332. [Google Scholar] [CrossRef]
- Wang, W.; Jiang, W.; Liu, J.; Li, Y.; Gai, J.; Li, Y. Genome-wide characterization of the Aldehyde dehydrogenase gene superfamily in soybean and its potential role in drought stress response. BMC Genom. 2017, 18, 518. [Google Scholar] [CrossRef]
- Zhang, X.; Zhong, J.; Cao, L.; Ren, C.; Yu, G.; Gu, Y.; Ruan, J.; Zhao, S.; Wang, L.; Ru, H.; et al. Genome-wide characterization of Aldehyde dehydrogenase gene family members in groundnut (Arachis hypogaea) and the analysis under saline-alkali stress. Front. Plant Sci. 2023, 14, 1097001. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, X.; Jiao, Y.; Qin, Y.; Liu, X.; He, K.; Chen, C.; Ma, L.; Wang, J.; Xiong, L.; et al. Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol. Biol. 2007, 63, 591–608. [Google Scholar] [CrossRef]
- Gao, C.; Han, B. Evolutionary and expression study of the Aldehyde dehydrogenase (ALDH) gene superfamily in rice (Oryza sativa). Gene 2009, 431, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zhang, Z.; Zhao, Z.; Xie, Y.; Li, H.; Ma, X.; Liu, Y.G.; Chen, L. The mitochondrial Aldehyde dehydrogenase OsALDH2b negatively regulates tapetum degeneration in rice. J. Exp. Bot. 2020, 71, 2551–2560. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple Functions of MYB Transcription Factors in Abiotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef]
- Guo, Z.; Dzinyela, R.; Yang, L.; Hwarari, D. bZIP Transcription Factors: Structure, Modification, Abiotic Stress Responses and Application in Plant Improvement. Plants 2024, 13, 2058. [Google Scholar] [CrossRef]
- Fang, L.; Su, L.; Sun, X.; Li, X.; Sun, M.; Karungo, S.K.; Fang, S.; Chu, J.; Li, S.; Xin, H. Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis. J. Exp. Bot. 2016, 67, 2829–2845. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Bartels, D. Comparative study of the Aldehyde dehydrogenase (ALDH) gene superfamily in the glycophyte Arabidopsis thaliana and Eutrema halophytes. Ann. Bot. 2015, 115, 465–479. [Google Scholar] [CrossRef]
- Zhang, Y.; Mao, L.; Wang, H.; Brocker, C.; Yin, X.; Vasiliou, V.; Fei, Z.; Wang, X. Genome-wide identification and analysis of grape Aldehyde dehydrogenase (ALDH) gene superfamily. PLoS ONE 2012, 7, e32153. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Mohtasim, M.; Islam, T.; Ghosh, A. Aldehyde dehydrogenase superfamily in sorghum: Genome-wide identification, evolution, and transcript profiling during development stages and stress conditions. BMC Plant Biol. 2022, 22, 316. [Google Scholar] [CrossRef]
- Kotchoni, S.O.; Jimenez-Lopez, J.C.; Gao, D.; Edwards, V.; Gachomo, E.W.; Margam, V.M.; Seufferheld, M.J. Modeling-dependent protein characterization of the rice Aldehyde dehydrogenase (ALDH) superfamily reveals distinct functional and structural features. PLoS ONE 2010, 5, e11516. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.L.; Zhang, Q.; Zhou, M.; Qi, L.P.; Yang, X.B.; Zhang, K.X.; Pang, J.F.; Zhu, X.M.; Shao, J.R.; Tang, Y.X.; et al. Aldehyde dehydrogenase protein superfamily in maize. Funct. Integr. Genom. 2012, 12, 683–691. [Google Scholar] [CrossRef]
- Gu, H.; Pan, Z.; Jia, M.; Fang, H.; Li, J.; Qi, Y.; Yang, Y.; Feng, W.; Gao, X.; Ditta, A.; et al. Genome-wide identification and analysis of the cotton ALDH gene family. BMC Genom. 2024, 25, 513. [Google Scholar] [CrossRef]
- Li, X.; Guo, R.; Li, J.; Singer, S.D.; Zhang, Y.; Yin, X.; Zheng, Y.; Fan, C.; Wang, X. Genome-wide identification and analysis of the Aldehyde dehydrogenase (ALDH) gene superfamily in apple (Malus × domestica Borkh.). Plant Physiol. Biochem. 2013, 71, 268–282. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Hasan, M.S.; Hasan, M.N.; Prodhan, S.H.; Islam, T.; Ghosh, A. Genome-wide identification, evolution, and transcript profiling of Aldehyde dehydrogenase superfamily in potato during development stages and stress conditions. Sci. Rep. 2021, 11, 18284. [Google Scholar] [CrossRef]
- Han, D.; Xu, T.; Han, J.; Liu, W.; Wang, Y.; Li, X.; Sun, X.; Wang, X.; Li, T.; Yang, G. Overexpression of MxWRKY53 increased iron and high salinity stress tolerance in Arabidopsis thaliana. In Vitr. Cell. Dev. Biol. Plant 2022, 58, 266–278. [Google Scholar] [CrossRef]
- Han, D.; Zhang, Z.-y.; Ding, H.; Chai, L.; Liu, W.; Li, H.; Yang, G. Isolation and characterization of MbWRKY2 gene involved in enhanced drought tolerance in transgenic tobacco. J. Plant Interact. 2018, 13, 163–172. [Google Scholar] [CrossRef]
- Li, W.; Li, P.; Chen, H.; Zhong, J.; Liang, X.; Wei, Y.; Zhang, L.; Wang, H.; Han, D. Overexpression of a Fragaria vesca 1R-MYB Transcription Factor Gene (FvMYB114) Increases Salt and Cold Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2023, 24, 5261. [Google Scholar] [CrossRef]
- Kotchoni, S.O.; Jimenez-Lopez, J.C.; Kayodé, A.P.; Gachomo, E.W.; Baba-Moussa, L. The soybean Aldehyde dehydrogenase (ALDH) protein superfamily. Gene 2012, 495, 128–133. [Google Scholar] [CrossRef]
- Wang, X.; Wu, M.; Yu, S.; Zhai, L.; Zhu, X.; Yu, L.; Zhang, Y. Comprehensive analysis of the Aldehyde dehydrogenase gene family in Phaseolus vulgaris L. and their response to saline-alkali stress. Front. Plant Sci. 2024, 15, 1283845. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, K.; Han, S.; Zhang, L.; Bai, H.; Bao, F.; Zeng, Y.; Wang, J.; Du, H.; Liu, Y.; et al. Constituents Isolated from the Leaves of Glycyrrhiza uralansis and Their Anti-Inflammatory Activities on LPS-Induced RAW264.7 Cells. Molecules 2019, 24, 1923. [Google Scholar] [CrossRef]
- Aipire, A.; Li, J.; Yuan, P.; He, J.; Hu, Y.; Liu, L.; Feng, X.; Li, Y.; Zhang, F.; Yang, J.; et al. Glycyrrhiza uralensis water extract enhances dendritic cell maturation and antitumor efficacy of HPV dendritic cell-based vaccine. Sci. Rep. 2017, 7, 43796. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef]
- Kim, A.R.; Min, J.H.; Lee, K.H.; Kim, C.S. PCA22 acts as a suppressor of atrzf1 to mediate proline accumulation in response to abiotic stress in Arabidopsis. J. Exp. Bot. 2017, 68, 1797–1809. [Google Scholar] [CrossRef][Green Version]
- Yang, H.; Zhang, D.; Li, H.; Dong, L.; Lan, H. Ectopic overexpression of the Aldehyde dehydrogenase ALDH21 from Syntrichia caninervis in tobacco confers salt and drought stress tolerance. Plant Physiol. Biochem. 2015, 95, 83–91. [Google Scholar] [CrossRef]
- Hobo, T.; Kowyama, Y.; Hattori, T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA 1999, 96, 15348–15353. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, K.; Todaka, D.; Mizoi, J.; Yoshida, T.; Kidokoro, S.; Matsukura, S.; Takasaki, H.; Sakurai, T.; Yamamoto, Y.Y.; Yoshiwara, K.; et al. Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 2012, 19, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Ishida, J.; Narusaka, M.; Fujita, M.; Nanjo, T.; Umezawa, T.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T.; et al. Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct. Integr. Genom. 2002, 2, 282–291. [Google Scholar] [CrossRef]
- Rabbani, M.A.; Maruyama, K.; Abe, H.; Khan, M.A.; Katsura, K.; Ito, Y.; Yoshiwara, K.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 2003, 133, 1755–1767. [Google Scholar] [CrossRef]
- Jones, A.M. A new look at stress: Abscisic acid patterns and dynamics at high-resolution. New Phytol. 2016, 210, 38–44. [Google Scholar] [CrossRef]
- Han, D.; Du, M.; Zhou, Z.; Wang, S.; Li, T.; Han, J.; Xu, T.; Yang, G. An NAC transcription factor gene from Malus baccata, MbNAC29, increases cold and high salinity tolerance in Arabidopsis. In Vitr. Cell. Dev. Biol. Plant 2020, 56, 588–599. [Google Scholar] [CrossRef]
- Han, D.; Hou, Y.; Ding, H.; Zhou, Z.; Li, H.; Yang, G. Isolation and Preliminary Functional Analysis of MbWRKY 4 Gene Involved in Salt Tolerance in Transgenic Tobacco. Int. J. Agric. Biol. 2018, 20, 2045–2052. [Google Scholar]
- Chen, C.; Wu, Y.; Xia, R. A painless way to customize Circos plot: From data preparation to visualization using TBtools. iMeta 2022, 1, e35. [Google Scholar] [CrossRef]
- Xian, Y.; Wang, F.; Xie, S.; Chen, X.; Shen, H.; Li, H. Identification and Expression Analysis of the CIPK Gene Family in Ural Licorice. J. Biol. 2022, 39, 62–68. [Google Scholar]


| Family | Gene Name | Gene ID | ORF(aa) | MW(kDa) | pI | Sub Localization |
|---|---|---|---|---|---|---|
| Family2 | GuALDH2B1 | Glycyrrhiza_uralensis_Fisch0104330.1 | 492 | 53.76 | 6.16 | Mitochondrion |
| GuALDH2C1 | Glycyrrhiza_uralensis_Fisch0121290.1 | 505 | 54.63 | 5.73 | Cytoplasm | |
| GuALDH2C2 | Glycyrrhiza_uralensis_Fisch0121300.1 | 498 | 54.13 | 5.99 | Cytoplasm | |
| GuALDH2B2 | Glycyrrhiza_uralensis_Fisch0205690.1 | 542 | 59.32 | 8.31 | Mitochondrion | |
| GuALDH2B3 | Glycyrrhiza_uralensis_Fisch0227020.1 | 494 | 53.51 | 5.74 | Cytoplasm | |
| GuALDH2B4 | Glycyrrhiza_uralensis_Fisch0269090.1 | 531 | 57.48 | 7.99 | Mitochondrion | |
| GuALDH2C3 | Glycyrrhiza_uralensis_Fisch0284140.1 | 501 | 54.43 | 6.35 | Cytoplasm | |
| GuALDH2C4 | Glycyrrhiza_uralensis_Fisch0284220.1 | 503 | 54.96 | 5.82 | Cytoplasm | |
| Family3 | GuALDH3H1 | Glycyrrhiza_uralensis_Fisch0061860.1 | 497 | 54.48 | 8.83 | Cytoplasm |
| GuALDH3I1 | Glycyrrhiza_uralensis_Fisch0091680.1 | 545 | 60.18 | 7.57 | Chloroplast | |
| GuALDH3H2 | Glycyrrhiza_uralensis_Fisch0114530.1 | 581 | 64.02 | 9.39 | Peroxisome | |
| GuALDH3F2 | Glycyrrhiza_uralensis_Fisch0197170.1 | 514 | 56.60 | 8.53 | Mitochondrion | |
| GuALDH3F3 | Glycyrrhiza_uralensis_Fisch0211780.1 | 451 | 50.64 | 8.45 | Cytoplasm | |
| GuALDH3F4 | Glycyrrhiza_uralensis_Fisch0253710.1 | 470 | 51.75 | 7.02 | Cytoplasm | |
| GuALDH3F5 | Glycyrrhiza_uralensis_Fisch0257550.1 | 594 | 65.54 | 6.26 | Chloroplast | |
| GuALDH3I2 | Glycyrrhiza_uralensis_Fisch0125250.1 | 545 | 60.07 | 6.74 | Chloroplast | |
| GuALDH3F1 | Glycyrrhiza_uralensis_Fisch0127260.1 | 449 | 50.18 | 8.88 | Nuclear | |
| Family10 | GuALDH10A2 | Glycyrrhiza_uralensis_Fisch0205450.1 | 503 | 54.48 | 5.18 | Peroxisome |
| GuALDH10A1 | Glycyrrhiza_uralensis_Fisch0104500.1 | 503 | 54.78 | 5.13 | Peroxisome | |
| Family6 | GuALDH6B1 | Glycyrrhiza_uralensis_Fisch0265940.1 | 510 | 54.78 | 6.34 | Mitochondrion |
| GuALDH6B2 | Glycyrrhiza_uralensis_Fisch0138040.1 | 483 | 52.19 | 7.09 | Chloroplast | |
| Family11 | GuALDH11A1 | Glycyrrhiza_uralensis_Fisch0065080.1 | 519 | 55.60 | 7.51 | Cytoplasm |
| GuALDH11A2 | Glycyrrhiza_uralensis_Fisch0113230.1 | 138 | 15.39 | 10.06 | Nuclear | |
| GuALDH11A3 | Glycyrrhiza_uralensis_Fisch0201600.1 | 496 | 53.14 | 7.08 | Cytoplasm | |
| Family12 | GuALDH12A1 | Glycyrrhiza_uralensis_Fisch0110500.1 | 727 | 79.45 | 6.87 | Cytoplasm |
| GuALDH12A2 | Glycyrrhiza_uralensis_Fisch0204970.1 | 544 | 60.45 | 6.48 | Mitochondrion |
| Primer Name | Primer Sequences F (5′-3′) | Primer Sequences R (5′-3′) |
|---|---|---|
| GuALDH3F4 | TCTCACCGAGCCTAAGGTCA | GACTGAACGCGTCAAACGAG |
| GuALDH6B1 | TTGGCATTGGAAGCTGGTCT | TTGCTGATGACCGGACCAAG |
| GuALDH12A1 | GGTCGCCAAAGGCTCAGATA | CCACCTCTTCCCACCCTAGA |
| GuALDH12A2 | GCCACGGTAGAAGCAGAAGA | TGACCTTGCCAGGAAACGAA |
| Gulectin | ctgatgcagagcttcaaatcgag | ttcggaaggaaggttgaggtaag |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, M.; Ouyang, X.; Cheng, L.; Li, Y.; Shi, N.; Ma, H.; Sun, Y.; Yao, H.; Shen, H. Identification of Aldehyde Dehydrogenase Gene Family in Glycyrrhiza uralensis and Analysis of Expression Pattern Under Drought Stress. Int. J. Mol. Sci. 2025, 26, 2333. https://doi.org/10.3390/ijms26052333
He M, Ouyang X, Cheng L, Li Y, Shi N, Ma H, Sun Y, Yao H, Shen H. Identification of Aldehyde Dehydrogenase Gene Family in Glycyrrhiza uralensis and Analysis of Expression Pattern Under Drought Stress. International Journal of Molecular Sciences. 2025; 26(5):2333. https://doi.org/10.3390/ijms26052333
Chicago/Turabian StyleHe, Mengyuan, Xu Ouyang, Linyuan Cheng, Yuetao Li, Nana Shi, Hongxia Ma, Yu Sun, Hua Yao, and Haitao Shen. 2025. "Identification of Aldehyde Dehydrogenase Gene Family in Glycyrrhiza uralensis and Analysis of Expression Pattern Under Drought Stress" International Journal of Molecular Sciences 26, no. 5: 2333. https://doi.org/10.3390/ijms26052333
APA StyleHe, M., Ouyang, X., Cheng, L., Li, Y., Shi, N., Ma, H., Sun, Y., Yao, H., & Shen, H. (2025). Identification of Aldehyde Dehydrogenase Gene Family in Glycyrrhiza uralensis and Analysis of Expression Pattern Under Drought Stress. International Journal of Molecular Sciences, 26(5), 2333. https://doi.org/10.3390/ijms26052333





