Genome-Wide Association Study and Genomic Prediction of Soybean Mosaic Virus Resistance
Abstract
1. Introduction
2. Results
2.1. SMV-G1 Resistance Phenotyping
2.2. Population Structure and GWAS
2.3. Candidate Gene Identification
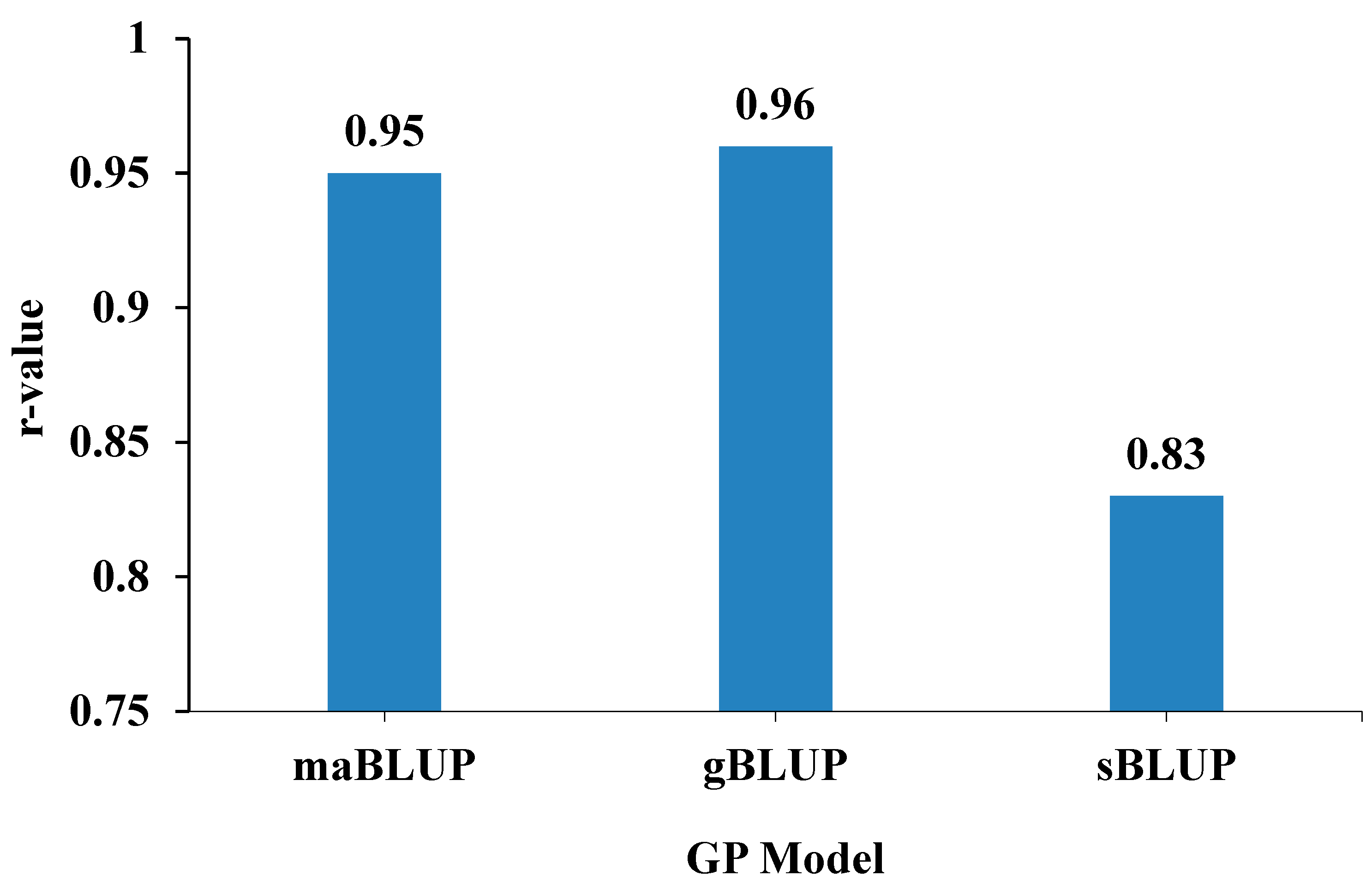
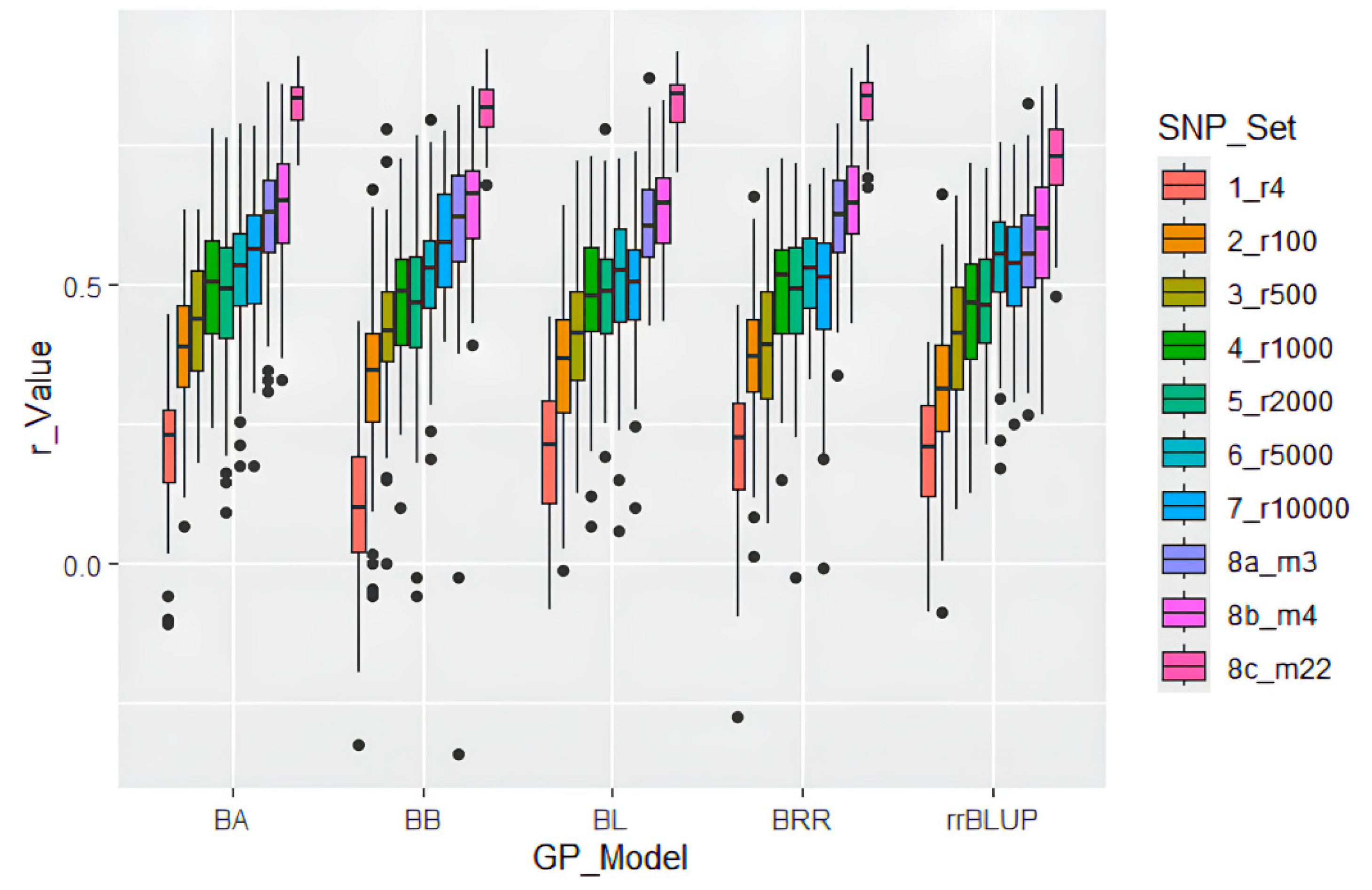
2.4. Genomic Prediction Accuracy
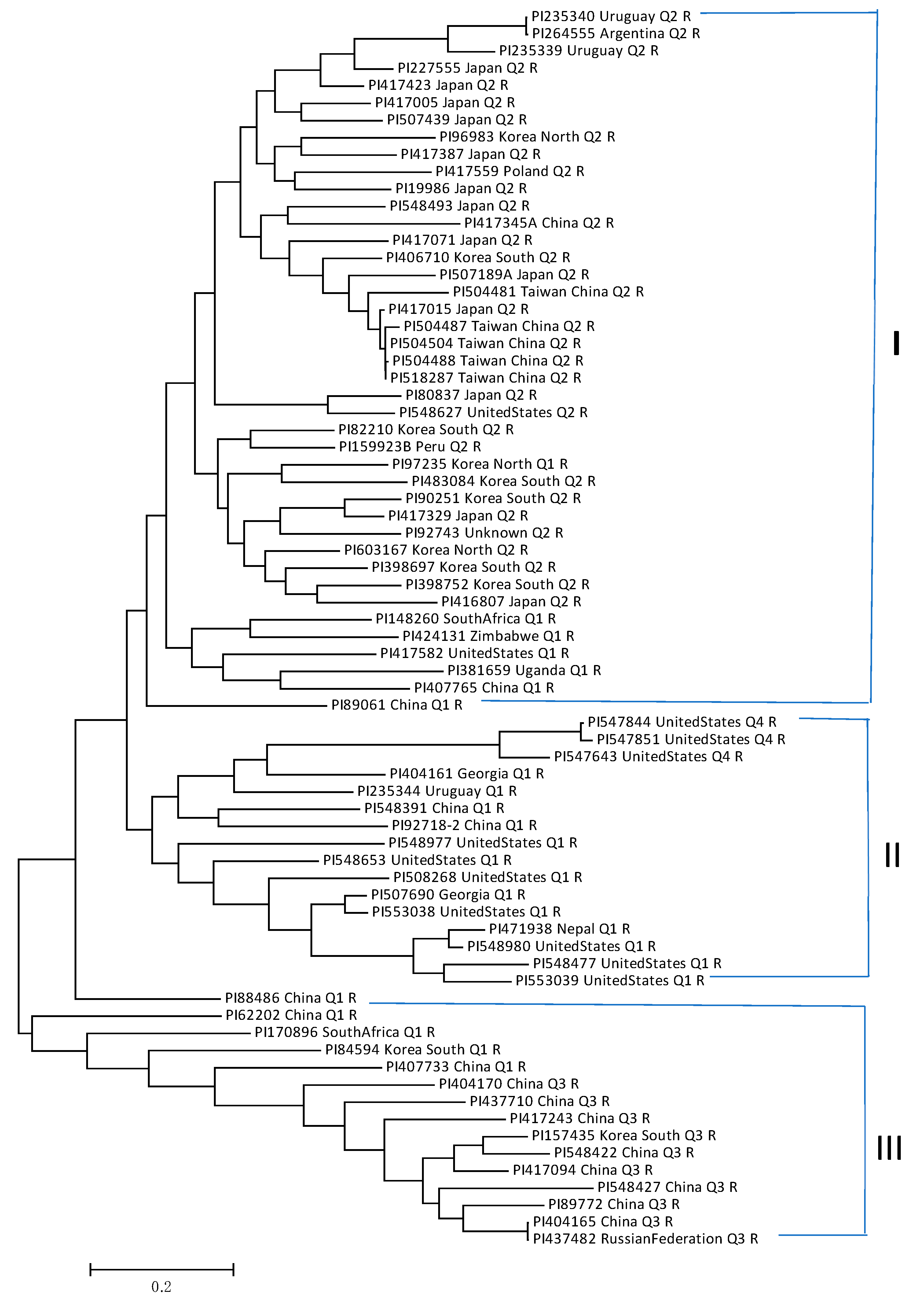
2.5. Genetic Diversity and Utilization of the SMV-Resistant Accessions
3. Discussion
3.1. Phenotypic Evaluation and Genetic Diversity of SMV-G1 Resistance
3.2. Genome-Wide Association Study
3.3. Functional Characterization of Candidate Genes
3.4. Genomic Prediction for Breeding Applications
4. Materials and Methods
4.1. Plant Material
4.2. Phenotyping and Genotyping
4.3. Principal Component Analysis and Genetic Diversity
4.4. Candidate Gene Annotation
4.5. Genomic Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bandara, A.Y.; Weerasooriya, D.K.; Bradley, C.A.; Allen, T.W.; Esker, P.D. Dissecting the economic impact of soybean diseases in the United States over two decades. PLoS ONE 2020, 15, e0231141. [Google Scholar] [CrossRef] [PubMed]
- Usovsky, M.; Chen, P.; Li, D.; Wang, A.; Shi, A.; Zheng, C.; Shakiba, E.; Lee, D.; Canella Vieira, C.; Lee, Y.C.; et al. Decades of genetic research on soybean mosaic virus resistance in soybean. Viruses 2022, 14, 1122. [Google Scholar] [CrossRef]
- Cho, E.K.; Goodman, R.M. Strains of soybean mosaic virus: Classification based on virulence in resistant soybean cultivars. Phytopathology 1979, 69, 467–470. [Google Scholar] [CrossRef]
- Pu, Y.; Yan, R.; Jia, D.; Che, Z.; Yang, R.; Yang, C.; Wang, H.; Cheng, H.; Yu, D. Identification of soybean mosaic virus strain SC7 resistance loci and candidate genes in soybean [Glycine max (L.) Merr.]. Mol. Genet. Genom. 2024, 299, 54. [Google Scholar] [CrossRef]
- Hajimorad, M.R.; Eggenberger, A.L.; Hill, J.H. Loss and Gain of Elicitor Function of Soybean Mosaic Virus G7 Provoking Rsv1-Mediated Lethal Systemic Hypersensitive Response Maps to P3. J. Virol. 2005, 79, 1215–1222. [Google Scholar] [CrossRef]
- Hayes, A.J.; Jeong, S.C.; Gore, M.A.; Yu, Y.G.; Buss, G.R.; Tolin, S.A.; Maroof, M.A. Recombination within a nucleotide-binding-site/leucine-rich-repeat gene cluster produces new variants conditioning resistance to soybean mosaic virus in soybeans. Genetics 2004, 166, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Du, H.; Zhao, T.; Liao, C.; Feng, T.; Qin, J.; Liu, B.; Kong, F.; Che, Z.; Chen, L. GmTOC1b negatively regulates resistance to Soybean mosaic virus. Crop J. 2023, 11, 1762–1773. [Google Scholar] [CrossRef]
- Wu, M.; Wu, W.P.; Liu, C.C.; Liu, Y.N.; Wu, X.Y.; Ma, F.F.; Zhu, A.Q.; Yang, J.Y.; Wang, B.; Chen, J.Q. A bean common mosaic virus (BCMV)—Resistance gene is fine-mapped to the same region as Rsv1-h in the soybean cultivar Suweon 97. Theor. Appl. Genet. 2018, 131, 1851–1860. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.-T.; Widyasari, K.; Seo, J.-K.; Kim, K.-H. Isolation and validation of a candidate Rsv3 gene from a soybean genotype that confers strain-specific resistance to soybean mosaic virus. Virology 2018, 513, 153–159. [Google Scholar] [CrossRef]
- Ishibashi, K.; Saruta, M.; Shimizu, T.; Shu, M.; Anai, T.; Komatsu, K.; Yamada, N.; Katayose, Y.; Ishikawa, M.; Ishimoto, M.; et al. Soybean antiviral immunity conferred by dsRNase targets the viral replication complex. Nat. Commun. 2019, 10, 4033. [Google Scholar] [CrossRef] [PubMed]
- Che, Y.; Gribenko, A.V.; Song, X.; Handke, L.D.; Efferen, K.S.; Tompkins, K.; Kodali, S.; Nunez, L.; Prasad, A.K.; Phelan, L.M.; et al. Rational design of a highly immunogenic prefusion-stabilized F glycoprotein antigen for a respiratory syncytial virus vaccine. Sci. Transl. Med. 2023, 15, eade6422. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lee, J.Y.; Yang, J.S.; Kim, J.W.; Kim, V.N.; Chang, H. The architecture of SARS-CoV-2 transcriptome. Cell 2020, 181, 914–921.e910. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Yan, H.; Wang, H.; Cheng, H.; Hu, Z.; Chu, S.; Zhang, G.; Yu, D. Detection and fine-mapping of SC7 resistance genes via linkage and association analysis in soybean. J. Integr. Plant Biol. 2015, 57, 722–729. [Google Scholar] [CrossRef]
- Seo, J.K.; Lee, S.H.; Kim, K.H. Strain-specific cylindrical inclusion protein of soybean mosaic virus elicits extreme resistance and a lethal systemic hypersensitive response in two resistant soybean cultivars. Mol. Plant Microbe Interact. 2009, 22, 1151–1159. [Google Scholar] [CrossRef] [PubMed]
- Che, Z.; Yan, H.; Liu, H.; Yang, H.; Du, H.; Yang, Y.; Liu, B.; Yu, D. Genome-wide association study for soybean mosaic virus SC3 resistance in soybean. Mol. Breed. 2020, 40, 69. [Google Scholar] [CrossRef]
- Tibbs Cortes, L.; Zhang, Z.; Yu, J. Status and prospects of genome-wide association studies in plants. Plant Genome 2021, 14, e20077. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Grosic, S.; Whitham, S.A.; Hill, J.H. The requirement of multiple defense genes in soybean Rsv1-mediated extreme resistance to soybean mosaic virus. Mol. Plant Microbe Interact. 2012, 25, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Adam Arsovski, A.; Yu, K.; Wang, A. Deep sequencing leads to the identification of eukaryotic translation initiation factor 5A as a key element in Rsv1-mediated lethal systemic hypersensitive response to Soybean mosaic virus infection in soybean. Mol. Plant Pathol. 2017, 18, 391–404. [Google Scholar] [CrossRef]
- Gore, M.A.; Hayes, A.J.; Jeong, S.C.; Yue, Y.G.; Buss, G.R.; Maroof, S. Mapping tightly linked genes controlling potyvirus infection at the Rsv1 and Rpv1 region in soybean. Genome 2002, 45, 592–599. [Google Scholar] [CrossRef]
- Hajimorad, M.R.; Hill, J.H. Rsv1-mediated resistance against soybean mosaic virus-N is hypersensitive response-independent at inoculation site, but has the potential to initiate a hypersensitive response-like mechanism. Mol. Plant Microbe Interact. 2001, 14, 587–598. [Google Scholar] [CrossRef]
- Maroof, M.A.S.; Tucker, D.M.; Skoneczka, J.A.; Bowman, B.C.; Tripathy, S.; Tolin, S.A. Fine mapping and candidate gene discovery of the soybean mosaic virus resistance gene, Rsv4. Plant Genome 2010, 3, 14–22. [Google Scholar] [CrossRef]
- Liao, L.; Chen, P.; Rajcan, I.; Buss, G.R.; Tolin, S.A. Genetic analysis of “8101” soybean containing three genes for resistance to soybean mosaic virus. Crop Sci. 2011, 51, 503–511. [Google Scholar] [CrossRef]
- Chowda-Reddy, R.V.; Sun, H.; Hill, J.H.; Poysa, V.; Wang, A. Simultaneous mutations in multi-viral proteins are required for soybean mosaic virus to gain virulence on soybean genotypes carrying different R genes. PLoS ONE 2011, 6, e28342. [Google Scholar] [CrossRef][Green Version]
- Chang, H.-X.; Lipka, A.E.; Domier, L.L.; Hartman, G.L. Characterization of disease resistance loci in the USDA soybean germplasm collection using genome-wide association studies. Phytopathology 2016, 106, 1139–1151. [Google Scholar] [CrossRef]
- Yang, Y.; Zheng, G.; Han, L.; Dagang, W.; Yang, X.; Yuan, Y.; Huang, S.; Zhi, H. Genetic analysis and mapping of genes for resistance to multiple strains of Soybean mosaic virus in a single resistant soybean accession PI 96983. Theor. Appl. Genet. 2013, 126, 1783–1791. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Wu, X.; Chen, Y.-X.; Liu, Y.-N.; Shao, Z.-Q.; Wu, P.; Wu, M.; Liu, C.-C.; Wu, W.-P.; Yang, J.-Y.; et al. Fine mapping of the Rsv1-h gene in the soybean cultivar Suweon 97 that confers resistance to two Chinese strains of the soybean mosaic virus. Theor. Appl. Genet. 2016, 129, 2227–2236. [Google Scholar] [CrossRef]
- Liu, Y.; Li, D.; Yang, N.; Zhu, X.; Han, K.; Gu, R.; Bai, J.; Wang, A.; Zhang, Y. Genome-wide identification and analysis of CC-NBS-LRR family in response to downy mildew and black rot in chinese cabbage. Int. J. Mol. Sci. 2021, 22, 4266. [Google Scholar] [CrossRef]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 2006, 7, 212. [Google Scholar] [CrossRef] [PubMed]
- van Ooijen, G.; Mayr, G.; Kasiem, M.M.; Albrecht, M.; Cornelissen, B.J.; Takken, F.L. Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J. Exp. Bot. 2008, 59, 1383–1397. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Li, S.; Cheng, X.; Zhi, H.; Yin, J.; Xu, K. Searching for plant NLR immune receptors conferring resistance to potyviruses. Crop J. 2024, 12, 28–44. [Google Scholar] [CrossRef]
- Shi, A.; Gepts, P.; Song, Q.; Xiong, H.; Michaels, T.E.; Chen, S. Genome-wide association study and genomic prediction for soybean cyst nematode resistance in USDA common bean (Phaseolus vulgaris) core collection. Front. Plant Sci. 2021, 12, 624156. [Google Scholar] [CrossRef]
- Wen, L.; Chang, H.X.; Brown, P.J.; Domier, L.L.; Hartman, G.L. Genome-wide association and genomic prediction identifies soybean cyst nematode resistance in common bean including a syntenic region to soybean Rhg1 locus. Hortic. Res. 2019, 6, 9. [Google Scholar] [CrossRef]
- Shi, A.; Bhattarai, G.; Xiong, H.; Avila, C.A.; Feng, C.; Liu, B.; Joshi, V.; Stein, L.; Mou, B.; du Toit, L.J.; et al. Genome-wide association study and genomic prediction of white rust resistance in USDA GRIN spinach germplasm. Hortic. Res. 2022, 9, uhac069. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.K.; Goodman, R.M. Evaluation of resistance in soybeans to soybean mosaic virus strains. Crop Sci. 1982, 22, 1133–1136. [Google Scholar] [CrossRef]
- Song, Q.; Hyten, D.L.; Jia, G.; Quigley, C.V.; Fickus, E.W.; Nelson, R.L.; Cregan, P.B. Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS ONE 2013, 8, e54985. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Hyten, D.L.; Jia, G.; Quigley, C.V.; Fickus, E.W.; Nelson, R.L.; Cregan, P.B. Fingerprinting soybean germplasm and its utility in genomic research. G3 Genes Genomes Genet. 2015, 5, 1999–2006. [Google Scholar] [CrossRef]
- Chen, P.; Buss, G.R.; Roane, C.W.; Tolin, S.A. Allelism among genes for resistance to soybean mosaic virus in strain-differential soybean cultivars. Crop Sci. 1991, 31, 305–309. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z. GAPIT version 3: Boosting power and accuracy for genomic association and prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Meher, P.; Sachin, R.; Anuj, K. Performance of Bayesian and BLUP alphabets for genomic prediction: Analysis, comparison and results. Heredity 2022, 128, 519–530. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Shi, A.; Chen, P.; Vierling, R.; Li, D.; Zheng, C. Identification of soybean mosaic virus resistance alleles in Jindou 1 soybean. Euphytica 2013, 192, 181–187. [Google Scholar] [CrossRef]
- Zhang, H.; Li, C.; Davis, E.L.; Wang, J.; Griffin, J.D.; Kofsky, J.; Song, B.H. Genome-wide association study of resistance to soybean cyst nematode (Heterodera glycines) HG type 2.5.7 in wild soybean (Glycine soja). Front. Plant Sci. 2016, 7, 1214. [Google Scholar] [CrossRef] [PubMed]
- Pérez, P.; de los Campos, G. Genome-wide regression and prediction with the BGLR statistical package. Genetics 2014, 198, 483–495. [Google Scholar] [CrossRef]
- Jarquín, D.; Kocak, K.; Posadas, L.; Hyma, K.; Jedlicka, J.; Graef, G.; Lorenz, A. Genotyping by sequencing for genomic prediction in a soybean breeding population. BMC Genom. 2014, 15, 740. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.J.; Bowman, B.C.; Jeong, N.; Yang, K.; Kastl, C.; Tolin, S.A.; Maroof, M.A.S.; Jeong, S.-C. The Rsv3 locus conferring resistance to soybean mosaic virus is associated with a cluster of coiled-coil nucleotide-binding leucine-rich repeat genes. Plant Genome 2011, 4, 55–64. [Google Scholar] [CrossRef]
- Krishnappa, G.; Savadi, S.; Tyagi, B.S.; Singh, S.K.; Mamrutha, H.M.; Kumar, S.; Mishra, C.N.; Khan, H.; Gangadhara, K.; Uday, G.; et al. Integrated genomic selection for rapid improvement of crops. Genomics 2021, 113, 1070–1086. [Google Scholar] [CrossRef]
- Qu, S.; Lu, S.; Liu, Y.; Li, M.; Chen, S. Accurate genomic selection using low-density SNP panels preselected by maximum likelihood estimation. Aquaculture 2024, 579, 740154. [Google Scholar] [CrossRef]



| SNP | Chr | Position | LOD [−log(p-Value)] | MAF% | Model |
|---|---|---|---|---|---|
| Gm13_29459954_ss715614803 | 13 | 29459954 | 10.58 | 19.5 | MLM |
| 14.53 | 19.5 | FarmCPU | |||
| Gm13_29751552_ss715614847 | 13 | 29751552 | 8.06 | 44 | MLM |
| 7.37 | 44 | MLMM | |||
| 10.76 | 44 | BLINK | |||
| Gm13_30293949_ss715614951 | 13 | 30293949 | 13.04 | 13.5 | MLM |
| 13.54 | 13.5 | MLMM | |||
| 14.09 | 13.5 | BLINK | |||
| Gm13_30724301_ss715615024 | 13 | 30724301 | 6.56 | 22.48 | MLMM |
| 9.21 | 22.48 | BLINK |
| SNP Set | GP Model r-Value | SE | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BA | BB | BL | BRR | rrBLUP | SNP Set Mean | BA | BB | BL | BRR | rrBLUP | SNP Set Mean | |
| r4 | 0.22 | 0.10 | 0.20 | 0.21 | 0.20 | 0.19 | 0.011 | 0.013 | 0.012 | 0.012 | 0.011 | 0.012 |
| r100 | 0.39 | 0.33 | 0.35 | 0.37 | 0.31 | 0.35 | 0.012 | 0.014 | 0.014 | 0.011 | 0.013 | 0.013 |
| r500 | 0.43 | 0.42 | 0.41 | 0.39 | 0.40 | 0.41 | 0.010 | 0.011 | 0.012 | 0.013 | 0.012 | 0.011 |
| r1000 | 0.50 | 0.47 | 0.48 | 0.50 | 0.45 | 0.48 | 0.011 | 0.011 | 0.012 | 0.011 | 0.012 | 0.012 |
| r2000 | 0.48 | 0.46 | 0.48 | 0.48 | 0.46 | 0.47 | 0.012 | 0.013 | 0.010 | 0.012 | 0.011 | 0.012 |
| r5000 | 0.52 | 0.51 | 0.51 | 0.52 | 0.54 | 0.52 | 0.012 | 0.010 | 0.012 | 0.009 | 0.011 | 0.011 |
| r10000 | 0.54 | 0.58 | 0.50 | 0.49 | 0.53 | 0.53 | 0.011 | 0.010 | 0.011 | 0.013 | 0.010 | 0.011 |
| m3 | 0.61 | 0.60 | 0.61 | 0.62 | 0.56 | 0.60 | 0.011 | 0.015 | 0.009 | 0.009 | 0.011 | 0.011 |
| m4 | 0.64 | 0.64 | 0.63 | 0.64 | 0.58 | 0.63 | 0.011 | 0.009 | 0.008 | 0.009 | 0.012 | 0.010 |
| m22 | 0.83 | 0.81 | 0.82 | 0.82 | 0.72 | 0.80 | 0.004 | 0.005 | 0.005 | 0.005 | 0.007 | 0.005 |
| Model Mean | 0.52 | 0.49 | 0.50 | 0.51 | 0.48 | 0.50 | 0.010 | 0.011 | 0.010 | 0.010 | 0.011 | 0.011 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, D.; Wu, X.; Liu, Z.; Yang, Q.; Shi, X.; Song, Q.; Shi, A.; Li, D.; Yan, L. Genome-Wide Association Study and Genomic Prediction of Soybean Mosaic Virus Resistance. Int. J. Mol. Sci. 2025, 26, 2106. https://doi.org/10.3390/ijms26052106
He D, Wu X, Liu Z, Yang Q, Shi X, Song Q, Shi A, Li D, Yan L. Genome-Wide Association Study and Genomic Prediction of Soybean Mosaic Virus Resistance. International Journal of Molecular Sciences. 2025; 26(5):2106. https://doi.org/10.3390/ijms26052106
Chicago/Turabian StyleHe, Di, Xintong Wu, Zhi Liu, Qing Yang, Xiaolei Shi, Qijian Song, Ainong Shi, Dexiao Li, and Long Yan. 2025. "Genome-Wide Association Study and Genomic Prediction of Soybean Mosaic Virus Resistance" International Journal of Molecular Sciences 26, no. 5: 2106. https://doi.org/10.3390/ijms26052106
APA StyleHe, D., Wu, X., Liu, Z., Yang, Q., Shi, X., Song, Q., Shi, A., Li, D., & Yan, L. (2025). Genome-Wide Association Study and Genomic Prediction of Soybean Mosaic Virus Resistance. International Journal of Molecular Sciences, 26(5), 2106. https://doi.org/10.3390/ijms26052106









