Cytosolic and Nucleosolic Calcium-Regulated Long Non-Coding RNAs and Their Target Protein-Coding Genes in Response to Hyperosmolarity and Salt Stresses in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
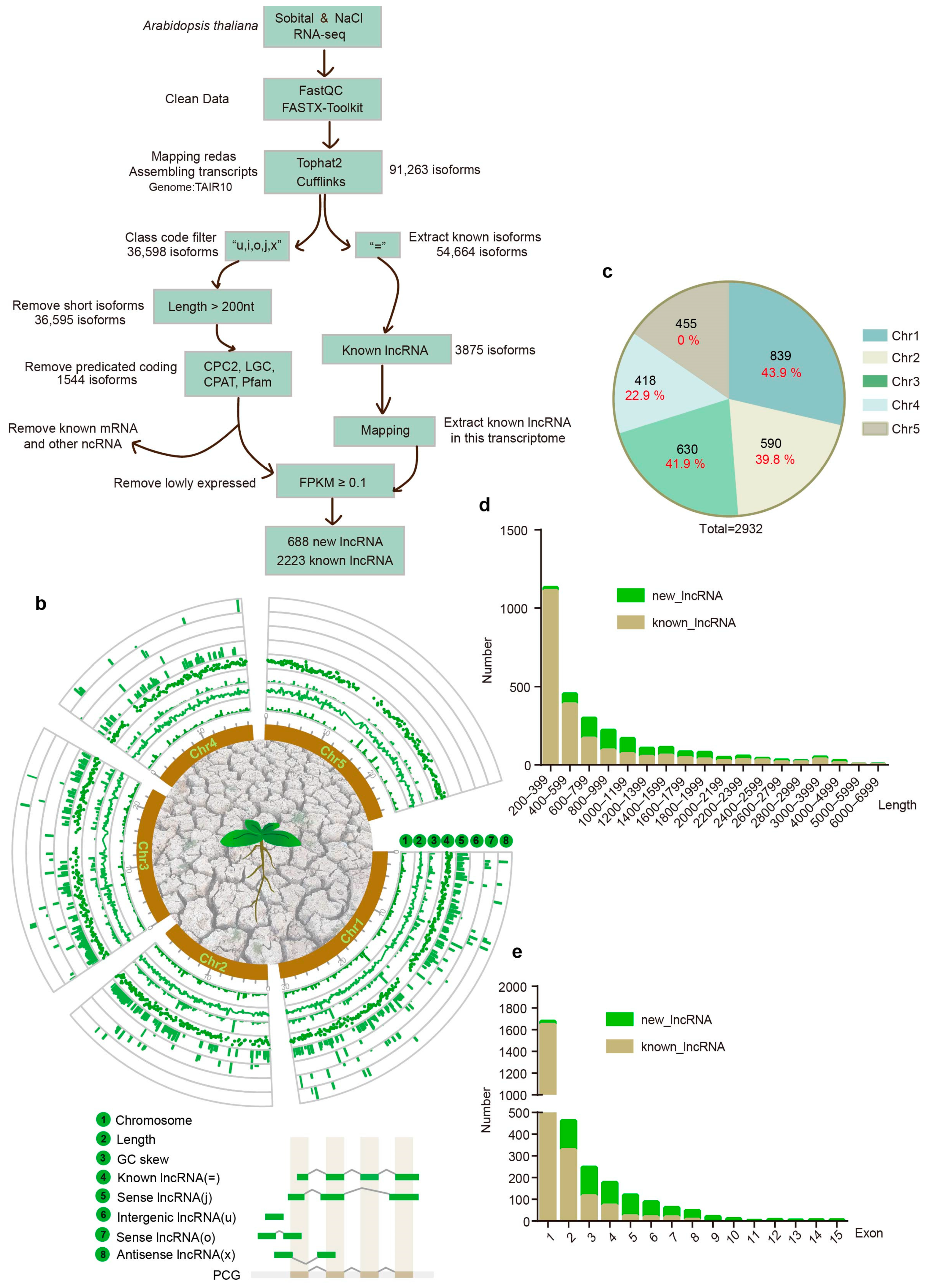
2.1. Transcriptome-Wide Profiles of lncRNAs in Response to Salt and Hyperosmolarity Stresses in Arabidopsis
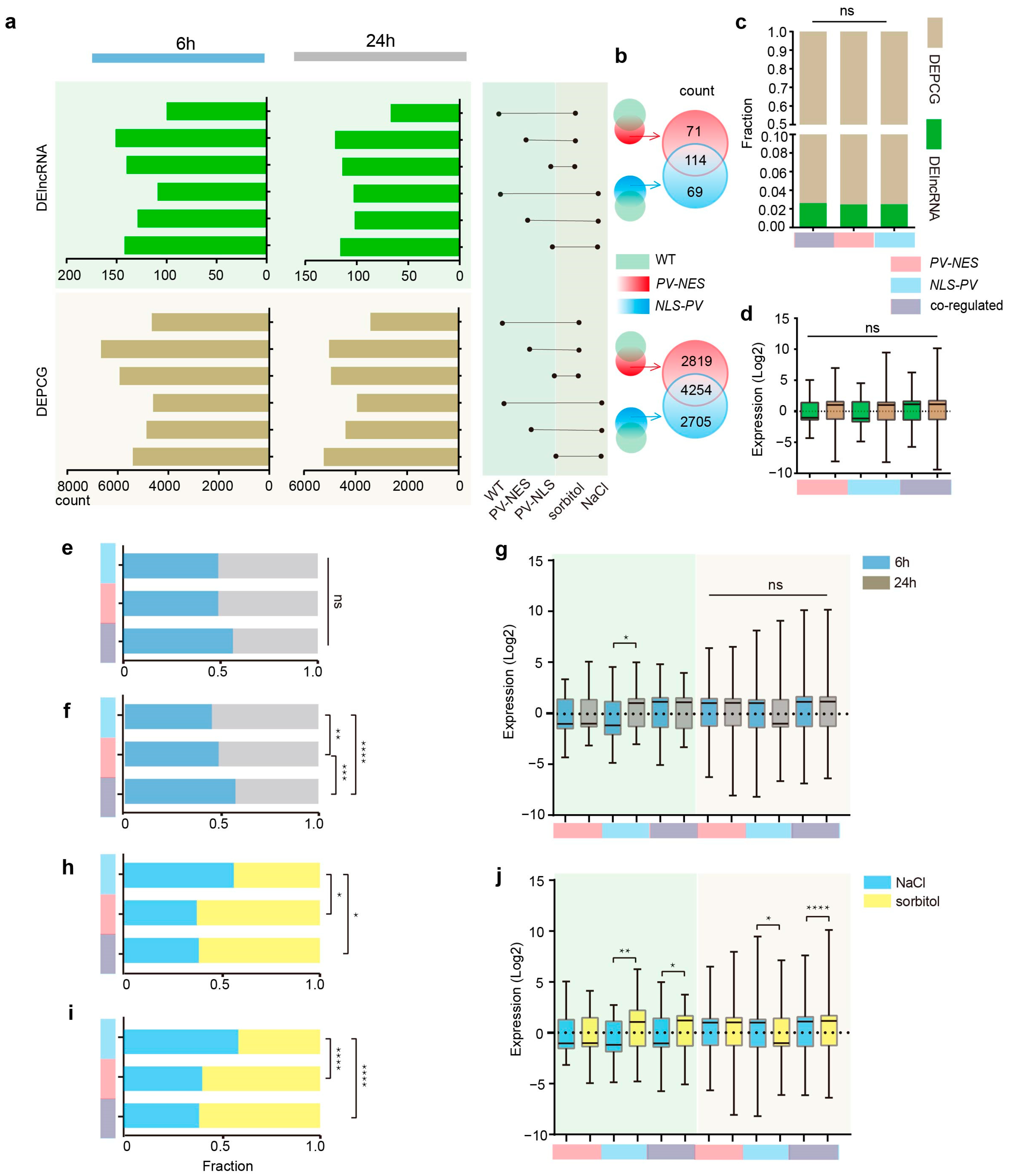
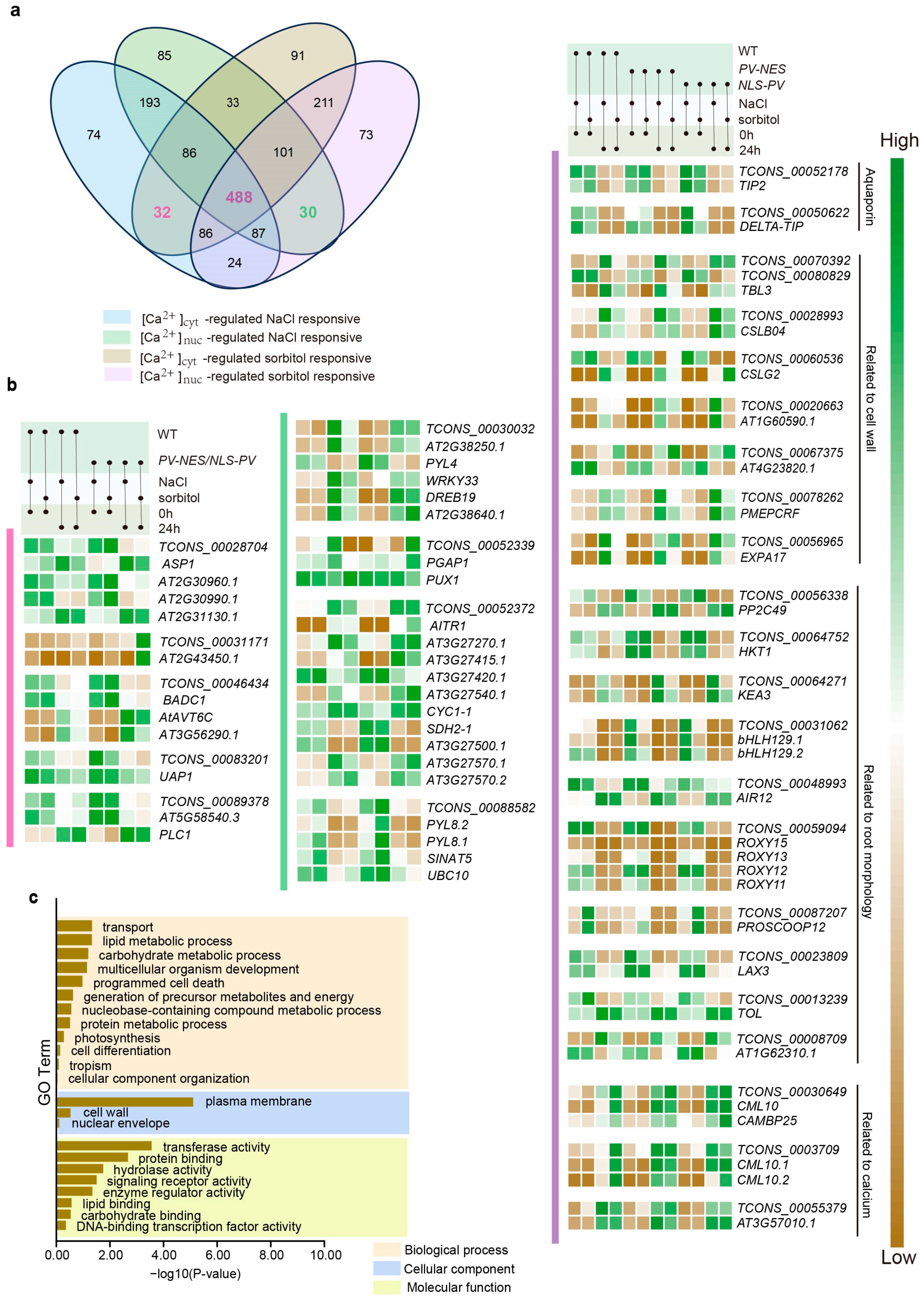
2.2. DElncRNAs and DEPCGs Respond to Cytosolic and Nucleosolic Calcium Signals in Different Ways in Terms of Quantity and Expression Level
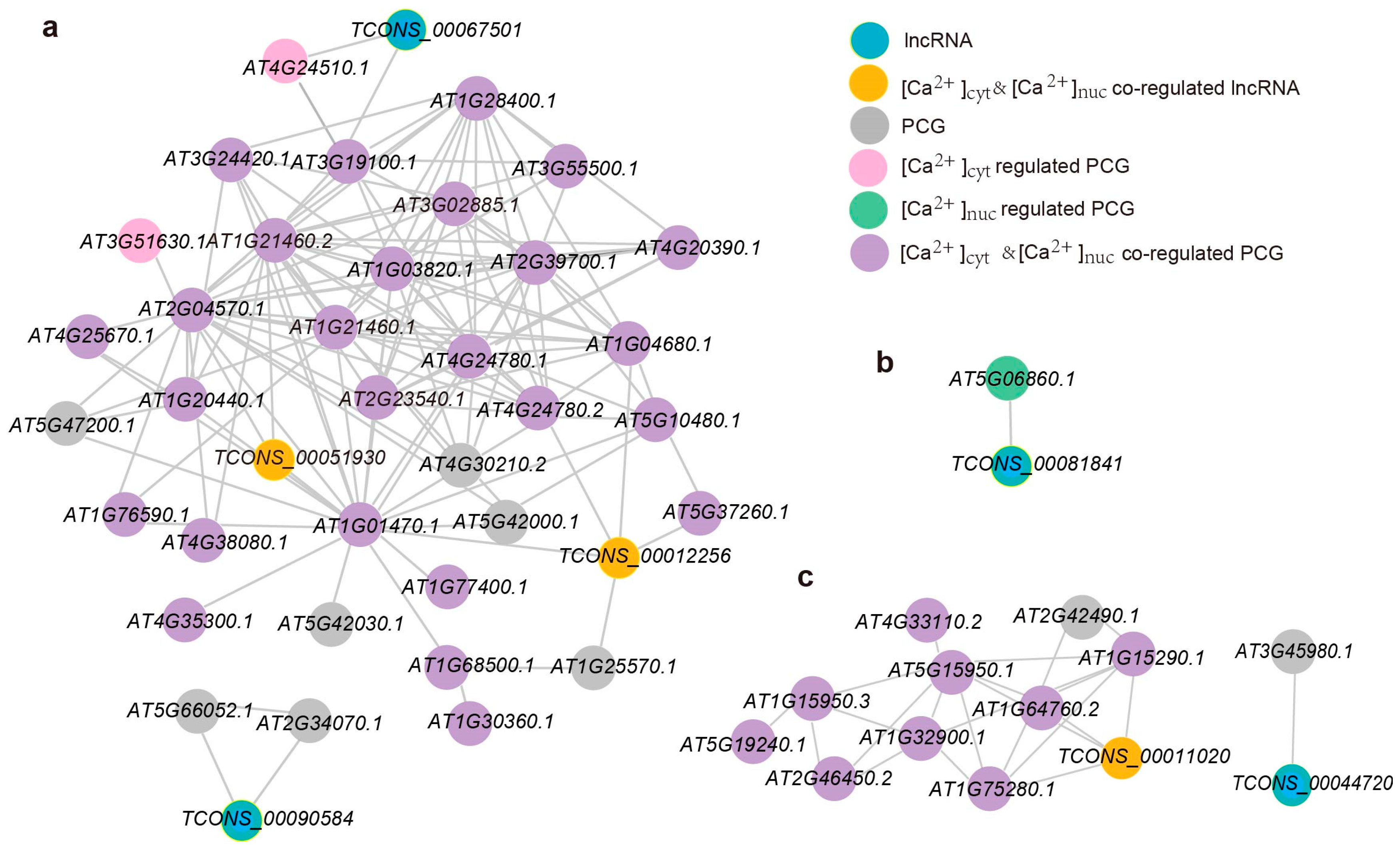
2.3. Co-Expression Network Reveals the Role of Calcium-Regulated lncRNAs and PCGs in Response to Salt Stress and Hyperosmotic Stress
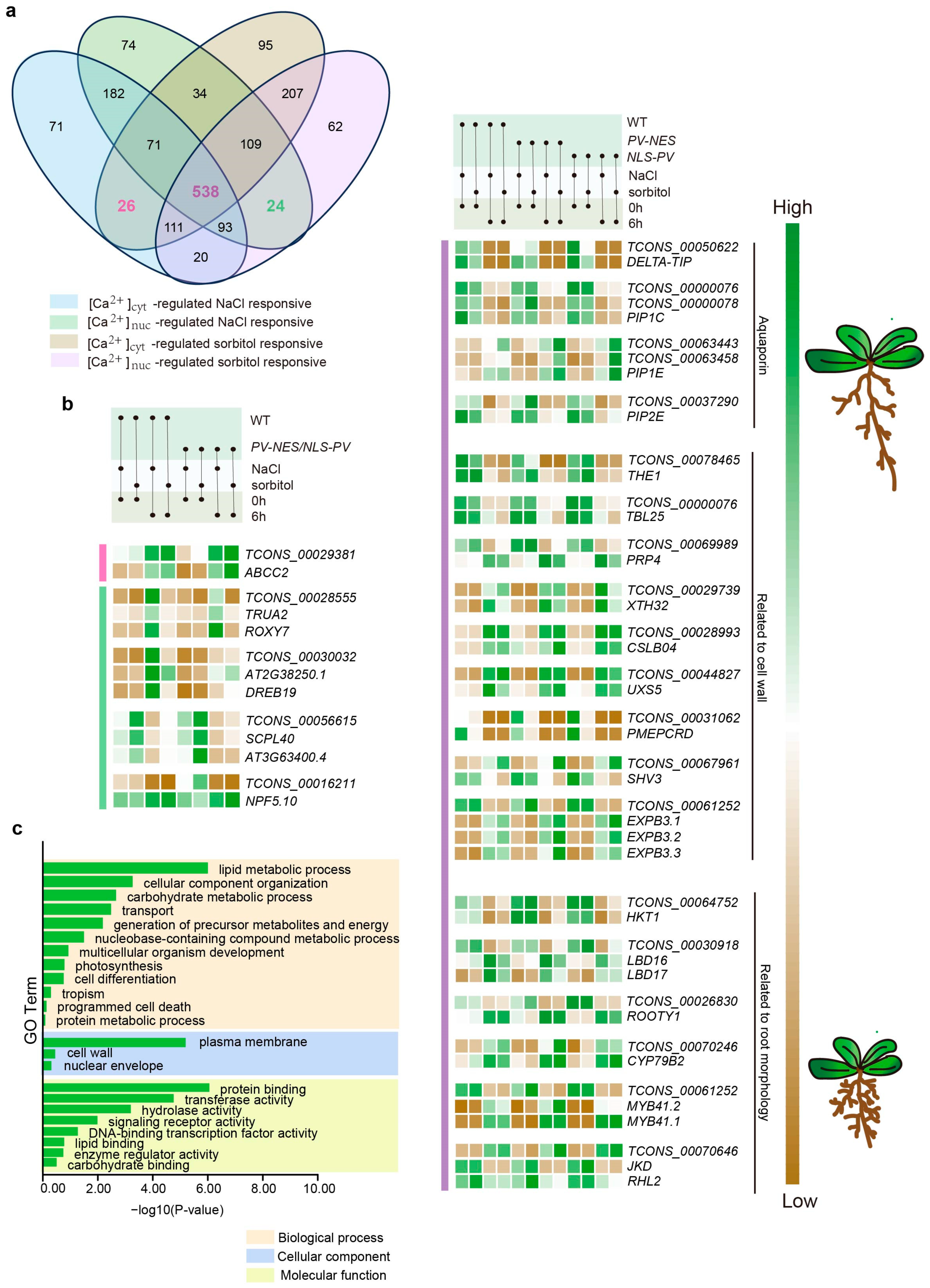
2.4. Ca2+-Regulated lncRNAs and Their Potential Cis-Regulated Target PCGs in Omostic Stress
3. Discussion
3.1. Regulation of Plant Cell Wall Strength by lncRNAs in Response to Salt Stress and Osmotic Stress
3.2. Calcium-Regulated lncRNAs Are Involved in the Regulation of Root Morphology Under Osmotic Stress in Arabidopsis
4. Materials and Methods
4.1. Plant Materials and Treatment
4.2. RNA Extraction and Real-Time Quantitative PCR
4.3. RNA-Seq and Transcriptome Assembly
4.4. Pipline of lncRNA Screening
- (1)
- Transcript Filtering: Transcripts with class code types “i”, “o”, “j”, “u”, and “x” were selected from the assembled transcriptome.
- (2)
- Length Filtering: Transcripts with lengths less than 200 nucleotides were excluded from further consideration.
- (3)
- Coding Potential Prediction: The subsequent step involved the application of the software CPC2, LGC, CPAT, and Pfam to predict the protein-coding potential of the remaining transcripts, thereby facilitating the exclusion of those possessing coding capabilities.
- (4)
- Blast Alignment: The transcripts that remained were aligned against a database of known mRNAs and other non-coding RNAs using Blast, with criteria set for high stringency (E-value < 10^ (−10), identity > 90%) to eliminate any matches.
- (5)
- Expression Level Screening: The final step involved the assessment of expression levels, with transcripts required to exhibit an FPKM value of at least 0.1 in at least one sample to be considered. This threshold was crucial for the identification of novel lncRNAs. For known lncRNAs, the same expression level filter was applied to ensure that only those with significant expression were included in the subsequent analysis.
4.5. Basic Characteristics of lncRNAs
4.6. Differential Expression Analysis
4.7. Prediction of Trans-Regulated Target PCGs of lncRNAs
4.8. Prediction of Cis-Regulated Target PCGs of lncRNAs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jha, U.C.; Nayyar, H.; Jha, R.; Khurshid, M.; Zhou, M.; Mantri, N.; Siddique, K.H.M. Long non-coding RNAs: Emerging players regulating plant abiotic stress response and adaptation. BMC Plant Biol. 2020, 20, 466–485. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lai, X.; Yu, X.; Xiong, Z.; Chen, J.; Lang, X.; Feng, H.; Wan, X.; Liu, K. Plant long noncoding RNAs: Recent progress in understanding their roles in growth, development, and stress responses. Biochem. Biophys. Res. Commun. 2023, 671, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, L.R.; Blom, S.; Henriques, R.; Bachem, C.W.B.; Immink, R.G.H. LncRNAs: The art of being influential without protein. Trends Plant Sci. 2024, 29, 770–785. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ariel, F.; Wang, D. Plant long non-coding RNAs: Biologically relevant and mechanistically intriguing. J. Exp. Bot. 2023, 74, 2364–2373. [Google Scholar] [CrossRef] [PubMed]
- Jampala, P.; Garhewal, A.; Lodha, M. Functions of long non-coding RNA in Arabidopsis thaliana. Plant Signal. Behav. 2021, 16, 1925440–1925451. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Zheng, K.; Min, Q.; Wang, Y.; Xue, X.; Li, W.; Zhao, H.; Qiao, F.; Han, S. Long noncoding RNAs in response to hyperosmolarity stress, but not salt stress, were mainly enriched in the rice roots. Int. J. Mol. Sci. 2024, 25, 6266. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Wu, X.; Xue, X.; Li, W.; Wang, Z.; Chen, J.; Zhang, Y.; Qiao, F.; Zhao, H.; Zhang, F.; et al. Transcriptome screening of long noncoding RNAs and their target protein-coding genes unmasks a dynamic portrait of seed coat coloration associated with anthocyanins in Tibetan Hulless Barley. Int. J. Mol. Sci. 2023, 24, 10587. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Zhan, C.; Yin, J.; Gong, S.; Ma, D.; Li, Y. Genome-wide identification of long intergenic non-coding RNAs for Ralstonia solanacearum resistance in tomato (Solanum lycopersicum). Front. Plant Sci. 2022, 13, 981281–981294. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, Y.; Chen, X.; Chen, Y. Plant noncoding RNAs: Hidden players in development and stress responses. Annu. Rev. Cell. Dev. Biol. 2019, 35, 407–431. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Fritschi, F.; Mittler, R. Global warming, climate change, and environmental pollution: Recipe for a multifactorial stress combination disaster. Trends Plant Sci. 2021, 26, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Gorgues, L.; Li, X.; Maurel, C.; Martinière, A.; Nacry, P. Root osmotic sensing from local perception to systemic responses. Stress Biol. 2022, 2, 36–56. [Google Scholar] [CrossRef]
- Bao, Y.; Aggarwal, P.; Robbins, N.E.; Sturrock, C.J.; Thompson, M.C.; Tan, H.Q.; Tham, C.; Duan, L.; Rodriguez, P.L.; Vernoux, T.; et al. Plant roots use a patterning mechanism to position lateral root branches toward available water. Proc. Natl. Acad. Sci. USA 2014, 111, 9319–9324. [Google Scholar] [CrossRef]
- Beatriz, O.P.; Leftley, N.; von Wangenheim, D.; Banda, J.; Srivastava, A.K.; Hill, K.; Truskina, J.; Bhosale, R.; Morris, E.; Srivastava, M.; et al. Root branching toward water involvesposttranslational modificationof transcription factor ARF7. Science 2018, 362, 1407–1410. [Google Scholar]
- Chandran, A.E.J.; Finkler, A.; Hait, T.A.; Kiere, Y.; David, S.; Metsada, P.C.; Shkolnik, D. Calcium regulation of the Arabidopsis Na+/K+ transporter HKT1;1 improves seed germination under salt stress. Plant Physiol. 2024, 194, 1834–1852. [Google Scholar] [CrossRef]
- Nongpiur, R.C.; Singla-Pareek, S.L.; Pareek, A. The quest for osmosensors in plants. J. Exp. Bot. 2020, 71, 595–607. [Google Scholar] [CrossRef]
- Gonneau, M.; Desprez, T.; Martin, M.; Doblas, V.G.; Bacete, L.; Miart, F.; Sormani, R.; Hématy, K.; Renou, J.; Landrein, B.; et al. Receptor kinase THESEUS1 is a rapid alkalinization factor 34 receptor in Arabidopsis. Curr. Biol. 2018, 28, 2452–2458. [Google Scholar] [CrossRef]
- Huang, F.; Luo, J.; Ning, T.; Cao, W.; Jin, X.; Zhao, H.; Wang, Y.; Han, S. Cytosolic and nucleosolic calcium signaling in response to osmotic and salt stresses are independent of each other in roots of Arabidopsis seedlings. Front. Plant Sci. 2017, 8, 1648–1660. [Google Scholar] [CrossRef] [PubMed]
- Amor, B.B.; Wirth, S.; Merchan, F.; Laporte, P.; d’Aubenton-Carafa, Y.; Hirsch, J.; Maizel, A.; Mallory, A.; Lucas, A.; Deragon, J.M.; et al. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009, 19, 57–69. [Google Scholar] [CrossRef]
- Qin, T.; Zhao, H.; Cui, P.; Albesher, N.; Xiong, L. A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. 2017, 175, 1321–1336. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dong, J.; Deng, F.; Wang, W.; Cheng, Y.; Song, L.; Hu, M.; Shen, J.; Xu, Q.; Shen, F. The long non-coding RNA lncRNA973 is involved in cotton response to salt stress. BMC Plant Biol. 2019, 19, 459–474. [Google Scholar] [CrossRef]
- Wang, D.; Huang, F.; Yan, P.; Nie, Y.; Chen, L.; Luo, J.; Zhao, H.; Wang, Y.; Han, S. Cytosolic and nucleosolic calcium-regulated molecular networks in response to long-term treatment with abscisic acid and methyl jasmonate in Arabidopsis thaliana. Genes 2022, 13, 524. [Google Scholar] [CrossRef]
- Kirsch, R.; Vurmaz, E.; Schaefer, C.; Eberl, F.; Sporer, T.; Haeger, W.; Pauchet, Y. Plants use identical inhibitors to protect their cell wall pectin against microbes and insects. Ecol. Evol. 2020, 10, 3814–3824. [Google Scholar] [CrossRef] [PubMed]
- Khadka, J.; Pesok, A.; Grafi, G. Plant Histone HTB (H2B) Variants in Regulating Chromatin Structure and Function. Plants 2020, 9, 1435. [Google Scholar] [CrossRef] [PubMed]
- Herman, A.B.; Tsitsipatis, D.; Gorospe, M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol. Cell 2022, 82, 2252–2266. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Behrens, C.E.; Smith, K.E.; Iancu, C.V.; Choe, J.Y.; Dean, J.V. Transport of anthocyanins and other flavonoids by the Arabidopsis ATP-Binding Cassette Transporter AtABCC2. Sci. Rep. 2019, 9, 437–451. [Google Scholar] [CrossRef]
- Péret, B.; Li, G.; Zhao, J.; Band, L.R.; Voß, U.; Postaire, O.; Luu, D.T.; Ines, O.D.; Casimiro, I.; Lucas, M.; et al. Auxin regulates aquaporin function to facilitate lateral root emergence. Nat. Cell Biol. 2012, 14, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S. Cell wall signaling in plant development and defense. Annu. Rev. Plant Biol. 2022, 73, 323–353. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Rose, J.K.; Braam, J.; Fry, S.C.; Nishitani, K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002, 43, 1421–1435. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Plant cell wall loosening by expansins. Annu. Rev. Cell Dev. Biol. 2024, 40, 329–352. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.B.; Cosgrove, D.J. Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol. 2015, 56, 180–194. [Google Scholar] [CrossRef]
- Han, Y.; Li, A.; Li, F.; Zhao, M.; Wang, W. Characterization of a wheat (Triticum aestivum L.) expansin gene, TaEXPB23, involved in the abiotic stress response and phytohormone regulation. Plant Physiol. Biochem. 2012, 54, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Lü, P.; Kang, M.; Jiang, X.; Dai, F.; Gao, J.; Zhang, C. RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta 2013, 237, 1547–1559. [Google Scholar] [CrossRef]
- Sun, X.; Xiong, H.; Jiang, C.; Zhang, D.; Yang, Z.; Huang, Y.; Zhu, W.; Ma, S.; Duan, J.; Wang, X.; et al. Natural variation of DROT1 confers drought adaptation in upland rice. Nat. Commun. 2022, 13, 4265–4281. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Wang, M.; Song, X.; Zhou, G.; Kong, Y. The evolving views of the simplest pectic polysaccharides: Homogalacturonan. Plant Cell Rep. 2022, 41, 2111–2123. [Google Scholar] [CrossRef] [PubMed]
- Konno, H.; Yamasaki, Y.; Sugimoto, M.; Takeda, K. Differential changes in cell wall matrix polysaccharides and glycoside-hydrolyzing enzymes in developing wheat seedlings differing in drought tolerance. J. Plant Physiol. 2008, 165, 745–754. [Google Scholar] [CrossRef] [PubMed]
- An, P.; Li, X.; Zheng, Y.; Matsuura, A.; Abe, J.; Eneji, A.E.; Tanimoto, E.; Inanaga, S. Effects of NaCl on root growth and cell wall composition of two soya bean cultivars with contrasting salt tolerance. J. Agron. Crop. Sci. 2014, 200, 212–218. [Google Scholar] [CrossRef]
- Karlova, R.; Boer, D.; Hayes, S.; Testerink, C. Root plasticity under abiotic stress. Plant Physiol. 2021, 187, 1057–1070. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Z.; Zhang, Z.; Li, Y.; Guo, J.; Liu, L.; Wang, C.; Fan, H.; Wang, B.; Han, G. Root hair development and adaptation to abiotic stress. J. Agric. Food. Chem. 2023, 71, 9573–9598. [Google Scholar] [CrossRef]
- Gandullo, J.; Ahmad, S.; Darwish, E.; Karlova, R.; Testerink, C. Phenotyping tomato root developmental plasticity in response to salinity in soil Rhizotrons. Plant Phenomics 2021, 2021, 2760532–2760545. [Google Scholar] [CrossRef] [PubMed]
- Julkowska, M.M.; Hoefsloot, H.C.; Mol, S.; Feron, R.; de Boer, G.J.; Haring, M.A.; Testerink, C. Capturing Arabidopsis root architecture dynamics withroot-fit reveals diversity in responses to salinity. Plant Physiol. 2014, 166, 1387–1402. [Google Scholar] [CrossRef]
- Galvis, V.C.; Strand, D.D.; Messer, M.; Thiele, W.; Bethmann, S.; Hübner, D.; Uflewski, M.; Kaiser, E.; Siemiatkowska, B.; Morris, B.A.; et al. H+ transport by K+ EXCHANGE ANTIPORTER3 promotes photosynthesis and growth in chloroplast ATP synthase mutants. Plant Physiol. 2020, 182, 2126–2142. [Google Scholar] [CrossRef] [PubMed]
- Shuai, B.; Reynaga-Peña, C.G.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Guo, H.; Dai, X.; Cheng, Y.; Zheng, K.; Wang, X.; Wang, S. An ABA down-regulated bHLH transcription repressor gene, bHLH129 regulates root elongation and ABA response when overexpressed in Arabidopsis. Sci. Rep. 2015, 5, 17587–17597. [Google Scholar] [CrossRef] [PubMed]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Zhu, J.; Du, X.; Cui, X. Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana. Planta 2012, 236, 1227–1237. [Google Scholar] [CrossRef]
- Shukla, V.; Han, J.; Cléard, F.; Lefebvre-Legendre, L.; Gully, K.; Flis, P.; Berhin, A.; Andersen, T.G.; Salt, D.E.; Nawrath, C.; et al. Suberin plasticity to developmental and exogenous cues is regulated by a set of MYB transcription factors. Proc. Natl. Acad. Sci. USA 2021, 118, e2101730118–e2101730128. [Google Scholar] [CrossRef]
- Zhang, L.; Merlin, I.; Pascal, S.; Bert, P.F.; Domergue, F.; Gambetta, G.A. Drought activates MYB41 orthologs and induces suberization of grapevine fine roots. Plant Direct 2020, 4, e00278–e00294. [Google Scholar] [CrossRef] [PubMed]
- Julkowska, M.M.; Koevoets, I.T.; Mol, S.; Hoefsloot, H.; Feron, R.; Tester, M.A.; Keurentjes, J.J.B.; Korte, A.; Haring, M.A.; de Boer, G.J.; et al. Genetic components of root architecture remodeling in response to salt stress. Plant Cell 2017, 29, 3198–3213. [Google Scholar] [CrossRef] [PubMed]
- Sorin, C.; Negroni, L.; Balliau, T.; Corti, H.; Jacquemot, M.P.; Davanture, M.; Sandberg, G.; Zivy, M.; Bellini, C. Proteomic analysis of different mutant genotypes of arabidopsis led to the identification of 11 proteins correlating with adventitious root development. Plant Physiol. 2006, 140, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Gibson, S.W.; Todd, C.D. Arabidopsis AIR12 influences root development. Physiol. Mol. Biol. Plants 2015, 21, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Korbei, B.; Moulinier-Anzola, J.; De-Araujo, L.; Lucyshyn, D.; Retzer, K.; Khan, M.A.; Luschnig, C. Arabidopsis TOL proteins act as gatekeepers for vacuolar sorting of PIN2 plasma membrane protein. Curr. Biol. 2013, 23, 2500–2505. [Google Scholar] [CrossRef] [PubMed]
- Walters, L.A.; Escobar, M.A. The AtGRXS3/4/5/7/8 glutaredoxin gene cluster on Arabidopsis thaliana chromosome 4 is coordinately regulated by nitrate and appears to control primary root growth. Plant Signal. Behav. 2016, 11, e1171450–e1171452. [Google Scholar] [CrossRef] [PubMed]
- Guillou, M.-C.; Vergne, E.; Aligon, S.; Pelletier, S.; Simonneau, F.; Rolland, A.; Chabout, S.; Mouille, G.; Gully, K.; Grappin, P.; et al. The peptide SCOOP12 acts on reactive oxygen species homeostasis to modulate cell division and elongation in Arabidopsis primary root. J. Exp. Bot. 2022, 73, 6115–6132. [Google Scholar] [CrossRef] [PubMed]
- Porco, S.; Larrieu, A.; Du, Y.; Gaudinier, A.; Goh, T.; Swarup, K.; Swarup, R.; Kuempers, B.; Bishopp, A.; Lavenus, J.; et al. Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulating auxin influx carrier LAX3. Development 2016, 143, 3340–3349. [Google Scholar] [CrossRef]
- Kaszler, N.; Benkő, P.; Bernula, D.; Szepesi, Á.; Fehér, A.; Gémes, K. Polyamine metabolism is involved in the direct regeneration of shoots from Arabidopsis lateral root primordia. Plants 2021, 10, 305. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Chen, X.; Zhang, S.; Zhu, J.; Tang, B.; Wang, A.; Dong, L.; Zhang, Z.; Yu, C.; Sun, Y.; et al. The genome sequence archive family: Toward explosive data growth and diverse data types. Genom. Proteom. Bioinform. 2021, 19, 578–583. [Google Scholar] [CrossRef] [PubMed]
- CNCB-NGDC Members and Partners. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids. Res. 2022, 50, D27–D38. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Zheng, K.; Long, W.; Zhao, L.; Li, W.; Xue, X.; Han, S. Cytosolic and Nucleosolic Calcium-Regulated Long Non-Coding RNAs and Their Target Protein-Coding Genes in Response to Hyperosmolarity and Salt Stresses in Arabidopsis thaliana. Int. J. Mol. Sci. 2025, 26, 2086. https://doi.org/10.3390/ijms26052086
Wang D, Zheng K, Long W, Zhao L, Li W, Xue X, Han S. Cytosolic and Nucleosolic Calcium-Regulated Long Non-Coding RNAs and Their Target Protein-Coding Genes in Response to Hyperosmolarity and Salt Stresses in Arabidopsis thaliana. International Journal of Molecular Sciences. 2025; 26(5):2086. https://doi.org/10.3390/ijms26052086
Chicago/Turabian StyleWang, Doudou, Kaifeng Zheng, Wenfen Long, Liang Zhao, Wanjie Li, Xiuhua Xue, and Shengcheng Han. 2025. "Cytosolic and Nucleosolic Calcium-Regulated Long Non-Coding RNAs and Their Target Protein-Coding Genes in Response to Hyperosmolarity and Salt Stresses in Arabidopsis thaliana" International Journal of Molecular Sciences 26, no. 5: 2086. https://doi.org/10.3390/ijms26052086
APA StyleWang, D., Zheng, K., Long, W., Zhao, L., Li, W., Xue, X., & Han, S. (2025). Cytosolic and Nucleosolic Calcium-Regulated Long Non-Coding RNAs and Their Target Protein-Coding Genes in Response to Hyperosmolarity and Salt Stresses in Arabidopsis thaliana. International Journal of Molecular Sciences, 26(5), 2086. https://doi.org/10.3390/ijms26052086






