PON-P3: Accurate Prediction of Pathogenicity of Amino Acid Substitutions
Abstract
1. Introduction
2. Results
2.1. Choice of the ML Algorithm
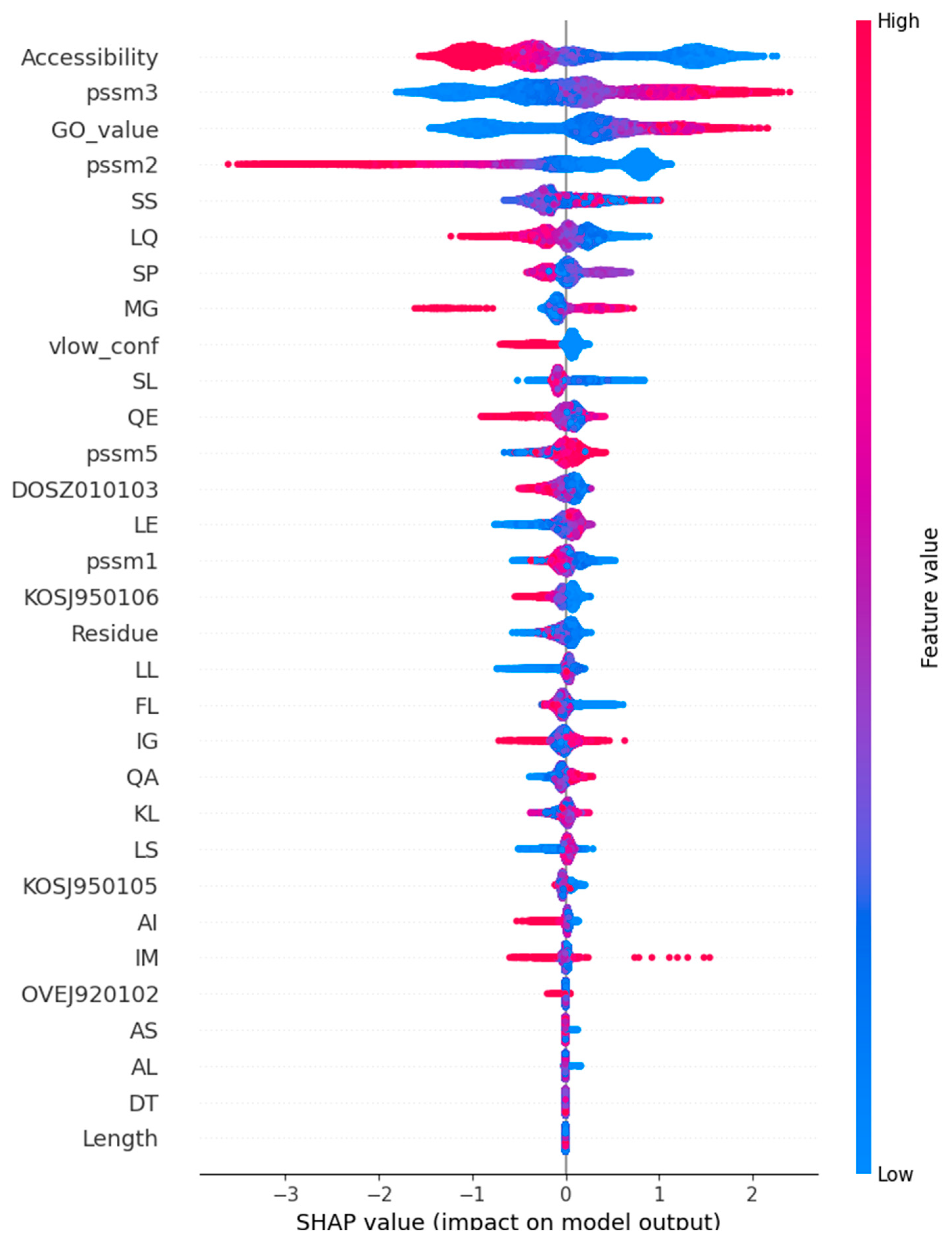
2.2. Feature Selection and Method Training
2.3. Benchmarking PON-P3 Performance
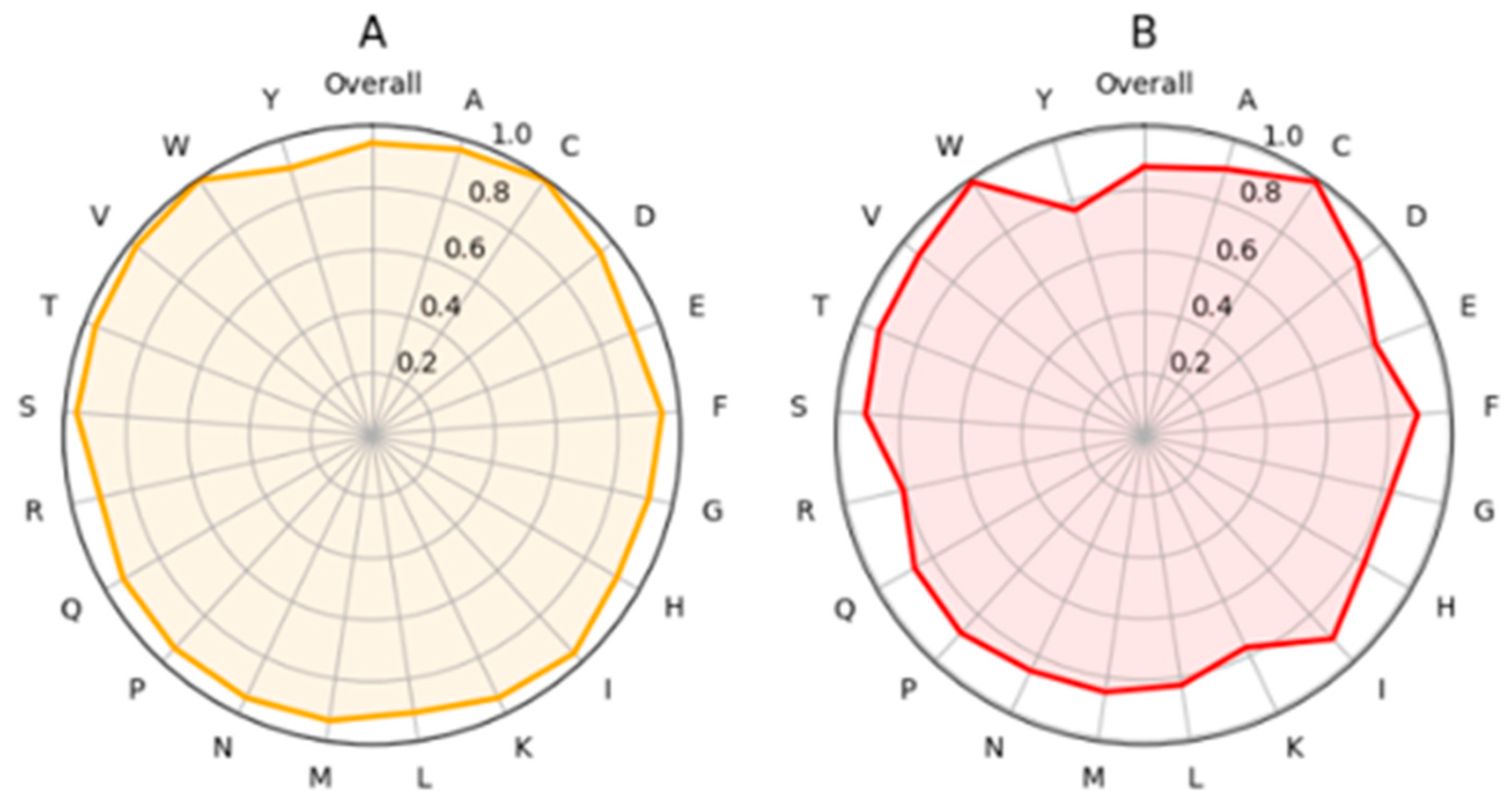
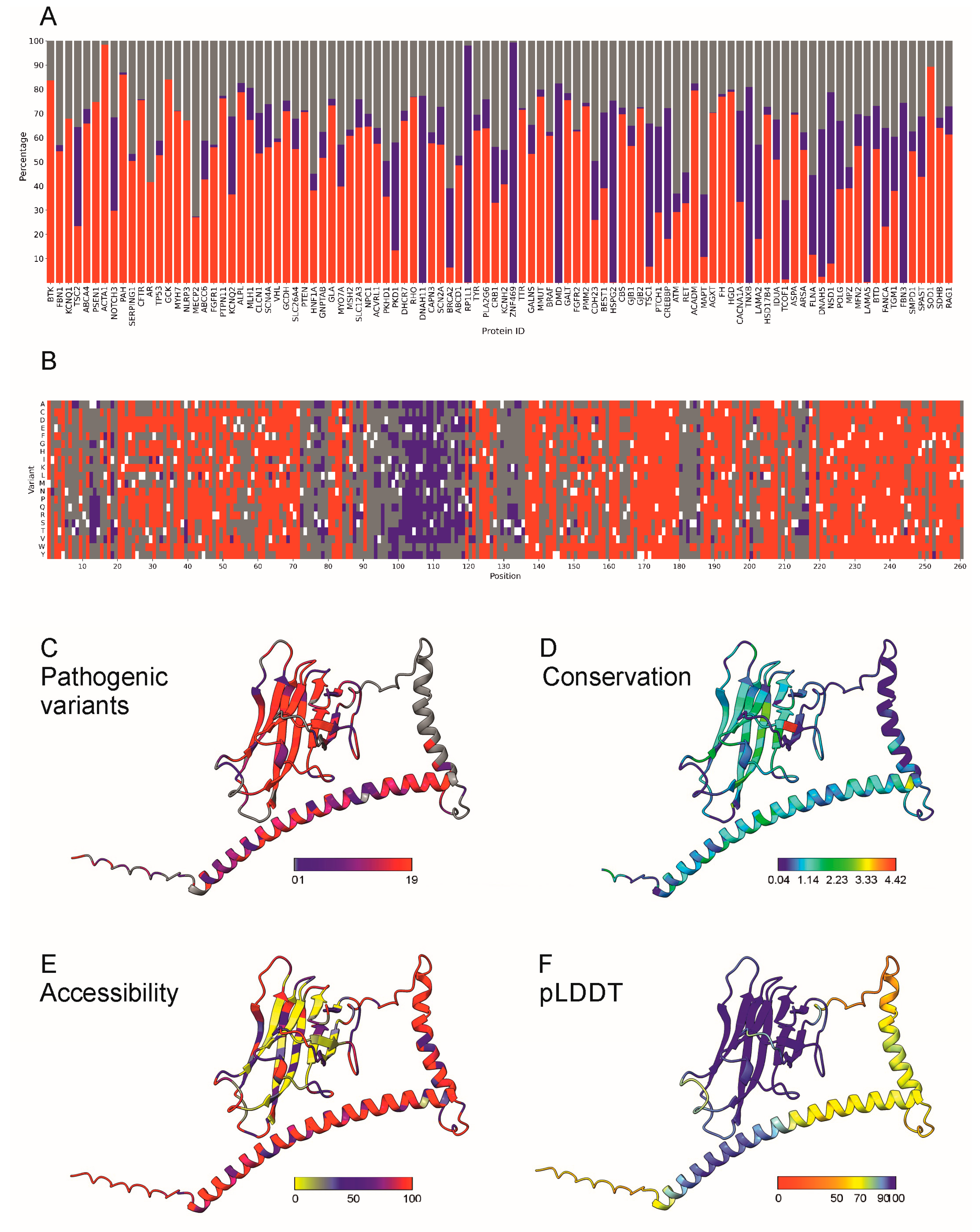
2.4. Examples of Applications of PON-P3
2.5. PON-P3 Server and Precalculated Results
3. Discussion
4. Materials and Methods
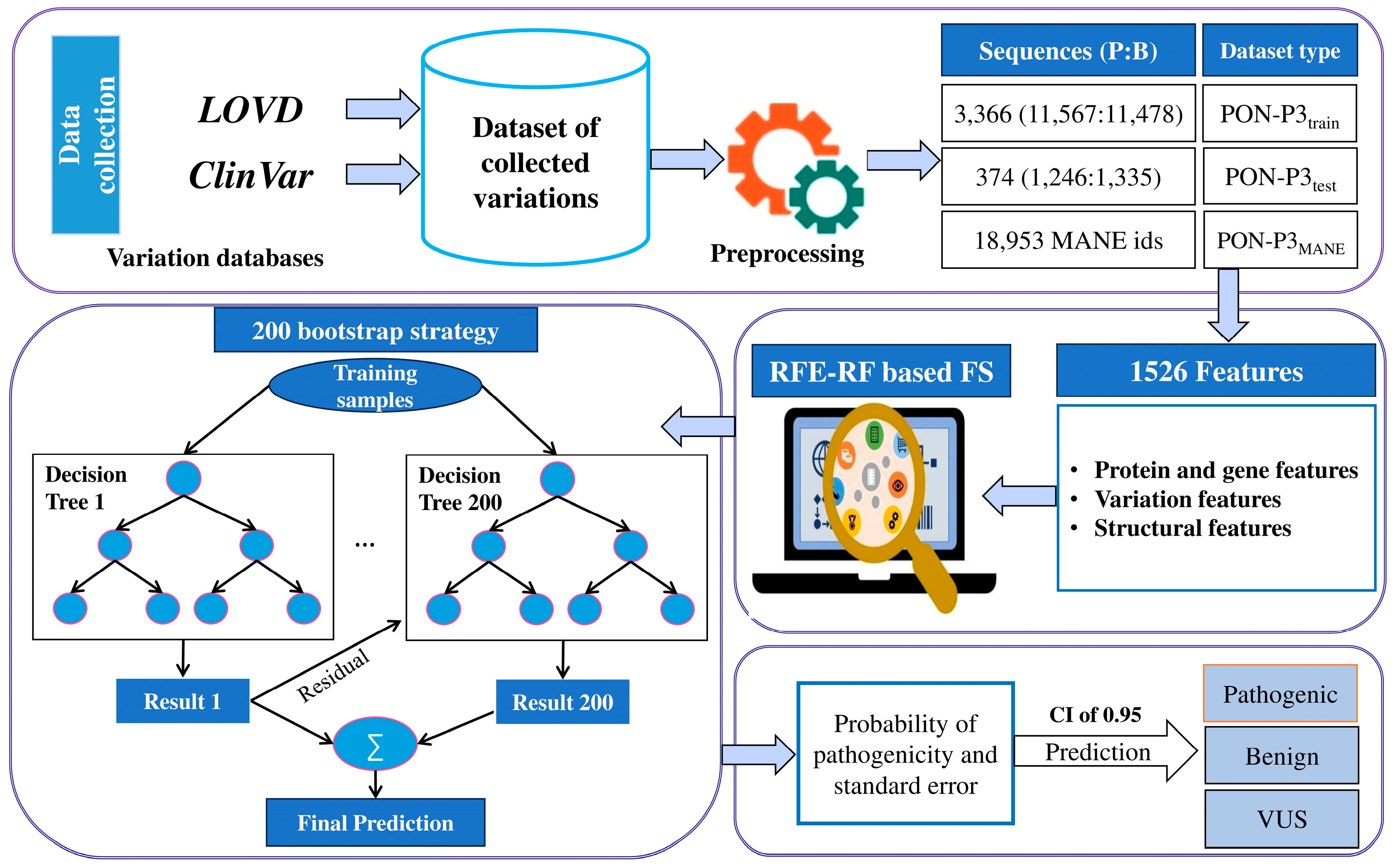
4.1. Datasets
4.2. Features
4.2.1. Protein and Gene Features
4.2.2. Variation Features
4.2.3. Structural Features
4.3. Functional Annotations
4.4. Feature Selection
4.5. Performance Assessment
4.6. Performance Evaluation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.J.; Menon, A.S.; Hu, Z.; Brenner, S.E. Variant Impact Predictor database (VIPdb), version 2: Trends from three decades of genetic variant impact predictors. Hum. Genom. 2024, 18, 90. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Chitipiralla, S.; Brown, G.R.; Chen, C.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; Kaur, K.; Liu, C.; et al. ClinVar: Improvements to accessing data. Nucleic Acids Res. 2020, 48, D835–D844. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kabir, M.; Ahmed, S.; Vihinen, M. There will always be variants of uncertain significance. Analysis of VUSes. NAR Genom. Bioinf. 2024, 6, lqae154. [Google Scholar]
- Vihinen, M. Poikilosis—Pervasive biological variation. F1000Research 2020, 9, 602. [Google Scholar] [CrossRef]
- Hu, J.; Ng, P.C. SIFT Indel: Predictions for the functional effects of amino acid insertions/deletions in proteins. PLoS ONE 2013, 8, e77940. [Google Scholar] [CrossRef]
- Shihab, H.A.; Gough, J.; Cooper, D.N.; Stenson, P.D.; Barker, G.L.; Edwards, K.J.; Day, I.N.; Gaunt, T.R. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 2013, 34, 57–65. [Google Scholar] [CrossRef]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef] [PubMed]
- Frazer, J.; Notin, P.; Dias, M.; Gomez, A.; Min, J.K.; Brock, K.; Gal, Y.; Marks, D.S. Disease variant prediction with deep generative models of evolutionary data. Nature 2021, 599, 91–95. [Google Scholar] [CrossRef]
- Brandes, N.; Goldman, G.; Wang, C.H.; Ye, C.J.; Ntranos, V. Genome-wide prediction of disease variant effects with a deep protein language model. Nat. Genet. 2023, 55, 1512–1522. [Google Scholar] [CrossRef]
- Niroula, A.; Urolagin, S.; Vihinen, M. PON-P2: Prediction method for fast and reliable identification of harmful variants. PLoS ONE 2015, 10, e0117380. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shao, A.; Vihinen, M. PON-All, amino acid substitution tolerance predictor for all organisms. Front. Mol. Biosci. 2022, 9, 867572. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Carter, H.; Douville, C.; Stenson, P.D.; Cooper, D.N.; Karchin, R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genom. 2013, 14 (Suppl. S3), S3. [Google Scholar] [CrossRef]
- Cheng, J.; Novati, G.; Pan, J.; Bycroft, C.; Žemgulytė, A.; Applebaum, T.; Pritzel, A.; Wong, L.H.; Zielinski, M.; Sargeant, T.; et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 2023, 381, eadg7492. [Google Scholar] [CrossRef]
- Dong, C.; Wei, P.; Jian, X.; Gibbs, R.; Boerwinkle, E.; Wang, K.; Liu, X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015, 24, 2125–2137. [Google Scholar] [CrossRef]
- Li, C.; Zhi, D.; Wang, K.; Liu, X. MetaRNN: Differentiating rare pathogenic and rare benign missense SNVs and InDels using deep learning. Genome Med. 2022, 14, 115. [Google Scholar] [CrossRef] [PubMed]
- Alirezaie, N.; Kernohan, K.D.; Hartley, T.; Majewski, J.; Hocking, T.D. ClinPred: Prediction Tool to Identify Disease-Relevant Nonsynonymous Single-Nucleotide Variants. Am. J. Hum. Genet. 2018, 103, 474–483. [Google Scholar] [CrossRef]
- Feng, B.J. PERCH: A Unified Framework for Disease Gene Prioritization. Hum. Mutat. 2017, 38, 243–251. [Google Scholar] [CrossRef]
- Olatubosun, A.; Väliaho, J.; Härkönen, J.; Thusberg, J.; Vihinen, M. PON-P: Integrated predictor for pathogenicity of missense variants. Hum. Mutat. 2012, 33, 1166–1174. [Google Scholar] [CrossRef]
- Niroula, A.; Vihinen, M. Predicting severity of disease-causing variants. Hum. Mutat. 2017, 38, 357–364. [Google Scholar] [CrossRef]
- Yang, Y.; Urolagin, S.; Niroula, A.; Ding, X.; Shen, B.; Vihinen, M. PON-tstab: Protein variant stability predictor. Importance of training data quality. Int. J. Mol. Sci. 2018, 19, 1009. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chong, Z.; Vihinen, M. PON-Fold: Prediction of Substitutions Affecting Protein Folding Rate. Int. J. Mol. Sci. 2023, 24, 13023. [Google Scholar] [CrossRef] [PubMed]
- Morales, J.; Pujar, S.; Loveland, J.E.; Astashyn, A.; Bennett, R.; Berry, A.; Cox, E.; Davidson, C.; Ermolaeva, O.; Farrell, C.M.; et al. A joint NCBI and EMBL-EBI transcript set for clinical genomics and research. Nature 2022, 604, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. LightGBM: A Highly Efficient Gradient Boosting Decision Tree; Neural Information Processing Systems: La Jolla, CA, USA, 2017. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the KDD’16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Fransisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, B.; Tan, G.; Vihinen, M.; Shen, B. Structure-based prediction of the effects of a missense variant on protein stability. Amino Acids 2013, 44, 847–855. [Google Scholar] [CrossRef]
- Batista, J.; Vikić-Topić, D.; Lučić, B. The difference between the accuracy of real and the corresponding random model is a useful parameter for validation of two-state classification model quality. Croat. Chem. Acta 2016, 89, 527–534. [Google Scholar] [CrossRef]
- Dosztányi, Z.; Torda, A.E. Amino acid similarity matrices based on force fields. Bioinformatics 2001, 17, 686–699. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Koshi, J.M.; Goldstein, R.A. Context-dependent optimal substitution matrices. Protein Eng. 1995, 8, 641–645. [Google Scholar] [CrossRef]
- Overington, J.; Donnelly, D.; Johnson, M.S.; Sali, A.; Blundell, T.L. Environment-specific amino acid substitution tables: Tertiary templates and prediction of protein folds. Protein Sci. 1992, 1, 216–226. [Google Scholar] [CrossRef]
- Liu, X.; Wu, C.; Li, C.; Boerwinkle, E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum. Mutat. 2016, 37, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Rogers, M.F.; Shihab, H.A.; Mort, M.; Cooper, D.N.; Gaunt, T.R.; Campbell, C. FATHMM-XF: Accurate prediction of pathogenic point mutations via extended features. Bioinformatics 2018, 34, 511–513. [Google Scholar] [CrossRef] [PubMed]
- Shihab, H.A.; Rogers, M.F.; Gough, J.; Mort, M.; Cooper, D.N.; Day, I.N.; Gaunt, T.R.; Campbell, C. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics 2015, 31, 1536–1543. [Google Scholar] [CrossRef]
- Malhis, N.; Jacobson, M.; Jones, S.J.M.; Gsponer, J. LIST-S2: Taxonomy based sorting of deleterious missense mutations across species. Nucleic Acids Res. 2020, 48, W154–W161. [Google Scholar] [CrossRef]
- Reva, B.; Antipin, Y.; Sander, C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011, 39, e118. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res. 2001, 11, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Adusumalli, S.; Leng, S.N.; Sikic, M.; Ng, P.C. SIFT missense predictions for genomes. Nat. Protoc. 2016, 11, 1–9. [Google Scholar] [CrossRef]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef]
- Raimondi, D.; Tanyalcin, I.; Ferté, J.; Gazzo, A.; Orlando, G.; Lenaerts, T.; Rooman, M.; Vranken, W. DEOGEN2: Prediction and interactive visualization of single amino acid variant deleteriousness in human proteins. Nucleic Acids Res. 2017, 45, W201–W206. [Google Scholar] [CrossRef]
- Jagadeesh, K.A.; Wenger, A.M.; Berger, M.J.; Guturu, H.; Stenson, P.D.; Cooper, D.N.; Bernstein, J.A.; Bejerano, G. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 2016, 48, 1581–1586. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, N.M.; Rothstein, J.H.; Pejaver, V.; Middha, S.; McDonnell, S.K.; Baheti, S.; Musolf, A.; Li, Q.; Holzinger, E.; Karyadi, D.; et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016, 99, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Vihinen, M. How to evaluate performance of prediction methods? Measures and their interpretation in variation effect analysis. BMC Genom. 2012, 13 (Suppl. S4), S2. [Google Scholar] [CrossRef] [PubMed]
- Vihinen, M. Guidelines for reporting and using prediction tools for genetic variation analysis. Hum. Mutat. 2013, 34, 275–282. [Google Scholar] [CrossRef]
- Schaafsma, G.C.P.; Vihinen, M. Large differences in proportions of harmful and benign amino acid substitutions between proteins and diseases. Hum. Mutat. 2017, 38, 839–848. [Google Scholar] [CrossRef]
- Ali, S.; Abuhmed, T.; El-Sappagh, S.; Muhammad, K.; Alonso-Moral, J.M.; Confalonieri, R.; Del Sar, J.; Diaz-Rodriguez, N.; Herrera, F. Explainable Artificial Intelligence (XAI): What we know and what is left to attain trustworthy artificial intelligence. Inf. Fusion 2023, 99, 101805. [Google Scholar] [CrossRef]
- Karim, M.R.; Islam, T.; Shajalal, M.; Beyan, O.; Lange, C.; Cochez, M.; Rebholz-Schuhmann, D.; Decker, S. Explainable AI for Bioinformatics: Methods, Tools and Applications. Brief. Bioinform. 2023, 24, bbad236. [Google Scholar] [CrossRef]
- Allen, B. The Promise of Explainable AI in Digital Health for Precision Medicine: A Systematic Review. J. Pers. Med. 2024, 14, 277. [Google Scholar] [CrossRef]
- Arbelaez Ossa, L.; Starke, G.; Lorenzini, G.; Vogt, J.E.; Shaw, D.M.; Elger, B.S. Re-focusing explainability in medicine. Digit. Health 2022, 8, 20552076221074488. [Google Scholar] [CrossRef]
- Banday, A.Z.; Nisar, R.; Patra, P.K.; Kaur, A.; Sadanand, R.; Chaudhry, C.; Bukhari, S.T.A.; Banday, S.Z.; Bhattarai, D.; Notarangelo, L.D. Clinical and Immunological Features, Genetic Variants, and Outcomes of Patients with CD40 Deficiency. J. Clin. Immunol. 2023, 44, 17. [Google Scholar] [CrossRef]
- Kornrumpf, K.; Kurz, N.S.; Drofenik, K.; Krauß, L.; Schneider, C.; Koch, R.; Beißbarth, T.; Dönitz, J. SeqCAT: Sequence Conversion and Analysis Toolbox. Nucleic Acids Res. 2024, 52, W116–W120. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef] [PubMed]
- Fokkema, I.; Kroon, M.; López Hernández, J.A.; Asscheman, D.; Lugtenburg, I.; Hoogenboom, J.; den Dunnen, J.T. The LOVD3 platform: Efficient genome-wide sharing of genetic variants. Eur. J. Hum. Genet. 2021, 29, 1796–1803. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Nair, P.S.; Vihinen, M. VariBench: A benchmark database for variations. Hum. Mutat. 2013, 34, 42–49. [Google Scholar] [CrossRef]
- Singh, A.K.; Amar, I.; Ramadasan, H.; Kappagantula, K.S.; Chavali, S. Proteins with amino acid repeats constitute a rapidly evolvable and human-specific essentialome. Cell Rep. 2023, 42, 112811. [Google Scholar] [CrossRef]
- Narasimhan, V.M.; Hunt, K.A.; Mason, D.; Baker, C.L.; Karczewski, K.J.; Barnes, M.R.; Barnett, A.H.; Bates, C.; Bellary, S.; Bockett, N.A.; et al. Health and population effects of rare gene knockouts in adult humans with related parents. Science 2016, 352, 474–477. [Google Scholar] [CrossRef] [PubMed]
- Saleheen, D.; Natarajan, P.; Armean, I.M.; Zhao, W.; Rasheed, A.; Khetarpal, S.A.; Won, H.H.; Karczewski, K.J.; O’Donnell-Luria, A.H.; Samocha, K.E.; et al. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature 2017, 544, 235–239. [Google Scholar] [CrossRef]
- Sulem, P.; Helgason, H.; Oddson, A.; Stefansson, H.; Gudjonsson, S.A.; Zink, F.; Hjartarson, E.; Sigurdsson, G.T.; Jonasdottir, A.; Jonasdottir, A.; et al. Identification of a large set of rare complete human knockouts. Nat. Genet. 2015, 47, 448–452. [Google Scholar] [CrossRef]
- Blake, J.A.; Baldarelli, R.; Kadin, J.A.; Richardson, J.E.; Smith, C.L.; Bult, C.J. Mouse Genome Database (MGD): Knowledgebase for mouse-human comparative biology. Nucleic Acids Res. 2021, 49, D981–D987. [Google Scholar] [CrossRef]
- Solomon, B.D.; Nguyen, A.D.; Bear, K.A.; Wolfsberg, T.G. Clinical genomic database. Proc. Natl. Acad. Sci. USA 2013, 110, 9851–9855. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJ. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Warwick Vesztrocy, A.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef]
- Capra, J.A.; Williams, A.G.; Pollard, K.S. ProteinHistorian: Tools for the comparative analysis of eukaryote protein origin. PLoS Comput. Biol. 2012, 8, e1002567. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, S.; Krishnamoorthy, B.; Ye, P. Neighborhood properties are important determinants of temperature sensitive mutations. PLoS ONE 2011, 6, e28507. [Google Scholar] [CrossRef]
- Kawashima, S.; Kanehisa, M. AAindex: Amino acid index database. Nucleic Acids Res. 2000, 28, 374. [Google Scholar] [CrossRef]
- Ruiz-Blanco, Y.B.; Paz, W.; Green, J.; Marrero-Ponce, Y. ProtDCal: A program to compute general-purpose-numerical descriptors for sequences and 3D-structures of proteins. BMC Bioinform. 2015, 16, 162. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Jorda, J.; Kajava, A.V. T-REKS: Identification of Tandem REpeats in sequences with a K-meanS based algorithm. Bioinformatics 2009, 25, 2632–2638. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Varabyou, A.; Sommer, M.J.; Erdogdu, B.; Shinder, I.; Minkin, I.; Chao, K.H.; Park, S.; Heinz, J.; Pockrandt, C.; Shumate, A.; et al. CHESS 3: An improved, comprehensive catalog of human genes and transcripts based on large-scale expression data, phylogenetic analysis, and protein structure. Genome Biol. 2023, 24, 249. [Google Scholar] [CrossRef]
- Heinig, M.; Frishman, D. STRIDE: A web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004, 32, W500–W502. [Google Scholar] [CrossRef]
- Mitternacht, S. FreeSASA: An open source C library for solvent accessible surface area calculations. F1000Research 2016, 5, 189. [Google Scholar] [CrossRef]
- Dobson, L.; Remenyi, I.; Tusnady, G.E. The human transmembrane proteome. Biol. Direct 2015, 10, 31. [Google Scholar] [CrossRef]
- Aspromonte, M.C.; Nugnes, M.V.; Quaglia, F.; Bouharoua, A.; Tosatto, S.C.E.; Piovesan, D. DisProt in 2024: Improving function annotation of intrinsically disordered proteins. Nucleic Acids Res. 2024, 52, D434–D441. [Google Scholar] [CrossRef]
- Darst, B.F.; Malecki, K.C.; Engelman, C.D. Using recursive feature elimination in random forest to account for correlated variables in high dimensional data. BMC Genet. 2018, 19, 65. [Google Scholar] [CrossRef]
| Training and CV | Blind Test Set | Total | ||||
|---|---|---|---|---|---|---|
| Variations | Proteins | Variations | Proteins | Variations | Proteins | |
| Pathogenic | 11,567 | 3366 | 1246 | 374 | 12,813 | 3740 |
| Benign | 11,478 | 1335 | 12,813 | |||
| Total | 23,045 | 2581 | 25,626 | |||
| Parameter | LightGBM a | RF | SVM | XGBoost |
|---|---|---|---|---|
| Accuracy | 0.933 (0.924) | 0.892 (0.889) | 0.926 (0.925) | 0.933 (0.925) |
| ΔAccuracy | 0.424 | 0.389 | 0.425 | 0.425 |
| MCC | 0.864 (0.854) | 0.785 (0.781) | 0.852 (0.851) | 0.864 (0.855) |
| PPV | 0.966 (0.975) | 0.917 (0.928) | 0.932 (0.939) | 0.966 (0.975) |
| NPV | 0.913 (0.883) | 0.874 (0.857) | 0.921 (0.913) | 0.913 (0.885) |
| Sensitivity | 0.871 (0.871) | 0.844 (0.844) | 0.91 (0.91) | 0.873 (0.873) |
| Specificity | 0.978 (0.978) | 0.934 (0.934) | 0.94 (0.94) | 0.977 (0.977) |
| OPM | 0.81 (0.81) | 0.711 (0.711) | 0.794 (0.794) | 0.81 (0.81) |
| Pathogenic as unknown | 4996 | 3971 | 4140 | 4871 |
| Neutral as unknown | 2.388 | 2631 | 3191 | 2362 |
| TP | 5722 (7916) | 6410 (7466) | 6758 (7541) | 5845 (7957) |
| TN | 8889 | 8264 | 7793 | 8910 |
| FP | 201 | 583 | 494 | 206 |
| FN | 849 (1174) | 1186 (1381) | 669 (746) | 851 (1159) |
| Coverage | 0.68 | 0.714 | 0.682 (0.693) | 0.686 |
| Parameter | All Features | woGO | wGO |
|---|---|---|---|
| Accuracy | 0.92 (0.92) a | 0.918 (0.913) | 0.945 (0.944) |
| ΔAccuracy | 0.420 | 0.413 | 0.444 |
| MCC | 0.84 (0.84) | 0.833 (0.828) | 0.889 (0.888) |
| PPV | 0.919 (0.926) | 0.937 (0.95) | 0.947 (0.953) |
| NPV | 0.922 (0.914) | 0.905 (0.881) | 0.942 (0.935) |
| Sensitivity | 0.913 (0.913) | 0.871 (0.871) | 0.934 (0.934) |
| Specificity | 0.927 (0.927) | 0.955 (0.955) | 0.954 (0.954) |
| OPM | 0.779 (0.779) | 0.771 (0.771) | 0.843 (0.843) |
| Pathogenic as unknown | 295 | 458 | 340 |
| Neutral as unknown | 281 | 322 | 308 |
| TP | 868 (962) | 686 (882) | 846 (959) |
| TN | 977 | 967 | 980 |
| FP | 77 | 46 | 47 |
| FN | 83 (92) | 102 (131) | 60 (68) |
| Coverage | 0.777 | 0.698 | 0.749 |
| Predictor | TP | TN | FP | FN | PPV | NPV | Sensitivity | Specificity | ACC | MCC | OPM | Coverage |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Evolutionary data/sequence information-based predictors | ||||||||||||
| ESM1b | 945 (1082) | 970 | 240 | 112 (128) | 0.797 (0.818) | 0.896 (0.883) | 0.894 (0.894) | 0.802 (0.802) | 0.845 (0.848) | 0.695 (0.699) | 0.607 (0.612) | 0.966 |
| EVE | 623 (439) | 549 | 48 | 225 (158) | 0.928 (0.901) | 0.709 (0.777) | 0.735 (0.735) | 0.92 (0.92) | 0.811 (0.827) | 0.646 (0.666) | 0.553 (0.576) | 1.0 |
| FATHMM | 851 (965) | 870 | 284 | 167 (189) | 0.75 (0.773) | 0.839 (0.822) | 0.836 (0.836) | 0.754 (0.754) | 0.792 (0.795) | 0.589 (0.592) | 0.501 (0.504) | 0.925 |
| FATHMM-MKL | 1059 (1242) | 546 | 717 | 18 (21) | 0.596 (0.634) | 0.968 (0.963) | 0.983 (0.983) | 0.432 (0.432) | 0.686 (0.708) | 0.484 (0.498) | 0.395 (0.412) | 0.997 |
| FATHMM-XF | 967 (1108) | 819 | 355 | 58 (66) | 0.731 (0.757) | 0.934 (0.925) | 0.943 (0.944) | 0.698 (0.698) | 0.812 (0.821) | 0.653 (0.662) | 0.56 (0.57) | 0.937 |
| LIST-S2 | 841 (936) | 647 | 378 | 80 (89) | 0.69 (0.712) | 0.89 (0.879) | 0.913 (0.913) | 0.631 (0.631) | 0.765 (0.772) | 0.562 (0.567) | 0.471 (0.477) | 1.0 |
| MutationAssessor | 846 (966) | 827 | 278 | 122 (139) | 0.753 (0.777) | 0.871 (0.856) | 0.874 (0.874) | 0.748 (0.748) | 0.807 (0.811) | 0.623 (0.628) | 0.533 (0.538) | 0.883 |
| PROVEAN | 842 (950) | 937 | 210 | 175 (197) | 0.8 (0.819) | 0.843 (0.826) | 0.828 (0.828) | 0.817 (0.817) | 0.822 (0.823) | 0.644 (0.645) | 0.555 (0.557) | 0.922 |
| SIFT | 947 (1067) | 769 | 377 | 70 (79) | 0.715 (0.739) | 0.917 (0.907) | 0.931 (0.931) | 0.671 (0.671) | 0.793 (0.801) | 0.617 (0.624) | 0.523 (0.532) | 0.922 |
| SIFT 4G | 918 (1028) | 890 | 268 | 116 (130) | 0.774 (0.793) | 0.885 (0.873) | 0.888 (0.888) | 0.769 (0.769) | 0.825 (0.828) | 0.658 (0.661) | 0.568 (0.572) | 0.934 |
| Multiple features utilizing predictors | ||||||||||||
| CADD | 1069 (1261) | 443 | 827 | 8 (9) | 0.564 (0.604) | 0.982 (0.98) | 0.993 (0.993) | 0.349 (0.349) | 0.644 (0.671) | 0.432 (0.447) | 0.353 (0.371) | 1.0 |
| DEOGEN2 | 796 (935) | 993 | 136 | 165 (194) | 0.854 (0.873) | 0.858 (0.837) | 0.828 (0.828) | 0.88 (0.88) | 0.856 (0.854) | 0.71 (0.709) | 0.625 (0.624) | 0.89 |
| M-CAP | 1064 (457) | 219 | 242 | 10 (4) | 0.815 (0.654) | 0.956 (0.982) | 0.991 (0.991) | 0.475 (0.475) | 0.836 (0.733) | 0.599 (0.545) | 0.531 (0.451) | 1.0 |
| PolyPhen2 Hvar | 119 (522) | 884 | 155 | 118 (517) | 0.434 (0.771) | 0.882 (0.631) | 0.502 (0.502) | 0.851 (0.851) | 0.786 (0.677) | 0.334 (0.377) | 0.323 (0.324) | 0.544 |
| PolyPhen2 Hdiv | 865 (910) | 749 | 231 | 67 (70) | 0.789 (0.798) | 0.918 (0.915) | 0.928 (0.929) | 0.764 (0.764) | 0.844 (0.846) | 0.700 (0.702) | 0.612 (0.615) | 0.933 |
| PON-P3 woGO | 686 (882) | 967 | 46 | 102 (131) | 0.937 (0.95) | 0.905 (0.881) | 0.871 (0.871) | 0.955 (0.955) | 0.918 (0.913) | 0.833 (0.828) | 0.771 (0.771) | 0.698 |
| PON-P3 wGO | 840 (961) | 981 | 50 | 61 (70) | 0.944 (0.951) | 0.941 (0.933) | 0.932 (0.932) | 0.952 (0.952) | 0.943 (0.942) | 0.885 (0.884) | 0.837 (0.837) | 0.749 |
| VEST4 | 997 (1140) | 947 | 232 | 34 (39) | 0.811 (0.831) | 0.965 (0.96) | 0.967 (0.967) | 0.803 (0.803) | 0.88 (0.885) | 0.773 (0.781) | 0.694 (0.704) | 1.0 |
| Structural data-based predictor | ||||||||||||
| Alpha Missense | 827 (1005) | 1,071 | 72 | 114 (138) | 0.92 (0.933) | 0.904 (0.886) | 0.879 (0.879) | 0.937 (0.937) | 0.911 (0.908) | 0.82 (0.818) | 0.754 (0.75) | 0.888 |
| Metapredictors | ||||||||||||
| BayesDel | 1030 (1208) | 1,121 | 142 | 47 (55) | 0.879 (0.895) | 0.96 (0.953) | 0.956 (0.956) | 0.888 (0.888) | 0.919 (0.922) | 0.841 (0.846) | 0.78 (0.786) | 0.997 |
| ClinPred | 1018 (1185) | 1,244 | 8 | 58 (67) | 0.992 (0.993) | 0.955 (0.949) | 0.946 (0.946) | 0.994 (0.994) | 0.972 (0.97) | 0.944 (0.941) | 0.918 (0.914) | 0.992 |
| MetaLR | 965 (1120) | 1,092 | 157 | 111 (129) | 0.86 (0.877) | 0.908 (0.894) | 0.897 (0.897) | 0.874 (0.874) | 0.885 (0.886) | 0.769 (0.771) | 0.693 (0.694) | 0.991 |
| MetaRNN | 643 (702) | 734 | 4 | 33 (36) | 0.994 (0.994) | 0.957 (0.953) | 0.951 (0.951) | 0.995 (0.995) | 0.974 (0.973) | 0.948 (0.947) | 0.924 (0.922) | 1.0 |
| MetaSVM | 970 (1126) | 1,129 | 120 | 106 (123) | 0.89 (0.904) | 0.914 (0.902) | 0.901 (0.902) | 0.904 (0.904) | 0.903 (0.903) | 0.805 (0.805) | 0.735 (0.736) | 0.991 |
| REVEL | 825 (1128) | 1,001 | 160 | 24 (33) | 0.838 (0.876) | 0.977 (0.968) | 0.972 (0.972) | 0.862 (0.862) | 0.908 (0.917) | 0.824 (0.839) | 0.757 (0.776) | 1.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kabir, M.; Ahmed, S.; Zhang, H.; Rodríguez-Rodríguez, I.; Najibi, S.M.; Vihinen, M. PON-P3: Accurate Prediction of Pathogenicity of Amino Acid Substitutions. Int. J. Mol. Sci. 2025, 26, 2004. https://doi.org/10.3390/ijms26052004
Kabir M, Ahmed S, Zhang H, Rodríguez-Rodríguez I, Najibi SM, Vihinen M. PON-P3: Accurate Prediction of Pathogenicity of Amino Acid Substitutions. International Journal of Molecular Sciences. 2025; 26(5):2004. https://doi.org/10.3390/ijms26052004
Chicago/Turabian StyleKabir, Muhammad, Saeed Ahmed, Haoyang Zhang, Ignacio Rodríguez-Rodríguez, Seyed Morteza Najibi, and Mauno Vihinen. 2025. "PON-P3: Accurate Prediction of Pathogenicity of Amino Acid Substitutions" International Journal of Molecular Sciences 26, no. 5: 2004. https://doi.org/10.3390/ijms26052004
APA StyleKabir, M., Ahmed, S., Zhang, H., Rodríguez-Rodríguez, I., Najibi, S. M., & Vihinen, M. (2025). PON-P3: Accurate Prediction of Pathogenicity of Amino Acid Substitutions. International Journal of Molecular Sciences, 26(5), 2004. https://doi.org/10.3390/ijms26052004









