Circulating Tumour DNA for Ovarian Cancer Diagnosis and Treatment Monitoring: What Perspectives for Clinical Use?
Abstract
1. Introduction
2. CtDNA in Ovarian Cancer
2.1. Diagnostic and Predictive Potential of ctDNA
2.2. Prognosis and Monitoring of Treatment Response Using ctDNA
2.3. Identifying Therapeutic Resistance Using ctDNA
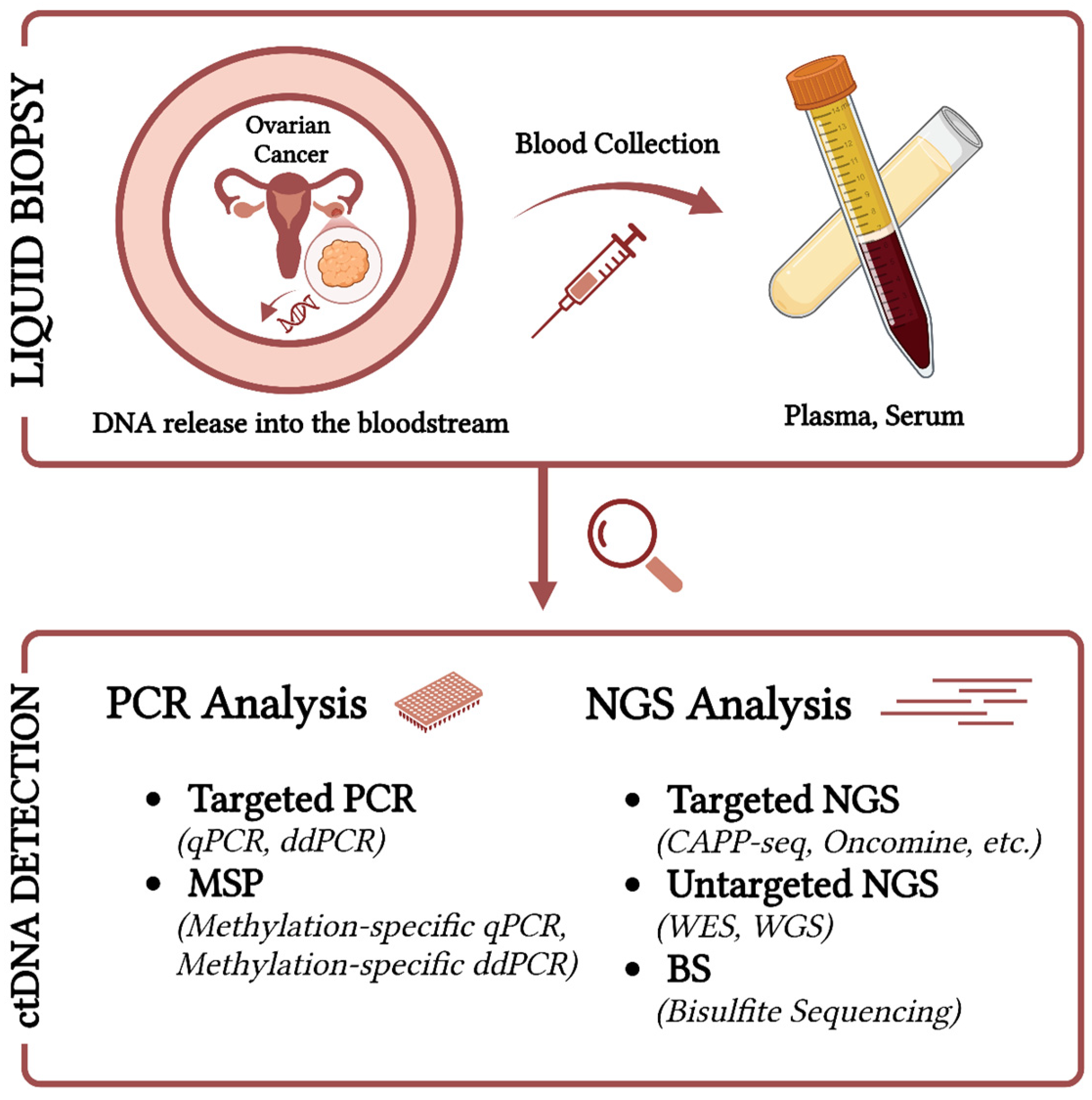
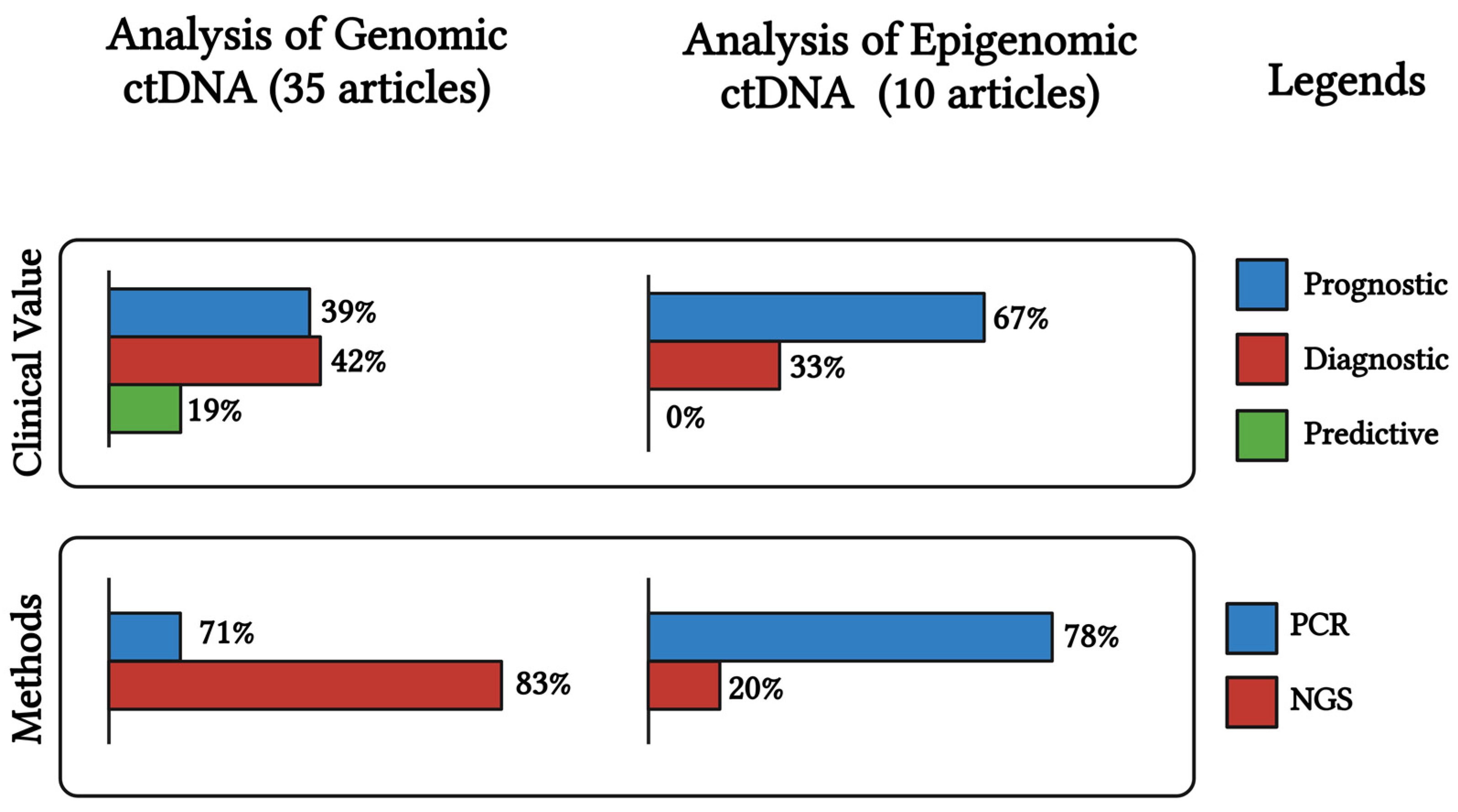
2.4. Detection Techniques of ctDNA and Challenges in Ovarian Cancer
3. Discussion
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kurman, R.J.; Shih, I.M. The Dualistic Model of Ovarian Carcinogenesis: Revisited, Revised, and Expanded. Am. J. Pathol. 2016, 186, 733–747. [Google Scholar] [CrossRef]
- Webb, P.M.; Jordan, S.J. Global Epidemiology of Epithelial Ovarian Cancer. Nat. Rev. Clin. Oncol. 2024, 21, 389–400. [Google Scholar] [CrossRef]
- Mazidimoradi, A.; Momenimovahed, Z.; Allahqoli, L.; Tiznobaik, A.; Hajinasab, N.; Salehiniya, H.; Alkatout, I. The Global, Regional and National Epidemiology, Incidence, Mortality, and Burden of Ovarian Cancer. Health Sci. Rep. 2022, 5, e936. [Google Scholar] [CrossRef] [PubMed]
- Reid, B.M.; Permuth, J.B.; Sellers, T.A. Epidemiology of Ovarian Cancer: A Review. Cancer Biol. Med. 2017, 14, 9–32. [Google Scholar] [CrossRef] [PubMed]
- Koutras, A.; Perros, P.; Prokopakis, I.; Ntounis, T.; Fasoulakis, Z.; Pittokopitou, S.; Samara, A.A.; Valsamaki, A.; Douligeris, A.; Mortaki, A.; et al. Advantages and Limitations of Ultrasound as a Screening Test for Ovarian Cancer. Diagnostics 2023, 13, 2078. [Google Scholar] [CrossRef] [PubMed]
- Charkhchi, P.; Cybulski, C.; Gronwald, J.; Wong, F.O.; Narod, S.A.; Akbari, M.R. CA125 and Ovarian Cancer: A Comprehensive Review. Cancers 2020, 12, 3730. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J.; Bonfrer, J.M.; Kulpa, J.; Rustin, G.J.S.; Soletormos, G.; Torre, G.C.; Tuxen, M.K.; Zwirner, M. CA125 in Ovarian Cancer: European Group on Tumor Markers Guidelines for Clinical Use. Int. J. Gynecol. Cancer 2005, 15, 679–691. [Google Scholar] [CrossRef] [PubMed]
- Elattar, A.; Bryant, A.; Winter-Roach, B.A.; Hatem, M.; Naik, R. Optimal Primary Surgical Treatment for Advanced Epithelial Ovarian Cancer. Cochrane Database Syst. Rev. 2011, 2011, CD007565. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabières, C.; Marchetti, D.; Lang, J.E. Liquid Biopsy: From Concept to Clinical Application. Sci. Rep. 2023, 13, 21685. [Google Scholar] [CrossRef]
- Stejskal, P.; Goodarzi, H.; Srovnal, J.; Hajdúch, M.; van ’t Veer, L.J.; Magbanua, M.J.M. Circulating Tumor Nucleic Acids: Biology, Release Mechanisms, and Clinical Relevance. Mol. Cancer 2023, 22, 15. [Google Scholar] [CrossRef]
- Asante, D.B.; Calapre, L.; Ziman, M.; Meniawy, T.M.; Gray, E.S. Liquid Biopsy in Ovarian Cancer Using Circulating Tumor DNA and Cells: Ready for Prime Time? Cancer Lett. 2020, 468, 59–71. [Google Scholar] [CrossRef]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical Relevance of Blood-Based CtDNA Analysis: Mutation Detection and Beyond. Br. J. Cancer 2021, 124, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Kallio, H.M.L.; Savolainen, K.; Virtanen, T.; Ryyppö, L.; Selin, H.; Martikainen, P.; Staff, S.; Kivinummi, K.; Sipola, J.; Vuorinen, J.; et al. Sensitive Circulating Tumor DNA-Based Residual Disease Detection in Epithelial Ovarian Cancer. Life Sci. Alliance 2024, 7, e202402658. [Google Scholar] [CrossRef] [PubMed]
- Kutz, O.; Drukewitz, S.; Krüger, A.; Aust, D.; William, D.; Oster, S.; Schröck, E.; Baretton, G.; Link, T.; Wimberger, P.; et al. Exploring Evolutionary Trajectories in Ovarian Cancer Patients by Longitudinal Analysis of CtDNA. Clin. Chem. Lab. Med. 2024, 62, 2070–2081. [Google Scholar] [CrossRef]
- Marchi, G.; Rajavuori, A.; Nguyen, M.T.N.; Huhtinen, K.; Oksa, S.; Hietanen, S.; Hautaniemi, S.; Hynninen, J.; Oikkonen, J. Extensive Mutational CtDNA Profiles Reflect High-Grade Serous Cancer Tumors and Reveal Emerging Mutations at Recurrence. Transl. Oncol. 2024, 39, 101814. [Google Scholar] [CrossRef]
- Heo, J.; Kim, Y.N.; Shin, S.; Lee, K.; Lee, J.H.; Lee, Y.J.; Choi, Z.; Park, J.; Min, S.; Kim, S.W.; et al. Serial Circulating Tumor DNA Analysis with a Tumor-Naïve Next-Generation Sequencing Panel Detects Minimal Residual Disease and Predicts Outcome in Ovarian Cancer. Cancer Res. 2024, 84, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Medina, J.E.; Annapragada, A.V.; Lof, P.; Short, S.; Bartolomucci, A.L.; Mathios, D.; Koul, S.; Niknafs, N.; Noë, M.; Foda, Z.H.; et al. Early Detection of Ovarian Cancer Using Cell-Free DNA Fragmentomes and Protein Biomarkers. Cancer Discov. 2024, 15, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Calapre, L.; Giardina, T.; Beasley, A.B.; Reid, A.L.; Stewart, C.; Amanuel, B.; Meniawy, T.M.; Gray, E.S. Identification of TP53 Mutations in Circulating Tumour DNA in High Grade Serous Ovarian Carcinoma Using next Generation Sequencing Technologies. Sci. Rep. 2023, 13, 278. [Google Scholar] [CrossRef] [PubMed]
- Chao, A.; Chen, S.J.; Chen, H.C.; Tan, K.T.; Hsiao, W.; Jung, S.M.; Yang, L.Y.; Huang, K.G.; Chou, H.H.; Huang, H.J.; et al. Mutations in Circulating Tumor DNA Detected in the Postoperative Period Predict Poor Survival in Patients with Ovarian Cancer. Biomed. J. 2023, 46, 100563. [Google Scholar] [CrossRef] [PubMed]
- Dobilas, A.; Chen, Y.; Brueffer, C.; Leandersson, P.; Saal, L.H.; Borgfeldt, C. Preoperative CtDNA Levels Are Associated With Poor Overall Survival in Patients With Ovarian Cancer. Cancer Genom. Proteom. 2023, 20, 763–770. [Google Scholar] [CrossRef]
- Kim, Y.N.; Shim, Y.; Seo, J.; Choi, Z.; Lee, Y.J.; Shin, S.; Kim, S.W.; Kim, S.; Choi, J.R.; Lee, J.Y.; et al. Investigation of PARP Inhibitor Resistance Based on Serially Collected Circulating Tumor DNA in Patients With BRCA-Mutated Ovarian Cancer. Clin. Cancer Res. 2023, 29, 2725–2734. [Google Scholar] [CrossRef] [PubMed]
- Lheureux, S.; Prokopec, S.D.; Oldfield, L.E.; Gonzalez-Ochoa, E.; Bruce, J.P.; Wong, D.; Danesh, A.; Torti, D.; Torchia, J.; Fortuna, A.; et al. Identifying Mechanisms of Resistance by Circulating Tumor DNA in EVOLVE, a Phase II Trial of Cediranib Plus Olaparib for Ovarian Cancer at Time of PARP Inhibitor Progression. Clin. Cancer Res. 2023, 29, 3706–3716. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.W.; Wong, F.; Szymiczek, A.; Ene, G.E.V.; Zhang, S.; May, T.; Narod, S.A.; Kotsopoulos, J.; Akbari, M.R. Evaluating the Utility of CtDNA in Detecting Residual Cancer and Predicting Recurrence in Patients with Serous Ovarian Cancer. Int. J. Mol. Sci. 2023, 24, 14388. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ma, R.; Luo, C.; Xie, Q.; Ning, X.; Sun, K.; Meng, F.; Zhou, M.; Sun, J. Noninvasive Early Differential Diagnosis and Progression Monitoring of Ovarian Cancer Using the Copy Number Alterations of Plasma Cell-Free DNA. Transl. Res. 2023, 262, 12–24. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, X.; Liu, Q.; Yang, J.; Bai, J.; Yin, M.; Cao, D.; Zhang, Q.; Zheng, L. Can Circulating Cell Free DNA Be a Promising Marker in Ovarian Cancer?—A Genome-Scale Profiling Study in a Single Institution. J. Ovarian Res. 2023, 16, 11. [Google Scholar] [CrossRef]
- Stergiopoulou, D.; Markou, A.; Giannopoulou, L.; Buderath, P.; Balgkouranidou, I.; Xenidis, N.; Kakolyris, S.; Kasimir-Bauer, S.; Lianidou, E. Detection of ESR1 Mutations in Primary Tumors and Plasma Cell-Free DNA in High-Grade Serous Ovarian Carcinoma Patients. Cancers 2022, 14, 3790. [Google Scholar] [CrossRef] [PubMed]
- Vanderstichele, A.; Busschaert, P.; Landolfo, C.; Olbrecht, S.; Coosemans, A.; Froyman, W.; Loverix, L.; Concin, N.; Braicu, E.I.; Wimberger, P.; et al. Nucleosome Footprinting in Plasma Cell-Free DNA for the Pre-Surgical Diagnosis of Ovarian Cancer. NPJ Genom. Med. 2022, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.Y.; Chapman, J.S.; Kalashnikova, E.; Pierson, W.; Smith-McCune, K.; Pineda, G.; Vattakalam, R.M.; Ross, A.; Mills, M.; Suarez, C.J.; et al. Circulating Tumor DNA Monitoring for Early Recurrence Detection in Epithelial Ovarian Cancer. Gynecol. Oncol. 2022, 167, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Jie, X.; Du, M.; Zhang, M.; Jin, X.; Cai, Q.; Xu, C.; Zhang, X. Mutation Analysis of Circulating Tumor DNA and Paired Ascites and Tumor Tissues in Ovarian Cancer. Exp. Ther. Med. 2022, 24. [Google Scholar] [CrossRef]
- Paracchini, L.; Mannarino, L.; Beltrame, L.; Landoni, F.; Fruscio, R.; Grassi, T.; Dalessandro, M.L.; D’Incalci, M.; Marchini, S. Targeted Mutational Analysis of Circulating Tumor DNA to Decipher Temporal Heterogeneity of High-Grade Serous Ovarian Cancer. Cancers 2022, 14, 3697. [Google Scholar] [CrossRef] [PubMed]
- Sharbatoghli, M.; Fattahi, F.; Aboulkheyr Es, H.; Akbari, A.; Akhavan, S.; Ebrahimi, M.; Asadi-Lari, M.; Totonchi, M.; Madjd, Z. Copy Number Variation of Circulating Tumor DNA (CtDNA) Detected Using NIPT in Neoadjuvant Chemotherapy-Treated Ovarian Cancer Patients. Front. Genet. 2022, 13, 938985. [Google Scholar] [CrossRef] [PubMed]
- Sabatier, R.; Garnier, S.; Guille, A.; Carbuccia, N.; Pakradouni, J.; Adelaide, J.; Provansal, M.; Cappiello, M.; Rousseau, F.; Chaffanet, M.; et al. Whole-Genome/Exome Analysis of Circulating Tumor DNA and Comparison to Tumor Genomics from Patients with Heavily Pre-Treated Ovarian Cancer: Subset Analysis of the PERMED-01 Trial. Front. Oncol. 2022, 12, 946257. [Google Scholar] [CrossRef] [PubMed]
- Braicu, E.I.; du Bois, A.; Sehouli, J.; Beck, J.; Prader, S.; Kulbe, H.; Eiben, B.; Harter, P.; Traut, A.; Pietzner, K.; et al. Cell-Free-DNA-Based Copy Number Index Score in Epithelial Ovarian Cancer-Impact for Diagnosis and Treatment Monitoring. Cancers 2021, 14, 168. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Shan, B.; Liang, S.; Zhang, J.; Yu, Y.; Zhang, Y.; Wang, G.; Bai, Y.; Qian, B.; Lu, J.; et al. Hybrid Capture-Based Genomic Profiling of Circulating Tumor DNA From Patients With Advanced Ovarian Cancer. Pathol. Oncol. Res. 2021, 27, 581534. [Google Scholar] [CrossRef] [PubMed]
- Paracchini, L.; Beltrame, L.; Grassi, T.; Inglesi, A.; Fruscio, R.; Landoni, F.; Ippolito, D.; Marchette, M.D.; Paderno, M.; Adorni, M.; et al. Genome-Wide Copy-Number Alterations in Circulating Tumor DNA as a Novel Biomarker for Patients with High-Grade Serous Ovarian Cancer. Clin. Cancer Res. 2021, 27, 2549–2560. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, Y.M.; Gao, Y.; Zhang, S.; Chu, W.; Wei, G.; Li, K.; He, X.; Chen, L.; Guo, L.; et al. A Novel Decision Tree Model Based on Chromosome Imbalances in Cell-Free DNA and CA-125 in the Differential Diagnosis of Ovarian Cancer. Int. J. Biol. Markers 2021, 36, 3–13. [Google Scholar] [CrossRef]
- Vitale, S.R.; Groenendijk, F.H.; van Marion, R.; Beaufort, C.M.; Helmijr, J.C.; Dubbink, H.J.; Dinjens, W.N.M.; Ewing-Graham, P.C.; Smolders, R.; van Doorn, H.C.; et al. TP53 Mutations in Serum Circulating Cell-Free Tumor DNA As Longitudinal Biomarker for High-Grade Serous Ovarian Cancer. Biomolecules 2020, 10, 415. [Google Scholar] [CrossRef]
- Waki, K.; Yokomizo, K.; Kawano, K.; Tsuda, N.; Komatsu, N.; Yamada, A. Integrity of Plasma DNA Is Inversely Correlated with Vaccine-Induced Antitumor Immunity in Ovarian Cancer Patients. Cancer Immunol. Immunother. 2020, 69, 2001–2007. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, T.; Iwahashi, N.; Sakai, K.; Matsuda, K.; Matsukawa, H.; Toujima, S.; Nishio, K.; Ino, K. Comprehensive Gene Mutation Profiling of Circulating Tumor DNA in Ovarian Cancer: Its Pathological and Prognostic Impact. Cancers 2020, 12, 3382. [Google Scholar] [CrossRef]
- Noguchi, T.; Sakai, K.; Iwahashi, N.; Matsuda, K.; Matsukawa, H.; Yahata, T.; Toujima, S.; Nishio, K.; Ino, K. Changes in the Gene Mutation Profiles of Circulating Tumor DNA Detected Using CAPP-Seq in Neoadjuvant Chemotherapy-Treated Advanced Ovarian Cancer. Oncol. Lett. 2020, 19, 2713–2720. [Google Scholar] [CrossRef] [PubMed]
- Jagelkova, M.; Zelinova, K.; Laucekova, Z.; Bobrovska, M.; Dankova, Z.; Grendar, M.; Dokus, K. Comparison of Somatic Mutation Profiles Between Formalin-Fixed Paraffin Embedded Tissues and Plasma Cell-Free DNA from Ovarian Cancer Patients Before and After Surgery. Biores. Open Access 2020, 9, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Han, M.R.; Lee, S.H.; Park, J.Y.; Hong, H.; Ho, J.Y.; Hur, S.Y.; Choi, Y.J. Clinical Implications of Circulating Tumor DNA from Ascites and Serial Plasma in Ovarian Cancer. Cancer Res. Treat. 2020, 52, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, A.; Hihara, T.; Shintani, D.; Yabuno, A.; Ikeda, Y.; Tai, K.; Fujiwara, K.; Watanabe, K.; Hasegawa, K. Evaluation of Circulating Tumor DNA in Patients with Ovarian Cancer Harboring Somatic PIK3CA or KRAS Mutations. Cancer Res. Treat. 2020, 52, 1219–1228. [Google Scholar] [CrossRef]
- Wang, N.N.; Yu, M.; Lu, J.; Wang, J.H. Detection of Plasma Circulating Tumor DNA in Ovarian and Its Clinical Significance. Indian. J. Pharm. Sci. 2020, 82, 145–151. [Google Scholar] [CrossRef]
- Stamenkovic, S.; Cheng, J.; Surowy, H.; Burwinkel, B.; Gündert, M. Circulating Cell-Free DNA Variables as Marker of Ovarian Cancer Patients: A Pilot Study. Cancer Biomark. 2020, 28, 159–167. [Google Scholar] [CrossRef]
- Lin, K.K.; Harrell, M.I.; Oza, A.M.; Oaknin, A.; Ray-Coquard, I.; Tinker, A.V.; Helman, E.; Radke, M.R.; Say, C.; Vo, L.T.; et al. BRCA Reversion Mutations in Circulating Tumor DNA Predict Primary and Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma. Cancer Discov. 2019, 9, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Oikkonen, J.; Zhang, K.; Salminen, L.; Schulman, I.; Lavikka, K.; Andersson, N.; Ojanperä, E.; Hietanen, S.; Grénman, S.; Lehtonen, R.; et al. Prospective Longitudinal CtDNA Workflow Reveals Clinically Actionable Alterations in Ovarian Cancer. JCO Precis. Oncol. 2019, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Herzog, C.; Jones, A.; Evans, I.; Reisel, D.; Olaitan, A.; Doufekas, K.; MacDonald, N.; Rådestad, A.F.; Gemzell-Danielsson, K.; Zikan, M.; et al. Plasma Cell-Free DNA Methylation Analysis for Ovarian Cancer Detection: Analysis of Samples from a Case-Control Study and an Ovarian Cancer Screening Trial. Int. J. Cancer 2024, 154, 679–691. [Google Scholar] [CrossRef]
- Werner, B.; Sjoquist, K.M.; Espinoza, D.; Yip, S.; Chang, G.; Cummins, M.M.; Mileshkin, L.; Ananda, S.; Shannon, C.; Friedlander, M.; et al. Cell-Free DNA in Plasma and Ascites as a Biomarker of Bevacizumab Response—A Translational Research Sub-Study of the REZOLVE (ANZGOG-1101) Clinical Trial. Transl. Oncol. 2024, 43, 101914. [Google Scholar] [CrossRef] [PubMed]
- Gahlawat, A.W.; Witte, T.; Sinn, P.; Schott, S. Circulating Cf-MiRNA as a More Appropriate Surrogate Liquid Biopsy Marker than CfDNA for Ovarian Cancer. Sci. Rep. 2023, 13, 5503. [Google Scholar] [CrossRef] [PubMed]
- Bahado-Singh, R.O.; Ibrahim, A.; Al-Wahab, Z.; Aydas, B.; Radhakrishna, U.; Yilmaz, A.; Vishweswaraiah, S. Precision Gynecologic Oncology: Circulating Cell Free DNA Epigenomic Analysis, Artificial Intelligence and the Accurate Detection of Ovarian Cancer. Sci. Rep. 2022, 12, 18625. [Google Scholar] [CrossRef] [PubMed]
- Faaborg, L.; Fredslund Andersen, R.; Waldstrøm, M.; Høgdall, E.; Høgdall, C.; Adimi, P.; Jakobsen, A.; Dahl Steffensen, K. Analysis of HOXA9 Methylated CtDNA in Ovarian Cancer Using Sense-Antisense Measurement. Clin. Chim. Acta 2021, 522, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Gupta, S.; Sachan, M. Evaluation of the Diagnostic Potential of Candidate Hypermethylated Genes in Epithelial Ovarian Cancer in North Indian Population. Front. Mol. Biosci. 2021, 8, 719056. [Google Scholar] [CrossRef] [PubMed]
- Rusan, M.; Andersen, R.F.; Jakobsen, A.; Steffensen, K.D. Circulating HOXA9-Methylated Tumour DNA: A Novel Biomarker of Response to Poly (ADP-Ribose) Polymerase Inhibition in BRCA-Mutated Epithelial Ovarian Cancer. Eur. J. Cancer 2020, 125, 121–129. [Google Scholar] [CrossRef]
- Singh, A.; Gupta, S.; Badarukhiya, J.A.; Sachan, M. Detection of Aberrant Methylation of HOXA9 and HIC1 through Multiplex MethyLight Assay in Serum DNA for the Early Detection of Epithelial Ovarian Cancer. Int. J. Cancer 2020, 147, 1740–1752. [Google Scholar] [CrossRef]
- Li, S.; Huang, W.; Li, Y.; Chen, B.; Li, D. A Study of HTERT Promoter Methylation in Circulating Tumour DNAs of Patients with Ovarian Magnificent Tumour. Onco Targets Ther. 2020, 13, 12317–12323. [Google Scholar] [CrossRef]
- Thomsen, C.B.; Andersen, R.F.; Steffensen, K.D.; Adimi, P.; Jakobsen, A. Delta Tocotrienol in Recurrent Ovarian Cancer. A Phase II Trial. Pharmacol. Res. 2019, 141, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hu, G.; Yang, Q.; Dong, R.; Xie, X.; Ma, D.; Shen, K.; Kong, B. A Multiplex Methylation-Specific PCR Assay for the Detection of Early-Stage Ovarian Cancer Using Cell-Free Serum DNA. Gynecol. Oncol. 2013, 130, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yu, L.; Luo, X.; Huang, L.; Li, Q.S.; Shao, X.S.; Liu, Y.; Fan, Y.; Yang, G.Z. Detection of OPCML Methylation, a Possible Epigenetic Marker, from Free Serum Circulating DNA to Improve the Diagnosis of Early-Stage Ovarian Epithelial Cancer. Oncol. Lett. 2017, 14, 217–223. [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V.; Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Smith, D.; Richards, D.; et al. Sensitive and Specific Multi-Cancer Detection and Localization Using Methylation Signatures in Cell-Free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Kfoury, M.; Hazzaz, R.E.; Sanson, C.; Durand, F.B.; Michels, J.; Blameble, E.C.; Tang, R.; Le Formal, A.; Lecerf, E.; Gouy, S.; et al. Circulating Tumor DNA from Ascites as an Alternative to Tumor Sampling for Genomic Profiling in Ovarian Cancer Patients. Biomark. Res. 2023, 11, 93. [Google Scholar] [CrossRef] [PubMed]
- Alatise, K.L.; Gardner, S.; Alexander-Bryant, A. Mechanisms of Drug Resistance in Ovarian Cancer and Associated Gene Targets. Cancers 2022, 14, 6246. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.N.; Tian, Q.; Teng, Q.X.; Wurpel, J.N.D.; Zeng, L.; Pan, Y.; Chen, Z.S. Understanding and Targeting Resistance Mechanisms in Cancer. MedComm 2023, 4, e265. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.A.; Bohlke, K.; Armstrong, D.K.; Bookman, M.A.; Cliby, W.A.; Coleman, R.L.; Dizon, D.S.; Kash, J.J.; Meyer, L.A.; Moore, K.N.; et al. Neoadjuvant Chemotherapy for Newly Diagnosed, Advanced Ovarian Cancer: Society of Gynecologic Oncology and American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 2016, 34, 3460–3473. [Google Scholar] [CrossRef]
- Brett, J.O.; Spring, L.M.; Bardia, A.; Wander, S.A. ESR1 Mutation as an Emerging Clinical Biomarker in Metastatic Hormone Receptor-Positive Breast Cancer. Breast Cancer Res. 2021, 23, 85. [Google Scholar] [CrossRef]
- Tébar Martínez, R.; Martín Arana, J.; Gimeno Valiente, F.; Tarazona, N.; Rentero Garrido, P.; Cervantes, A. Strategies for Improving Detection of Circulating Tumor DNA Using next Generation Sequencing. Cancer Treat. Rev. 2023, 119, 102595. [Google Scholar] [CrossRef]
- Kim, H.; Park, K.U. Clinical Circulating Tumor DNA Testing for Precision Oncology. Cancer Res. Treat. 2023, 55, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef] [PubMed]
- Lisio, M.A.; Fu, L.; Goyeneche, A.; Gao, Z.H.; Telleria, C. High-Grade Serous Ovarian Cancer: Basic Sciences, Clinical and Therapeutic Standpoints. Int. J. Mol. Sci. 2019, 20, 952. [Google Scholar] [CrossRef] [PubMed]
- Bell, D.; Berchuck, A.; Birrer, M.; Chien, J.; Cramer, D.W.; Dao, F.; Dhir, R.; Disaia, P.; Gabra, H.; Glenn, P.; et al. Integrated Genomic Analyses of Ovarian Carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Lee, J.S.; Kim, M.; Seong, M.W.; Kim, H.S.; Lee, Y.K.; Kang, H.J. Plasma vs. Serum in Circulating Tumor DNA Measurement: Characterization by DNA Fragment Sizing and Digital Droplet Polymerase Chain Reaction. Clin. Chem. Lab. Med. 2020, 58, 527–532. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Aucamp, J.; Pretorius, P.J. Cell-Free DNA: Preanalytical Variables. Clin. Chim. Acta 2015, 450, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Janku, F.; Huang, H.J.; Pereira, D.Y.; Kobayashi, M.; Chiu, C.H.; Call, S.G.; Woodbury, K.T.; Chao, F.; Marshak, D.R.; Chiu, R.Y.T. A Novel Method for Liquid-Phase Extraction of Cell-Free DNA for Detection of Circulating Tumor DNA. Sci. Rep. 2021, 11, 19653. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Yadav, P. Liquid Biopsy in Cancer Management: Integrating Diagnostics and Clinical Applications. Pract. Lab. Med. 2024, 43, e00446. [Google Scholar] [CrossRef] [PubMed]
- Giannopoulou, L.; Kasimir-Bauer, S.; Lianidou, E.S. Liquid Biopsy in Ovarian Cancer: Recent Advances on Circulating Tumor Cells and Circulating Tumor DNA. Clin. Chem. Lab. Med. 2018, 56, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Tivey, A.; Church, M.; Rothwell, D.; Dive, C.; Cook, N. Circulating Tumour DNA—Looking beyond the Blood. Nat. Rev. Clin. Oncol. 2022, 19, 600–612. [Google Scholar] [CrossRef]
- Rickard, B.P.; Conrad, C.; Sorrin, A.J.; Ruhi, M.K.; Reader, J.C.; Huang, S.A.; Franco, W.; Scarcelli, G.; Polacheck, W.J.; Roque, D.M.; et al. Malignant Ascites in Ovarian Cancer: Cellular, Acellular, and Biophysical Determinants of Molecular Characteristics and Therapy Response. Cancers 2021, 13, 4318. [Google Scholar] [CrossRef]
- Ding, S.C.; Lo, Y.M.D. Cell-Free DNA Fragmentomics in Liquid Biopsy. Diagnostics 2022, 12, 978. [Google Scholar] [CrossRef] [PubMed]
- Abdelwahab, O.; Belzile, F.; Torkamaneh, D. Performance Analysis of Conventional and AI-Based Variant Callers Using Short and Long Reads. BMC Bioinform. 2023, 24, 472. [Google Scholar] [CrossRef] [PubMed]
- Widman, A.J.; Shah, M.; Frydendahl, A.; Halmos, D.; Khamnei, C.C.; Øgaard, N.; Rajagopalan, S.; Arora, A.; Deshpande, A.; Hooper, W.F.; et al. Ultrasensitive Plasma-Based Monitoring of Tumor Burden Using Machine-Learning-Guided Signal Enrichment. Nat. Med. 2024, 30, 1655–1666. [Google Scholar] [CrossRef]
| OC Subtype & Stage | Patients and Controls | Age (yrs) | Plasma or Serum | Biomarker | Detection Method | Detection Rate (%) | Sensitivity and Specificity | Clinical Value | Refs |
|---|---|---|---|---|---|---|---|---|---|
| EOC/I–IV | P= 63 | 30–80 | Plasma | 21 gene panel, including TP53, CLDN19, ARID4B, etc. | NGS | Diagnosis = 93 Progression = 100 | NR | Prognostic: OS: HR = 6.60, p < 0.001 TTP: HR = 4.69, p < 0.001 | (Kallio et al., 2024) [13] |
| High- and low-grade serous EOC/I–IV | P = 15 (Low grade = 1; High grade = 14) | 47–82 | Plasma | 275 cancer-associated genes, including TP53, KMT2A, NOTCH1, KDM5C, ARID1B, etc. | NGS | 100 | - | Diagnostic/Predictive/Longitudinal monitoring | (Kutz et al., 2024) [14] |
| HGSOC/II–IV | P = 29 (Platinum resistant = 14; Platinum sensitive = 15) | Platinum resistant = 57–80 Platinum sensitive = 59–81 | Plasma | 700 cancer-associated genes, including TP53, KCNH2, JAK2, GRIN2A, FGFR3, ARID1B, etc. | NGS | Pre-treatment = 82.8, Relapse = 89.5 | NR | Prognostic: PFS: R = −0.72, p = 0.008 OS: R = −0.74, p = 0.005 | (Marchi et al., 2024) [15] |
| EOC/I–IV | P = 296 B = 95 | 58.1 ± 12.3 (median) | Plasma | 9 gene panel, including TP53, BRCA1/2, PTEN, etc. | NGS | 89.9 | Sn = 92% Sp = 84% | Prognostic: PFS: HR = 10.71, 95% CI: 4.43–25.9 | (Heo et al., 2024) [16] |
| EOC/I–IV | P = 591 HC = 204 B = 253 | P = 37–85 HC = 47–75 B = 19–92 | Plasma | cfDNA fragmentomes | WGS | NR | Sn = 72%, 69%, 87%, 100% Sp > 99% | Diagnostic | (Medina et al., 2024) [17] |
| HGSOC/III–IV | P = 10 | NR | Plasma | 10 gene panel, including TP53, AKT1, KRAS, PIK3CA EGFR, etc. | Two NGS platforms plus ddPCR | Accel = 60 Oncomine = 100 | Oncomine: Sn ≥ 90%, Sp ≥ 99% Accel: Sn = 90% Sp ≥ 99% | Longitudinal monitoring/Predictive | (Calapre et al., 2023) [18] |
| EOC/I–IV | P = 29 | 18–85 | Plasma | 50 gene panel, including TP53, KRAS, PIK3CA, etc. | NGS | 82.8 | NR | Prognostic: OS: HR = 6.56, 95% CI: 1.07–40.17 | (Chao et al., 2023) [19] |
| EOC/I–IV | P = 41 B = 6 BOT = 9 | NR | Plasma | Mutant genes, including TP53, KRAS, PIK3CA, etc. | dPCR | 58 | NR | Prognostic: Log-rank test: OS (p = 0.008) | (Dobilas et al., 2023) [20] |
| EOC/I–IV | P = 54 | 43–79 | Plasma | 531 gene panel, including TP53, BRCA1/2, CHEK2, etc. | NGS | 89.7 | NR | Prognostic: PFS (p = 0.0003) | (Kim et al., 2023) [21] |
| HGSOC | P = 30 | 63 (median) | Plasma | BRCA1/2, TP53, TP53, PALB2, CCNE1, etc. | NGS | 74.4 | NR | Prognostic: DFS: HR = 4.79, 95% CI: 1.84–12.5 | (Lheureux et al., 2023) [22] |
| HGSOC, LGSOC/I–III | P = 48 | 58.9 (median) | Plasma | 59 gene panel, including BRCA1/2, TP53, BRAF, RB1, etc. | NGS | 77.4 | NR | Prognostic: RFS: HR = 2.06, 95% CI: 0.60–7.10, p = 0.23 | (Zhu et al., 2023) [23] |
| EOC/II–IV | P = 44 HC = 17 | NR | Plasma | CNV | LC-WGS | NR | NR | Diagnostic/Predictive | (Chen et al., 2023) [24] |
| EOC/I–IV | P = 59 HC = 100 | 27–82 | Plasma | OC score (CNV, nucleosome footprint, 5′ end motifs, fragmentation profiles) | Low-pass WGS | I–II = 85.7 | Sn = 97.7% Sp = 94.7% | Diagnostic | (Zhou et al., 2023) [25] |
| HGSOC/I–IV | P = 80 HC = 11 | <39 and ≥41 (median) | Plasma | ESR1 mutations | ddPCR | 13.8 | NR | Diagnostic | (Stergiopoulou et al., 2022) [26] |
| EOC and non-EOC/I–IV | P = 271 B = 130 BOT = 41 Invasive = 92 Metastatic = 8 | B = 43–64, BOT = 37–63, Invasive = 57–73, Metastatic = 52–69 | Plasma | Nucleosome footprint and CNA | LC-WGS | NR | Nucleosome footprint: AUC = 0.71 (95% CI: 0.65–0.77) CNA: AUC = 0.72 (95% CI: 0.66–0.78) | Diagnostic | (Vanderstichele et al., 2022) [27] |
| EOC/I–IV | P = 69 (Cohort A, pre-surgical = 44; Cohort B, post-surgical and/or after therapy = 12; Cohort C, after completed treatments = 13) | 29–82 (55.5 median) | Plasma | TP53, ARID1A, KRAS, etc. | Tumour-informed multiplex PCR NGS | Cohort A = 73 Cohort B = 33 Cohort B and C = 23 | Sn = 100 Sp = 100 | Prognostic: RFS: HR = 7.34, 95% CI: 0.75–72.3, p = 0.087 | (Hou et al., 2022) [28] |
| EOC/II–IV | P = 18 (6 paired ascites and 8 paired tumour tissues) | 24–70 | Plasma | 333 cancer-related genes, including TGFBR2, ARID1A, ATR, BCR, KMT2C, etc. | NGS | 94.4 | NR | Diagnostic/Predictive | (Jie et al., 2022) [29] |
| HGSOC/III–IV | P = 18 | 48–79 | Plasma | 65 cancer-related genes, including TP53, BRCA1/2, POLE, MSH3, ATR, etc. | NGS | NR | NR | Diagnostic/Predictive | (Paracchini et al., 2022) [30] |
| EOC/II–IV | P = 6 | 38–78 | Plasma | CNA | WGS | NR | NR | Prognostic: OS (p < 0.0001) | (Sharbatoghli et al., 2022) [31] |
| HGSOC, LGSOC, Clear cell, endometrioid, carcinosarcoma | P = 24 | 21–71 | Plasma | Identified CNAs and mutations in other cancer-related genes such as TP53 | WGS, WES | 88 | NR | Prognostic: PFS (TMB: HR = 8.6, 95% CI: 1.4–52; GAF: HR = 8.9, 95% CI: 0.91–87) | (Sabatier et al., 2022) [32] |
| EOC/III–IV | P = 109 (Upfront group = 23; CTX group = 9; Follow-up group = 13) | 22–84 | Plasma | CNV (CNI scores) | WGS | Chemo naive = 78, Platinum-eligible recurrent = 83.3, Non-platinum-eligible recurrent = 82.6 | Sn: primary and recurrent = 87%, primary only = 78–91% Sp = 95–100% | Diagnostic | (Braicu et al., 2021) [33] |
| SOC/I, III, and IV | P = 138 | 31–81 | Plasma | 150 cancer gene panel, including TP53, KRAS, LRP1B, ZNF703, NF1, etc. | Hybrid capture-based NGS | 83 | NR | Diagnostic | (Shen et al., 2021) [34] |
| HGSOC/III–IV | P = 46 | 21–81 | Plasma | CNA | Shallow WGS | 14.47 | NR | Prognostic: PFS: HR = 3.31, 95% CI: 1.33–9.13, p = 0.011 | (Paracchini et al., 2021) [35] |
| EOC and non-EOC/I–IV | P = 80 (Malignant = 58 B = 66; BOT = 10) HC = 82 | Malignant = 12–77, B = 21–82, BOT = 27–62, HC = 22–36 | Plasma | CNA | WGS | NR | NR | Diagnostic | (Zhang et al., 2021) [36] |
| HGSOC/III–IV | P = 20 | 37–75 | Serum | Total of 51 genes: 41 for Ampliseq and 10 for Oncomine, including TP53, PIK3CA, ESR1, etc. | NGS, ddPCR | 85 | NR | Diagnostic/Predictive | (Vitale et al., 2020) [37] |
| EOCs/III–IV | P = 39 | NR | Plasma | ALU | qPCR | NR | NR | Diagnostic | (Waki et al., 2020) [38] |
| EOC/I–IV | P = 51 | 28–82 | Plasma | 197 cancer-related genes, including TP53, APC, KRAS, EGFR, MET, PIK3CA, etc. | Deep NGS (CAPP-Seq) | 94 | NR | Prognostic: PFS (p = 0.048) | (Noguchi, Iwahashi, et al., 2020) [39] |
| HGSOC and mucinous/III–IV | P = 10 | 44–74 | Plasma | 197 cancer-related genes, including TP53, APC, KRAS, EGFR, MET, PIK3CA, etc. | Deep NGS (CAPP-Seq) | 100 | NR | Diagnostic | (Noguchi, Sakai, et al., 2020) [40] |
| HGSOC/III–IV | P = 7 | 54–78 | Plasma | 26 genes, including TP53, APC, KRAS, PTEN, PIK3CA, etc. | NGS | 86 | NR | Diagnostic/Predictive | (Jagelkova et al., 2020) [41] |
| HGSOC/III–IV | P = 10 | 44–65 | Plasma | 88 genes, including TP53, PIK3CA, MYC, etc. | NGS | 60 | NR | Diagnostic | (Han et al., 2020) [42] |
| EOC/I–IV | P = 306 | 33–80 | Plasma | 2 genes: KRAS and PIK3CA | ddPCR | 27.1 | NR | Prognostic: PFS (p = 0.0001) OS (p = 0.017) | (Ogasawara et al., 2020) [43] |
| High- and low-grade serous EOC/I–IV | P = 70 | NR | Plasma | CNV | WGS | Stage (I and II) = 55.56 Stage (III and IV) = 85.71 | NR | Diagnostic/Predictive | (Wang et al., 2020) [44] |
| EOC/I–IV | P = 37 HC = 28 | HC = 30–63 OC = 37–80 | Plasma | ALU, LINE 1 | qRT-PCR | NR | NR | Diagnostic | (Stamenkovic et al., 2020) [45] |
| EOCs (96% HGSOC) | P = 112 | 33–82 | Plasma | 54 cancer-related genes, including BRAC1, BRAC2, TP53, etc. | NGS | TP53 = 96 | NR | Prognostic: PFS: HR = 0.12 (p < 0.0001) | (Lin et al., 2019) [46] |
| HGSOC/II–IV | P = 12 | 68 (median) | Plasma | CNV and >500 cancer-related genes, including TP53, PTEN, BRCA2, etc. | NGS | 100 for TP53 and variable for the other genes | NR | PFS (p < 0.01) | (Oikkonen et al., 2019) [47] |
| OC Subtype & Stage | Patient/ Controls | Age (yrs) | Plasma/ Serum | Biomarker | Detection Method | Detection Rate (%) | Sensitivity and Specificity | Clinical Value | Refs |
|---|---|---|---|---|---|---|---|---|---|
| EOC/I–IV | P = 70 B = 39 HC = 4 | OC = 52–64, Controls = 48–66 | Plasma | ZNF154, C2CD4D, and WNT6 | Bisulphite sequencing | 82.8 | Sn = 80% Sp = 97.6% | Diagnostic | (Herzog et al., 2024) [48] |
| EOCs | P = 19 | 38–86 | Plasma | Alu115, IFFO1, and CDH5 promoters | Methylation-specific qPCR | Alu115 = 100, IFFO1Me = 66, CDH5UM = 86 | NR | Prognostic: PFI: HR = 3.21, 95% CI: 1.15–9.00, p = 0.008 | (Werner, Sjoquist, et al., 2024) [49] |
| EOC/I–IV | P = 125 HC = 72 | Non-malignant = 25–85, Cancer = 21–83 | Plasma | miR-200c and miR-141 genes | Methylation-specific PCR | NR | NR | Prognostic: OS: HR = 1.53, 95% CI: 1.0–2.35, p = 0.049 | (Gahlawat et al., 2023) [50] |
| EOC/II–III | P = 5 HC = 12 | P = 66.2 ± 18.14; HC: 67.8 ± 12.96 (median) | Plasma | ANO2, ATP11A, AGAP1, ARFGEF2, BBS9, etc. | AI and GW-DMP | 100 | GW-DMP: Sn = 95%, Sp = 100%; AI: Sn = 100%, Sp = 88% | Diagnostic | (Bahado-Singh et al., 2022) [51] |
| EOC/I–IV | P = 79 HC = 64 | 25–86 | Plasma | HOXA9 | Methylation-specific ddPCR | 59.5 | Sn = 37.5–59.5% Sp = 95.3% | Diagnostic | (Faaborg et al., 2021) [52] |
| EOC/I–IV | P = 85 | 20–65 | Serum | RASSF1A, DAPK1, SOX1, HOXA9, HIC1, SPARC, and SFRP1 | Multiplex methylation-specific qPCR | 82.3–61.3 | Varies with each gene. Sn = 85.88–72.94% Sp = 88.57–77.14% | Diagnostic | (Singh et al., 2021) [53] |
| EOC/I–IV, platinum resistant | P = 32 | 46–70 | Plasma | HOXA9 | Methylation-specific ddPCR | 62 | NR | Prognostic: PFS (p < 0.0001) OS (p = 0.002) | (Rusan et al., 2020) [54] |
| EOC/I–IV | P = 44 | 18–70 | Serum | HOXA9 and HLC1 | Multiplex methylation-specific qPCR | HOXA9 = 62.2 HLC1 = 71.1 | Sn = 88.9% Sp = 100% | Diagnostic | (Singh et al., 2020) [55] |
| EOC/I–III | P = 17 B = 15 HC = 15 A = OC group (P) B = Control group (B and HC) | 32–68 | Plasma | hTERT | Methylation-specific PCR | Group A = 70.6 Group B = 20 | Group A: Sn = 76.9% Sp = 50%; Group B: Sn = 50% Sp = 90.9% | Diagnostic | (Li et al., 2020) [56] |
| EOC/I, III, and IV, platinum resistant | P = 23 | 41–81 | Plasma | HOXA9 | Methylation-specific dPCR | NR | NR | Prognostic: OS (p = 0.01) | (Thomsen et al., 2019) [57] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asante, D.-B.; Tierno, D.; Grassi, G.; Scaggiante, B. Circulating Tumour DNA for Ovarian Cancer Diagnosis and Treatment Monitoring: What Perspectives for Clinical Use? Int. J. Mol. Sci. 2025, 26, 1889. https://doi.org/10.3390/ijms26051889
Asante D-B, Tierno D, Grassi G, Scaggiante B. Circulating Tumour DNA for Ovarian Cancer Diagnosis and Treatment Monitoring: What Perspectives for Clinical Use? International Journal of Molecular Sciences. 2025; 26(5):1889. https://doi.org/10.3390/ijms26051889
Chicago/Turabian StyleAsante, Du-Bois, Domenico Tierno, Gabriele Grassi, and Bruna Scaggiante. 2025. "Circulating Tumour DNA for Ovarian Cancer Diagnosis and Treatment Monitoring: What Perspectives for Clinical Use?" International Journal of Molecular Sciences 26, no. 5: 1889. https://doi.org/10.3390/ijms26051889
APA StyleAsante, D.-B., Tierno, D., Grassi, G., & Scaggiante, B. (2025). Circulating Tumour DNA for Ovarian Cancer Diagnosis and Treatment Monitoring: What Perspectives for Clinical Use? International Journal of Molecular Sciences, 26(5), 1889. https://doi.org/10.3390/ijms26051889








