Exploring the Multifunctional Role of Paenibacillus Metabolites in Various Fields
Abstract
1. Introduction
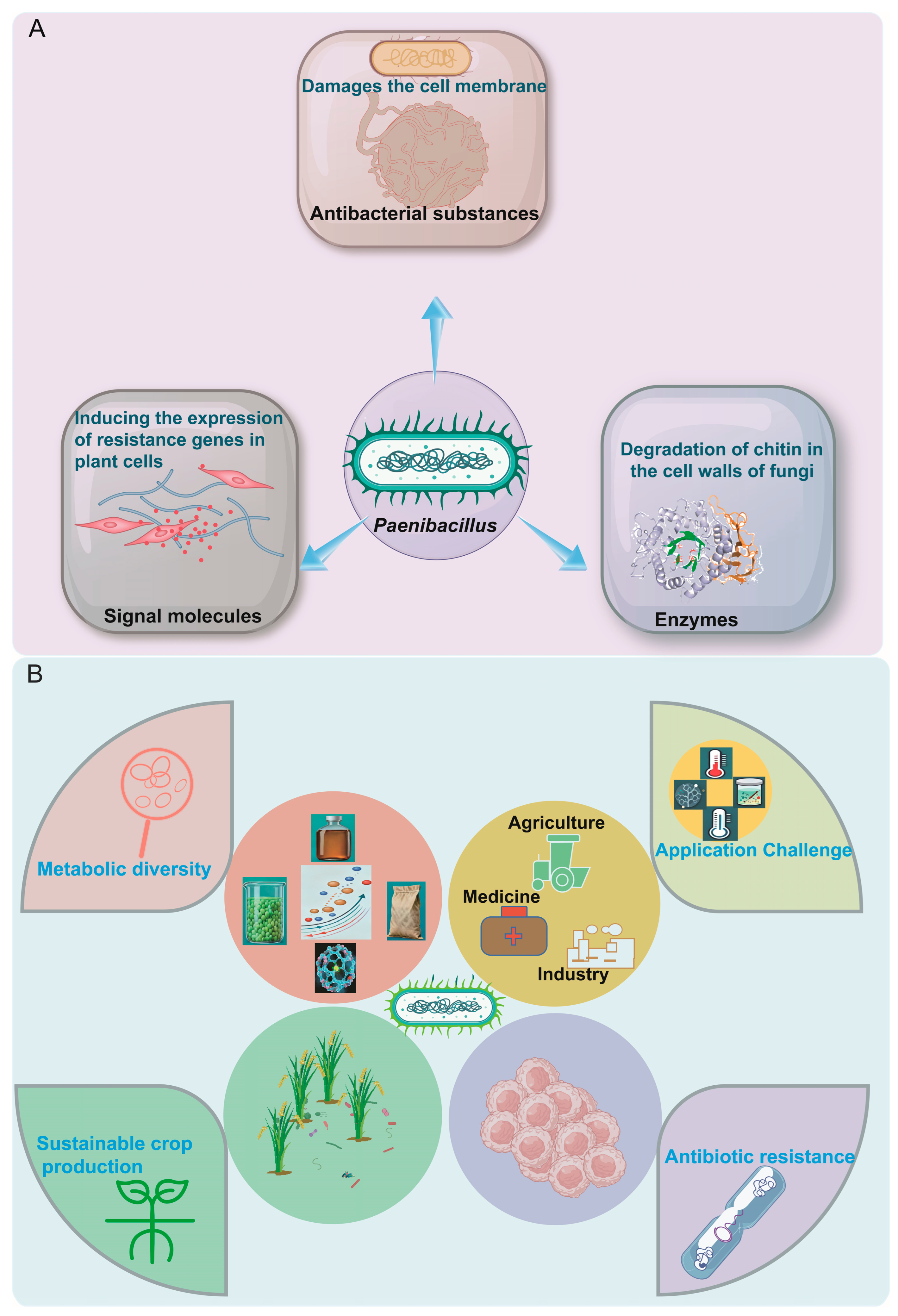
2. The Diversity and Mechanism of Action of Metabolites Produced by Paenibacillus
3. Research on the Application of Paenibacillus in Different Fields
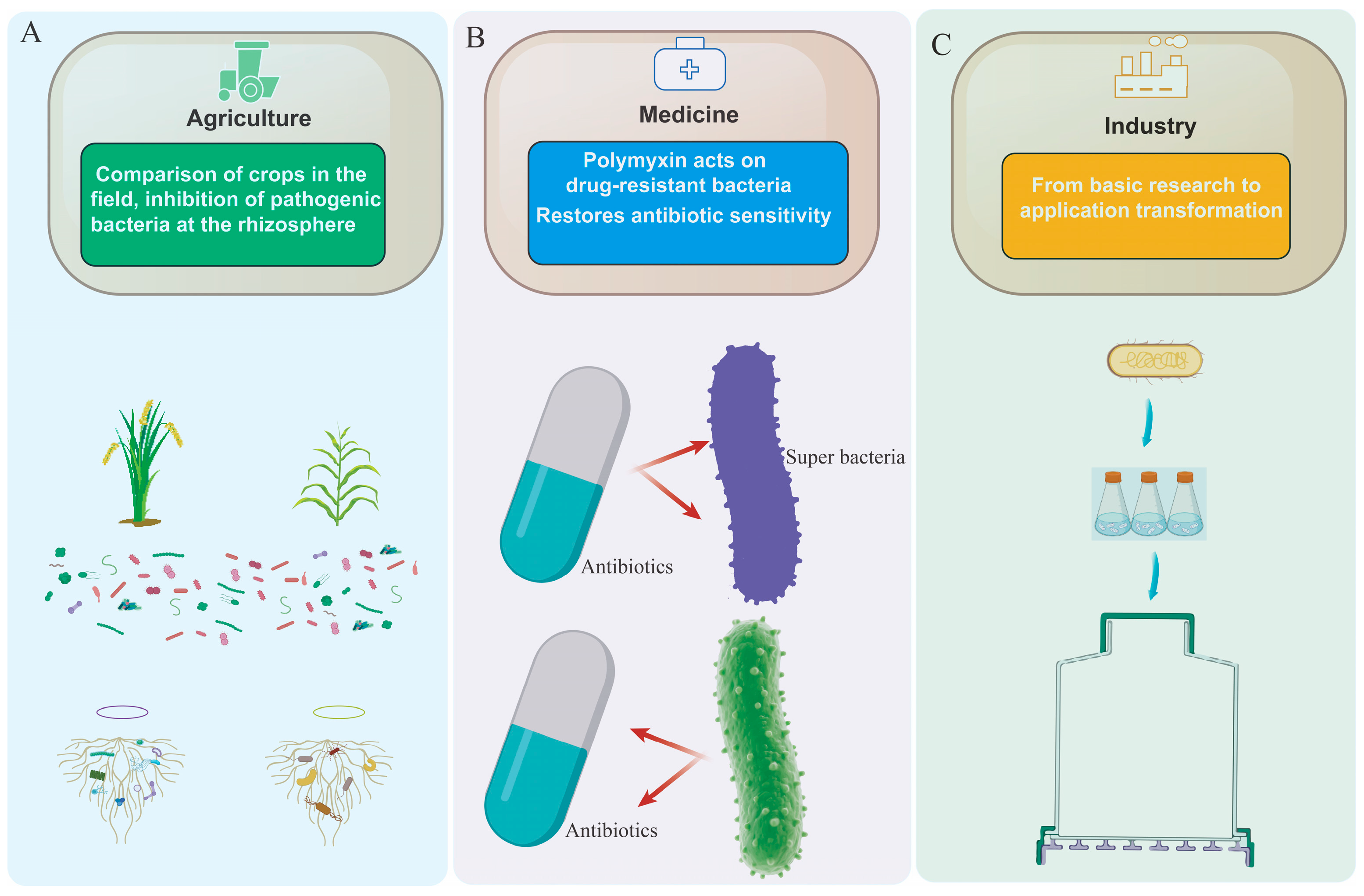
3.1. Application of Paenibacillus in the Field of Agriculture
3.2. Application of Paenibacillus in the Field of Medicine
3.3. Application of Paenibacillus in the Industrial Field
4. The Research Challenges and Future Prospects of Paenibacillus
4.1. The Research Challenges of Paenibacillus
4.2. The Future Prospects of Paenibacillus
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yuan, C.; Ruan, L.; Yang, Y.; Wang, L.; Ke, Z.; He, J. Paenibacillus wuxiensis sp. nov., a novel bacterium capable of producing Indole-3-Acetic Acid (IAA) and inhibiting Rhizoctonia solani Kühn isolated from agricultural soil. Curr. Microbiol. 2025, 82, 514. [Google Scholar] [CrossRef]
- Wu, Z.; Dou, W.; Yang, X.; Niu, T.; Han, Z.; Yang, L.; Wang, R.; Wang, Z. Novel glycosidase from Paenibacillus lactis 154 hydrolyzing the 28-O-β-D-glucopyranosyl ester bond of oleanane-type saponins. Appl. Microbiol. Biotechnol. 2024, 108, 282. [Google Scholar] [CrossRef]
- Xie, J.B.; Bai, L.Q.; Wang, L.Y.; Chen, S.F. Phylogeny of 16S rRNA and nifH genes and regulation of nitrogenase activity by oxygen and ammonium in the genus Paenibacillus. Mikrobiologiia 2012, 81, 760–767. [Google Scholar] [CrossRef] [PubMed]
- Miral, A.; Fournet, S.; Porte, C.; Sauvager, A.; Montarry, J.; Tomasi, S.; Tranchimand, S. Volatile organic compounds from a lichen-associated bacterium, Paenibacillus etheri, interact with plant-parasitic Cyst nematodes. ACS Omega 2022, 7, 43084–43091. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Cheng, Y.; Zhao, L.; Cao, R. Improvement of the catalytic performance of chitosanase Csn-PD from Paenibacillus dendritiformis by semi-rational design. Int. J. Biol. Macromol. 2024, 264, 130753. [Google Scholar] [CrossRef] [PubMed]
- Songnaka, N.; Ratanaphan, A.; Sermkaew, N.; Sawatdee, S.; Krobthong, S.; Aonbangkhen, C.; Yingchutrakul, Y.; Atipairin, A. Discovery of a novel antimicrobial peptide from Paenibacillus sp. Na14 with potent activity against Gram-negative bacteria and genomic insights into its biosynthetic pathway. Antibiotics 2025, 14, 805. [Google Scholar] [CrossRef]
- Huang, X.Y.; Ye, X.P.; Hu, Y.Y.; Tang, Z.X.; Zhang, T.; Zhou, H.; Zhou, T.; Bai, X.L.; Pi, E.X.; Xie, B.H.; et al. Exopolysaccharides of Paenibacillus polymyxa: A review. Int. J. Biol. Macromol. 2024, 261, 129663. [Google Scholar] [CrossRef]
- Pandey, A.K.; Barbetti, M.J.; Lamichhane, J.R. Paenibacillus polymyxa. Trends Microbiol. 2023, 31, 657–659. [Google Scholar] [CrossRef]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Fact. 2016, 15, 203. [Google Scholar] [CrossRef]
- Wang, W.; He, H.; Zhang, P.; Yan, J.; He, H.; Chen, X.; Wang, H.; Zhu, W.; Cui, Z.; Yuan, X. Industrial-scale aerobic composting with the addition of Paenibacillus mucilaginosus: Improving product quality and removing antibiotic resistance genes. J. Environ. Manag. 2025, 376, 124187. [Google Scholar] [CrossRef]
- Yuan, P.; Chen, Z.; Xu, M.; Cai, W.; Liu, Z.; Sun, D. Microbial cell factories using Paenibacillus: Status and perspectives. Crit. Rev. Biotechnol. 2024, 44, 1386–1402. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Shen, M.; Mao, C.; Wang, C.; Yuan, P.; Wang, T.; Sun, D. A type I restriction modification system influences genomic evolution driven by horizontal gene transfer in Paenibacillus polymyxa. Front. Microbiol. 2021, 12, 709571. [Google Scholar] [CrossRef] [PubMed]
- Pawlowski, A.C.; Westman, E.L.; Koteva, K.; Waglechner, N.; Wright, G.D. The complex resistomes of Paenibacillaceae reflect diverse antibiotic chemical ecologies. ISME J. 2018, 12, 885–897. [Google Scholar] [CrossRef]
- Costa, A.; Corallo, B.; Amarelle, V.; Stewart, S.; Pan, D.; Tiscornia, S.; Fabiano, E. Paenibacillus sp. Strain UY79, isolated from a root nodule of arachis villosa, displays a broad spectrum of antifungal activity. Appl. Environ. Microbiol. 2022, 88, e0164521. [Google Scholar] [CrossRef]
- Kai, H.; Yamashita, M.; Takase, S.; Hashimoto, M.; Muramatsu, H.; Nakamura, I.; Yoshikawa, K.; Kanasaki, R.; Ezaki, M.; Nitta, K.; et al. Identification of ten KB425796-A congeners from Paenibacillus sp. 530603 using an antifungal assay against Aspergillus fumigatus in combination with micafungin. J. Antibiot. 2013, 66, 473–478. [Google Scholar] [CrossRef]
- Yang, F.; Dai, W.; Xue, H.; Chen, W.; Liu, C.; Tian, Y.; Cheng, W.; Zhang, J. Efficacy of 2-undecanol produced by Paenibacillus polymyxa KM2501-1 in controlling Meloidogyne incognita. Microbiol. Spectr. 2025, 13, e0306224. [Google Scholar] [CrossRef]
- Hertlein, G.; Seiffert, M.; Gensel, S.; Garcia-Gonzalez, E.; Ebeling, J.; Skobalj, R.; Kuthning, A.; Süssmuth, R.D.; Genersch, E. Biological role of paenilarvins, iturin-like lipopeptide secondary metabolites produced by the honey bee pathogen Paenibacillus larvae. PLoS ONE 2016, 11, e0164656. [Google Scholar] [CrossRef]
- Lebedeva, J.; Jukneviciute, G.; Čepaitė, R.; Vickackaite, V.; Pranckutė, R.; Kuisiene, N. Genome mining and characterization of biosynthetic gene clusters in two cave strains of Paenibacillus sp. Front. Microbiol. 2021, 11, 612483. [Google Scholar] [CrossRef]
- Olishevska, S.; Nickzad, A.; Déziel, E. Bacillus and Paenibacillus secreted polyketides and peptides involved in controlling human and plant pathogens. Appl. Microbiol. Biotechnol. 2019, 103, 1189–1215. [Google Scholar] [CrossRef]
- Kajimura, Y.; Kaneda, M. Fusaricidin A, a new depsipeptide antibiotic produced by Bacillus polymyxa KT-8 taxonomy, fermentation, isolation, structure elucidation and biological activity. J. Antibiot. 1996, 49, 129–135. [Google Scholar] [CrossRef]
- Mousa, W.K.; Raizada, M.N. Biodiversity of genes encoding anti-microbial traits within plant associated microbes. Front. Plant Sci. 2015, 6, 231. [Google Scholar] [CrossRef]
- Vater, J.; Niu, B.; Dietel, K.; Borriss, R. Characterization of novel fusaricidins produced by Paenibacillus polymyxa-M1 using MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 2015, 26, 1548–1558. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, S. Fusaricidin produced by Paenibacillus polymyxa WLY78 induces systemic resistance against Fusarium wilt of cucumber. Int. J. Mol. Sci. 2019, 20, 5240. [Google Scholar] [CrossRef] [PubMed]
- Le, K.D.; Kim, J.; Yu, N.H.; Kim, B.; Lee, C.W.; Kim, J.C. Biological control of tomato bacterial wilt, kimchi cabbage soft rot, and red pepper bacterial leaf spot using Paenibacillus elgii JCK-5075. Front. Plant Sci. 2020, 11, 775. [Google Scholar] [CrossRef]
- Li, X.; Wang, J.; Lv, Y.; Zhao, L.; Jiang, W.; Lv, J.; Xu, X.; Yu, Y.; Liu, Y.; Chen, X.; et al. Screening and identification of Paenibacillus polymyxa GRY-11 and its biological control potential against apple replant disease. Folia Microbiol. 2025, 70, 475–487. [Google Scholar] [CrossRef]
- Xie, S.; Si, H.; Xue, Y.; Zhou, R.; Wang, S.; Duan, Y.; Niu, J.; Wang, Z. Efficacy of rhizobacteria Paenibacillus polymyxa SY42 for the biological control of Atractylodes chinensis root rot. Microb. Pathog. 2024, 187, 106517. [Google Scholar] [CrossRef]
- Abdallah, Y.; Yang, M.; Zhang, M.; Masum, M.M.I.; Ogunyemi, S.O.; Hossain, A.; An, Q.; Yan, C.; Li, B. Plant growth promotion and suppression of bacterial leaf blight in rice by Paenibacillus polymyxa Sx3. Lett. Appl. Microbiol. 2019, 68, 423–429. [Google Scholar] [CrossRef]
- Wang, D.; Poinsot, V.; Li, W.; Lu, Y.; Liu, C.; Li, Y.; Xie, K.; Sun, L.; Shi, C.; Peng, H.; et al. Genomic insights and functional analysis reveal plant growth promotion traits of Paenibacillus mucilaginosus G78. Genes 2023, 14, 392. [Google Scholar] [CrossRef]
- Wang, K.; Lin, Z.; Dou, J.; Jiang, M.; Shen, N.; Feng, J. Identification and surveys of promoting plant growth VOCs from biocontrol bacteria Paenibacillus peoriae GXUN15128. Microbiol. Spectr. 2023, 11, e0434622. [Google Scholar] [CrossRef]
- Li, X.; Ma, S.; Meng, Y.; Wei, W.; Peng, C.; Ling, C.; Fan, S.; Liu, Z. Characterization of antagonistic bacteria Paenibacillus polymyxa ZYPP18 and the effects on plant growth. Plants 2023, 12, 2504. [Google Scholar] [CrossRef]
- Chiu, S.; Hancock, A.M.; Schofner, B.W.; Sniezek, K.J.; Soto-Echevarria, N.; Leon, G.; Sivaloganathan, D.M.; Wan, X.; Brynildsen, M.P. Causes of polymyxin treatment failure and new derivatives to fill the gap. J. Antibiot. 2022, 75, 593–609. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; E, W.; Zhao, D.; Liu, H.; Pei, J.; Du, B.; Liu, K.; Zhu, X.; Wang, C. Response of Paenibacillus polymyxa SC2 to the stress of polymyxin B and a key ABC transporter YwjA involved. Appl. Microbiol. Biotechnol. 2024, 108, 17. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xu, Y.; Gao, R.; Lin, J.; Wei, W.; Srinivas, S.; Li, D.; Yang, R.S.; Li, X.P.; Liao, X.P.; et al. Deciphering MCR-2 colistin resistance. mBio 2017, 8, e00625-17. [Google Scholar] [CrossRef] [PubMed]
- Di Conza, J.A.; Radice, M.A.; Gutkind, G.O. MCR-1: Rethinking the origin. Int. J. Antimicrob. Agents. 2017, 50, 737. [Google Scholar]
- Cochrane, S.A.; Vederas, J.C. Lipopeptides from Bacillus and Paenibacillus spp.: A gold mine of antibiotic candidates. Med. Res. Rev. 2016, 36, 4–31. [Google Scholar]
- Itoh, T. Structures and functions of carbohydrate-active enzymes of chitinolytic bacteria Paenibacillus sp. str. FPU-7. Biosci. Biotechnol. Biochem. 2021, 85, 1314–1323. [Google Scholar] [CrossRef]
- Wang, X.; Xu, W.; Dai, Q.; Liu, X.; Guang, C.; Zhang, W.; Mu, W. Characterization of a thermostable PL-31 family alginate lyase from Paenibacillus ehimensis and its application for alginate oligosaccharides bioproduction. Enzym. Microb. Technol. 2023, 166, 110221. [Google Scholar] [CrossRef]
- Luo, Y.; Cheng, Y.; Yi, J.; Zhang, Z.; Luo, Q.; Zhang, D.; Li, Y. Complete genome sequence of industrial biocontrol strain Paenibacillus polymyxa HY96-2 and further analysis of its biocontrol mechanism. Front. Microbiol. 2018, 9, 1520. [Google Scholar] [CrossRef]
- Edith Ayala-Rodríguez, A.; Valdés-Rodríguez, S.; Enrique Olalde-Mathieu, V.; Arias-Padró, M.; Reyes-Moreno, C.; Olalde-Portugal, V. Extracellular ligninases production and lignin degradation by Paenibacillus polymyxa. J. Gen. Appl. Microbiol. 2024, 70, 1–10. [Google Scholar] [CrossRef]
- Xu, S.; Zhan, J.; Li, L.; Zhu, Y.; Liu, J.; Guo, X. Total petroleum hydrocarbons and influencing factors in co-composting of rural sewage sludge and organic solid wastes. Environ. Pollut. 2023, 319, 120911. [Google Scholar]
- Bampidis, V.; Azimonti, G.; de Lourdes Bastos, M.; Christensen, H.; Durjava, M.; Dusemund, B.; Kouba, M.; López-Alonso, M.; López Puente, S.; Marcon, F.; et al. Safety and efficacy of a feed additive consisting of endo-1,4-β-d-mannanase produced by Paenibacillus lentus DSM 33618 (Hemicell® HT/HT-L) for chickens and turkeys for fattening, chickens reared for laying, turkeys reared for breeding, minor poultry species to point of lay, pigs for fattening, weaned piglets and minor porcine species (Elanco GmbH). EFSA J. 2023, 21, e07878. [Google Scholar]
- Dickel, F.; Bos, N.M.P.; Hughes, H.; Martín-Hernández, R.; Higes, M.; Kleiser, A.; Freitak, D. The oral vaccination with Paenibacillus larvae bacterin can decrease susceptibility to American Foulbrood infection in honey bees-A safety and efficacy study. Front. Vet. Sci. 2022, 9, 946237. [Google Scholar] [CrossRef] [PubMed]
- Gopal, N.; Hill, C.; Ross, P.R.; Beresford, T.P.; Fenelon, M.A.; Cotter, P.D. The prevalence and control of Bacillus and related spore-forming bacteria in the dairy industry. Front. Microbiol. 2015, 6, 1418. [Google Scholar] [CrossRef] [PubMed]
- Pacher, N.; Burtscher, J.; Johler, S.; Etter, D.; Bender, D.; Fieseler, L.; Domig, K.J. Ropiness in Bread-A Re-Emerging Spoilage Phenomenon. Foods 2022, 11, 3021. [Google Scholar] [CrossRef] [PubMed]
- Ravagnan, G.; Meliawati, M.; Schmid, J. CRISPR-Cas9-mediated genome editing in Paenibacillus polymyxa. In Synthetic Biology: Methods and Protocols; Methods in Molecular Biology; Springer: New York, NY, USA, 2024; Volume 2760, pp. 267–280. [Google Scholar]
- Meliawati, M.; Teckentrup, C.; Schmid, J. CRISPR-Cas9-mediated large cluster deletion and multiplex genome editing in Paenibacillus polymyxa. ACS Synth. Biol. 2022, 11, 77–84. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, P.; Song, Z.; Zhang, L.; Pan, Z.; Chen, X. Exploring the Multifunctional Role of Paenibacillus Metabolites in Various Fields. Int. J. Mol. Sci. 2025, 26, 12089. https://doi.org/10.3390/ijms262412089
Yuan P, Song Z, Zhang L, Pan Z, Chen X. Exploring the Multifunctional Role of Paenibacillus Metabolites in Various Fields. International Journal of Molecular Sciences. 2025; 26(24):12089. https://doi.org/10.3390/ijms262412089
Chicago/Turabian StyleYuan, Panhong, Zonghui Song, Langjie Zhang, Zhengsheng Pan, and Xiaolong Chen. 2025. "Exploring the Multifunctional Role of Paenibacillus Metabolites in Various Fields" International Journal of Molecular Sciences 26, no. 24: 12089. https://doi.org/10.3390/ijms262412089
APA StyleYuan, P., Song, Z., Zhang, L., Pan, Z., & Chen, X. (2025). Exploring the Multifunctional Role of Paenibacillus Metabolites in Various Fields. International Journal of Molecular Sciences, 26(24), 12089. https://doi.org/10.3390/ijms262412089





